Abstract
In recent years, the study of marine viromes has become one of the most relevant areas of geoecology. Viruses are the most numerous, genetically diverse and pervasive biological entities on Earth, including in aquatic ecosystems. Information about viral diversity in aquatic ecosystems remains limited and requires more research. This work provides the first-ever look at the current DNA virome of the Northern Caspian Sea. A comparison with other freshwater and marine viromes revealed that the North Caspian Sea virome has the greatest similarity with those of the Baltic Sea and Lake Baikal. The study described in this article expands the knowledge about aquatic viromes and provides key data for a more comprehensive analysis of viruses circulating in the Caspian Sea, the largest inland body of water on Earth.
1. Introduction
The diversity of living organisms in ecosystems is one of the most important biotope characteristics [1]. Unfortunately, methods of visual observation or microbial isolation cannot provide a complete picture as no more than 5% of the organisms making up an ecosystem can be cultured. New generation metagenomic sequencing has revolutionized the study of viruses, revealing greater diversity, scope and structure of the virosphere, as the study of diversity is no longer bound to imperfect cultivation methods [2]. With the rapid development of high-throughput sequencing technology and bioinformatic analysis, metagenomics has become an important and powerful tool to better understand the structure, diversity and variability of the viral community [3,4,5,6]. Water samples from different geographical regions are among the most popular environmental samples for metagenomics applications. The marine environment is a rich source of viruses. Bacteriophages are significantly more abundant in aquatic ecosystems than other life forms [7], with an approximate concentration of approximately 10 billion particles per liter in surface waters [8,9], although their abundance varies depending on ocean depth, temperature [10], latitude [11] and phytoplankton bloom development [12]. Marine viruses play an important role in regulating bacterial abundance, forming the structure of the microbial community and modifying the genetic diversity of microorganisms through horizontal transfer of genes [13]. Viral lysis mediates the transfer of organic matter between living biomass and the pool of dissolved organic carbon through a viral shunt [14,15]. An estimated 10 billion tons of carbon are released daily through the viral shunt; this organic matter constitutes a fundamental part of the nutrient cycle that sustains the productivity of oceans [16,17]. The relationship between virus abundance and host abundance is introduced by the Kill-the-Winner theory, which postulates that the higher the growth rate of a microorganism, the more likely it is to become a target for lytic viral infection. This feature allows slow-growing prokaryotes to reach higher numbers than fast-growing ones because they are less susceptible to lytic infections [18,19]. However, Piggyback-the-Winner theory postulates that when the host population is high, viruses adhere to lysogenic infection and integrate into the host genome (as prophages). Thereby, the prophage protects the host cell from being infected by a closely related bacteriophage by eliminating superinfection, a process in which various proteins block other phages from establishing a productive infection. The prophage also ensures improved fitness through lysogenic conversion, i.e., the prophage expresses genes that change the physiology of the lysogen [10,20,21]. During the past two decades, studies have been conducted to characterize viral communities from various marine sites using metagenomics. Many of them were part of major expeditions and data collection projects such as the Global Ocean Sampling Expedition, Pacific Ocean Virome, Tara Ocean Expedition, Malaspina Expedition, Tara Polar Circle Ocean Expedition (TOPC), etc. [11,22,23,24,25,26,27]. Recently, the Global Ocean Viromes 2.0 (GOV 2.0) dataset derived from 145 marine metagenomic virus samples identified a total of 195,728 viral populations, about 12 times the viral populations from the original Tara Ocean and Malaspina expeditions [11,26,27]. To date, there have been many reviews on phages present in the surface layers of marine environments [28,29,30,31,32,33]. However, our knowledge of the diversity of DNA-containing viruses infecting microbes in inland water reservoirs is still very limited. One such reservoir is the Caspian Sea. The Caspian Sea is the largest endorheic body of water on Earth with a volume of 78,000 km3 and a surface area of 371,000 km2 [34]. As with the Aral and Black Seas, it was once part of the world oceans Tethys and Paratethys but has been an isolated reservoir for the last five million years [35]. The depth of the Caspian Sea reaches a maximum of 20 m in the northern part and a maximum of 1025 m in the southern part [36]. Approximately 130 rivers flow into the Caspian Sea, with the Volga taking the bulk of the inflow [37]. Because of the freshwater inflow, the salinity of the Caspian Sea is about one-third that of the world’s oceans, making it a brackish lake [38]. Unfortunately, despite considerable progress in the study of the zoological and botanical diversity of the region [39], only sporadic studies of the composition of the Caspian Sea’s microbiome have been made [40,41].
This work presents a metagenomic study of viral diversity and community structure in three surface seawater samples from the North Caspian Sea, and a comparative analysis of the North Caspian virome and other marine and freshwater viromes. The datasets used for comparison (from North Sea, Baltic Sea, Mediterranean Sea, Ionian Sea, Red Sea, Arabian Sea and Lake Baikal were downloaded from the NCBI SRA database (further detailed in the Materials and Methods section).
2. Materials and Methods
2.1. Study Site and Sampling
Sampling sites in the North Caspian region were determined according to the Kaz Hydromet regulatory standards developed for the systematic monitoring of water quality (Figure 1).
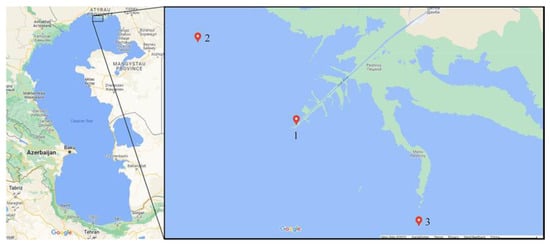
Figure 1.
Water sampling sites in the Northern Caspian Sea. (1) Maritime shipping channel (Morskoy sudokhodnyy kanal)—46°50′05.9″ N 51°32′18.0″ E (46.834972, 51.538320), (2) Seashore of the Ural River (Vzmor’ye reki Ural)—46°55′59.2″ N 51°22′02.5″ E (46.933123, 51.367352), (3) Shalygi Bay Islands (Ostrova zaliva Shalygi)—46°42′52.7″ N 51°45′04.3″ E (46.714651, 51.751187). All studied samples from the Northern Caspian were taken on 21 May 2021.
A total of 10 L of water was collected into sterile containers at a depth of up to 1 m from each sampling site. The following quality indicators were determined in each of the obtained samples: pH, dissolved oxygen, nitrite nitrogen, nitrate nitrogen, ammonium salt, total nitrogen, phosphates and total phosphorus.
The samples were labelled as follows: (1) Maritime shipping channel—MSC; (2) Seashore of the Ural River—SUR; (3) Shalygi Bay Islands—SBI.
2.2. Sample Processing
The seawater samples were sequentially filtered through 3 μm and 0.22 μm pore size filters to remove any zooplankton, phytoplankton and bacteria. Then, they were subjected to a two-step tangential flow filtration (TFF) with a 30 kDa cartridge (Vivaflow 200, Sartorius, Göttingen, Germany) to concentrate the samples to the final volume of 500 mL. After that, the samples were centrifuged at 100,000 g for 2 h at 4 °C using the Avanti J30I ultracentrifuge (Beckman Coulter, Brea, CA, USA) [42]. Then, the pellet was resuspended in a minimal volume of phosphate-buffered saline (250 µL) and used for DNA extraction.
2.3. Transmission Electron Microscopy
To study the morphology of viruses, after sedimentation by ultracentrifugation, the concentrated aqueous samples were applied on mesh. The samples were spotted on formvar coated copper grid (Polysciences, Inc., Hirschberg an der Bergstrasse, Germany) for 5 min. Then, 3% phosphotungstic acid (pH 6.8) was used to stain the samples, and excess solution was removed.
2.4. Isolation of Nucleic Acids and Sequencing
To remove extracellular bacterial DNA, the obtained samples were treated with DNase 50 U mL−1 (Thermo Fisher Scientific, Waltham, MA, USA) at 37 °C for 30 min. Then, 20 μL of 50 mM EDTA was added to inactivate DNase and the solution was incubated for 10 min at 65 °C. To confirm the removal of bacterial DNA from the sample after the treatment with the enzyme, real-time PCR with primers was performed to detect 16S ribosomal RNA.
Total DNA was isolated from the obtained samples using the PureLink Genomic DNA MiniKit (Invitrogen, Waltham, MA, USA) according to the manufacturer’s protocol.
Quantitative measurements were performed using the Qubit dsDNA HS kit (High Sensitivity, Invitrogen, Waltham, MA, USA) according to the instructions for the Qubit 3.0 fluorimeter. The A260–A280 ratio was measured with a Tecan multi-reader using a NanoQuant plate for measuring micro quantities of nucleic acids (Invitrogen, Waltham, MA, USA).
DNA libraries were prepared from 1 ng DNA using the Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA, USA) in accordance with the instructions. In the course of the preparation of the libraries, enzymatic fragmentation of DNA, ligation of sequence adapters, preliminary amplification of the library, selection of fractions of the desired length and clonal amplification of the selected library were performed. The purification of genomic libraries and the selection of fractions of the required length were carried out using the Agencourt AMPure XP paramagnetic beads system (Beckman Coulter, Brea, CA, USA). Excessive primers, nucleotides, salts and enzymes were removed by washing with freshly prepared 80% C2H5OH. The quality of genomic libraries was determined using the Agilent 2100 bioanalyzer with a DNA 7500 Kit.
High-throughput sequencing was performed by using Illumina MiSeq (Paired-end sequencing, 2 × 300 bp, MiSeq Kit v3).
2.5. Initial Shotgun Metagenomic Data on DNA Viruses in Marine and Freshwater Samples
For comparative analysis, we also used the NCBI SRA datasets (Table 1) of viral communities of other marine and freshwater ecosystems. The datasets for these relatively similar ecosystems were obtained using similar library preparation techniques and sequencing platform (Illumina) as those in our study.

Table 1.
Description of sequencing datasets used for comparative analysis.
2.6. Primary Processing of Virome Reads
The quality of the virome datasets (paired reads) was assessed using FastQC. Then, quality trimming of reads was performed with Trimmomatic [47]. Trimming removed Illumina adapters from the reads (if they were found in NCBI SRA data). Reads with an average quality of 25 or more units and a length of 140 or more base pairs were selected for further analysis. To make analyzing a large number of samples technically possible (for de novo cross-assembly), the size of third-party fastq data after trimming was reduced to the number of nucleotides corresponding to the samples from the Caspian Sea (reduced to ≈1000 Mbp in forward and reverse reads) using a proprietary R script. If the NCBI SRA sample contained more than 1000 Mbp, random pairs of reads were selected from the fastq file until the data size reached 1000 Mbp. If the NCBI SRA sample contained less than 1000 Mbp, the number of reads was not reduced. The R script used is provided in Supplementary File S1.
2.7. Taxonomic Analysis of Original Genome Reads
Taxonomic classification was performed through the k-mers analysis in the Kraken2 software [48] with default options according to the NCBI nr (nonredundant) nucleotide database.
2.8. Assembly of Virome Reads
All the genomic data (Table 1) were aggregated into one array for de novo cross-assembly as recommended [46]. For cross-assembly, the MEGAHIT assembler was used with default options [49]. The Bowtie2 [50] and SAMtools [51] software was used to map paired-end reads on scaffolds and calculate the total coverage of the scaffolds in the assembly and coverage of the scaffolds by reads from each sample. The scaffolds with a total coverage of more than five and a length of ≥2500 nucleotides were used for further analysis.
2.9. Identification of Viral Scaffolds
We identified the viral scaffolds and open reading frames (ORFs) (typically multiple ORFs per scaffold) in them using the VirSorter tool [52] on the CYVERSE Discovery Environment webserver (https://de.cyverse.org/de/; accessed on 20 December 2022). The Bowtie2 and SAMtools results were used to determine the number of reads mapped on each predicted viral scaffold from each sample. In Bowtie2, reads from fastq files of each sample were mapped to cross-assembled scaffolds separately. The mapping data were converted to a count table of viral scaffold representation per samples. TPM (transcripts per million) normalization recommended for metagenomics was used to normalize the count table for scaffold length and number of reads per samples [53]. For all the predicted viral proteins (ORFs) in viral scaffolds, counts per samples were defined as the TPM of the scaffolds containing the given proteins.
2.10. Taxonomic Assignment of Viral Scaffolds
Taxonomic identification for the viral scaffolds was carried out by comparing the predicted viral proteins in the scaffolds with the NCBI RefSeq [54] complete viral proteome database. The comparison was carried out by the DIAMOND [55] algorithm using the More-Sensitive option, e-value ≤ 0.00001 and bit score ≥ 50. During the analysis, all the proteins from all the scaffolds were matched against the NCBI RefSeq complete viral proteome. If the analyzed scaffold contained one predicted protein and it matched any protein from the databases, the taxonomic identifier of this protein was chosen as the most closely related virus taxon (virotype) of this scaffold. If the scaffold contained several proteins and they all matched different NCBI RefSeq taxa, the best match in terms of the bit score value was selected and its taxon was chosen as the most closely related virus taxon (virotype) of this scaffold. If the analyzed scaffold contained several proteins and two or more of them were associated with one NCBI RefSeq taxon, this taxon was chosen as the most closely related virus taxon (virotype) of this scaffold regardless of the bit score value. If none of the predicted viral scaffold proteins matched NCBI RefSeq, such a scaffold was considered to be a fragment of an unclassified virus genome. TPM value for scaffold per samples was transformed into a table of TPM values per virotype (the sum of TPM value for different scaffolds with the same virotype) in samples.
2.11. Functional Assignment of Viral Communities
For functional assignment, the predicted viral proteins (ORFs) were matched with functional motifs of proteins in the KOfam database [56] using the KofamScan [57] software with default options. All VirSorter predicted viral proteins (ORFs) (ORFs from scaffolds both classified and not classified to virotype) were used in the analysis. All the KofamScan results were transformed to KO (KEGG pathway classification functional groups) anthologies [58] with the KEGGREST package [59] for R. The TPM count of the predicted viral proteins in samples was transformed by summation into per samples TPM counts of the KEGG pathway classification groups. Based on the KEGG pathway classification, the TPM counts of AMGs (auxiliary metabolic genes) viral proteins were isolated for further analysis.
2.12. Statistical Analysis of Taxonomic and Functional Diversity
The potential (underestimated) number of virotypes (species richness) in communities was evaluated using the Chao1 [60] and ACE [61] indices. The Shannon and Simpson indices [62] of virotypes’ biodiversity were also calculated. The similarity in the taxonomic composition of the samples (similarity in virus scaffold count table per samples) was visualized using hierarchical cluster analysis (Bray–Curtis distances with the average clustering method) with bootstrap support (1000 bootstrap replicas) calculation of clustering in the pvclust [63] package for R and the nonmetric multidimensional scaling (NMDS) ordination method (based on Bray–Curtis distances). Biodiversity analysis and NMDS were carried out in the vegan package for R [64]. The first 30 dominant virotypes in samples were visualized with the heat map using the gplots [65] package for R. The TPM counts of KEGG pathway classification groups and AMGs TPM counts were visualized with the heat map using the gplots package for the R with column (sample) clustering and were grouped in similarity order (Euclidean distance with the average clustering method).
3. Results
3.1. Environmental Characteristics
The biogenic organic matter load of water reservoirs is one of the key parameters in net eutrophication balance models. The biogenic organic matter content in bodies of water, primarily the total phosphorus and nitrogen content, is an objective indicator of the trophic state of the reservoirs. Analysis of nutrient content in the samples showed that the selected water samples belonged to the oligotrophic type (Table 2).

Table 2.
Hydrochemical parameters of the samples.
3.2. Morphology of Viruses Discovered in the North Caspian Sea
The morphology of viruses from aquatic samples observed with a transmission electron microscope revealed that viruses with typical head and tail structures, such as sipho-, myo- and podo viruses belonging to the Caudoviricetes class, were the most common (Figure 2).
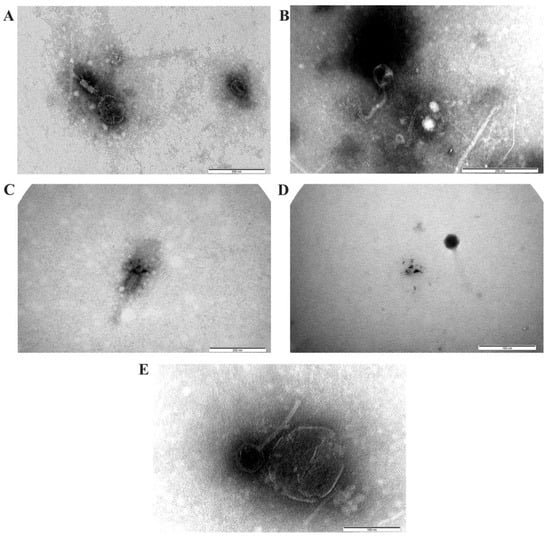
Figure 2.
Diverse viruses in the surface water of the North Caspian Sea observed with a transmission electron microscope. (A,C)—morphology structure typical of myoviruses group; (B,E)—morphology structure typical of siphoviruses group, (D)—morphology structure typical of podoviruses group.
3.3. Identification of Viral Reads
In this study, high-throughput sequencing of North Caspian samples (SUR, MSC, SBI) produced three datasets containing 2,326,049; 2,220,155; and 2,331,660 reads, respectively. After sequence adapters and low-quality reads were removed, the sets of nucleotide sequences of SUR, MSC and SBI samples made 1,970,178 (mean read length 248), 1,882,933 (mean read length 249) and 1,942,399 (mean read length 250) reads, respectively. The proportion of sequences identified as viral with significant matches to known sequences (e-value < 10−5, bit score ≥ 50) ranged from 11.96 to 43.19% (471,151–1,677,937 reads); these values are comparable to the number of viral reads in datasets from other ecosystems (Table 3).

Table 3.
General statistics and indices of viral diversity in the studied datasets.
3.4. Taxonomic Composition of Viromes of the Caspian Sea
Sequence annotation of the samples revealed 12 families of DNA-containing viruses affecting bacteria, algae, fish, insects, etc., where 11 had the dsDNA genome and one family possessed the ssDNA genome (Table 4). In the Caspian Sea samples, the qualitative composition of viral families was similar, but the proportions of families differed. It was found that among the viral sequences belonging to dsDNA viruses, the majority belonged to the tailed myoviruses, siphoviruses and podoviruses of the Caudoviricetes class. Thus, in the SUR and MSC samples, sequences belonging to siphoviruses dominated, while podoviruses dominated in the SBI sample. The highest number of sequences belonging to myoviruses was present in the MSC sample, the lowest in SBI. In the SUR and MSC samples, the quantitative distribution of podoviruses was approximately the same, about 22%. Bacteriophages with the ssDNA genome of the Inoviridae family accounted for 0.01–0.02% of the reads in the list of virotypes. Among the normalized number of reads, sequences similar to microalgae viruses (Phycodnaviridae) also dominated: their number prevailed in the MSC sample and was equal to 1.45%; in the SUR sample, their number was 1.22%, while in the SBI sample, their number did not exceed 0.42%. Additionally, sequences of virophages of the Lavidaviridae family were identified in all samples (from 0.26% to 0.86%). These satellite viruses infect protozoa, but only when co-infected with other viruses belonging to the Mimiviridae family [66]. Mimiviruses also represented an insignificant part of the viral sequences; their number varied within 0.03–0.14%. Among the others were viral families whose known representatives affect archaea, insects and fish.

Table 4.
Viral families identified in the studied viromes from the North Caspian.
3.5. Diversity of Virotypes in the North Caspian Sea Datasets
Taxonomic identification of viral scaffolds by comparing predicted viral proteins with the NCBI RefSeq database identified 621, 631 and 612 different virotypes in the SUR, MSC, and SBI samples, respectively (Supplementary Table S1). The number of virotypes and viral diversity indices in the Caspian samples were approximately the same or often higher than in other compared samples (Table 3). We assume that the virus recovery was incomplete (this was not assessed in this study) because loss of viruses is inevitable at different stages of extraction and concentration of virus-like particles (during filtration, TFF or ultracentrifugation). However, statistical data showed that the viral fraction was sufficiently enriched and we were able to detect a wide variety of viruses in the samples using the chosen protocol.
A large number of sequences in the Caspian samples belonged to the virotype infecting representatives of the proteobacteria of the Alphaproteobacteria class (Sulfitobacter phage pCB2047-A), as well as to the virotype infecting unicellular cyanobacteria, Synechococcus sp. (Synechococcus phage S-CBS4). In the studied samples, basically all the identified cyanophages belonged to siphoviruses.
Viruses of the Alphaproteobacteria (Pelagibacter phage HTVC010P, Puniceispirillum phage HMO-2011, Dinoroseobacter phage vB_DshS-R5C, Agrobacterium phage Atu_ph07, Sphingobium phage Lacusarx), Gammaproteobacteria (Psychrobacter phage Psymv2, Idiomarinaceae phage 1N2-2, Marinomonas phage P12026, Shigella phage), Betaproteobacteria (Rhodoferax phage P26218, Burkholderia phage KS9, Bordetella virus BPP1), Actinobacteria (Rhodococcus phage REQ1, Streptomyces virus phiBT1) classes and of FCB-group bacteria (Cellulophaga phage phi38: 1, Nonlabens phage P12024L) were among the top 30 dominant virotypes of North Caspian viromes (Figure 3). More than 1% of the sequences belonged to the virus of Archaea of the halobacteria class (Salicola phage CGphi29). Despite the similar composition of the viromes, the number of certain virotypes differed in the studied samples: in the SBI sample, 44% of the viral sequences belonged to the Sulfitobacter phage pCB2047-A, whereas in the SUR and MSC samples, the sequences of cyanobacteria viruses (Synechococcus phages) dominated. In the SBI virome, there was a higher content of the Salicola phage CGphi29 and the Marinomonas phages compared to other samples. Virotypes affecting the Idiomarinaceae families were more abundant in the SUR sample. In addition, it was found that a greater number of sequences belonging to virotypes for which pathogenic microorganism genera, such as Burkholderia and Bordetella, are potential hosts were contained in the SUR and MSC samples. The Burkholderia phage KS9 sequences were represented more in the SUR and MSC samples, and the Bordetella virus BPP1 sequences dominated only in the SUR sample.
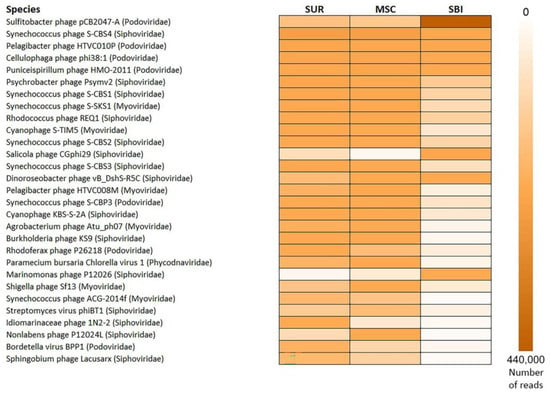
Figure 3.
Heat map of the first 30 dominant virotypes comprising 50% of all the sequences in the North Caspian Sea viromes.
Eukaryotic viruses were less represented in the studied datasets (<1%) than prokaryotic viruses. The dominant virotype in all the samples was the Paramecium bursaria Chlorella virus 1 of the Phycodnaviridae family, with a quantitative predominance in the MSC sample.
3.6. Analysis of Assembled Reads
Metagenomic assembly of all analyzed datasets (including previously published ones, Table 1) using the MEGAHIT program resulted in 15,784 viral scaffolds ranging in length from 2500 bp to 98,420 bp (Supplementary Table S2). The number of scaffolds covered by reads from the samples of the Caspian Sea was 2933; of these, taxonomic affiliation (as a virotype) was assigned to 1093 (78.9%) scaffolds. Obtained scaffolds and predicted open reading frames are available in the Supplementary Materials (Supplementary Table S3). Table 5 presents a set of viral scaffolds most represented in the North Caspian viromes (the first ten scaffolds were selected and arranged in descending order by the number of reads per scaffold in each sample). The similarity of the predicted ORF scaffolds to the data from the RefSeq protein database averaged from 31.5 to 64.4% (Table 5). The largest number of scaffolds belonged to phages infecting blue-green algae Synechococcus spp. and Prochlorococcus spp. As can be seen from Table 5, the composition of predominant scaffolds varied in the studied samples. The longest scaffold, k141_561861, which belonged to the Paracoccus phage Shpa, was predominant in the SUR and MSC samples based on the number of reads assigned to the scaffold. Virotypes such as the Marinomonas phage P12026, Sulfitobacter phage pCB2047-A and Ralstonia phage RP12 were mostly represented in the SBI virome. The number of reads per scaffold assigned to such virotypes as the Idiomarinaceae phage Phi1M2-2 (k141_1342719), Pseudoalteromonas phage H103 (k141_3192492), Acinetobacter phage Loki (k141_2823432) and Salicola phage CGphi29 (k141_275) also overwhelmingly prevailed in the SBI dataset. An equal ratio of detected ORFs and predicted proteins was found only in the scaffold that had a maximum identity of 83.8% with proteins of the Synechococcus phage S-CBS4. A dominant number of reads belonging to this phage was detected in the SUR and MSC samples.

Table 5.
The most represented viral scaffolds in the samples of the Northern Caspian Sea.
3.7. Functional Analysis
Functional analysis according to the KEGG Orthology database revealed genes of the four highest hierarchical categories in each sample (Figure 4a and Figure 5a), including genes for proteins involved in global ecological processes (Figure 4b).
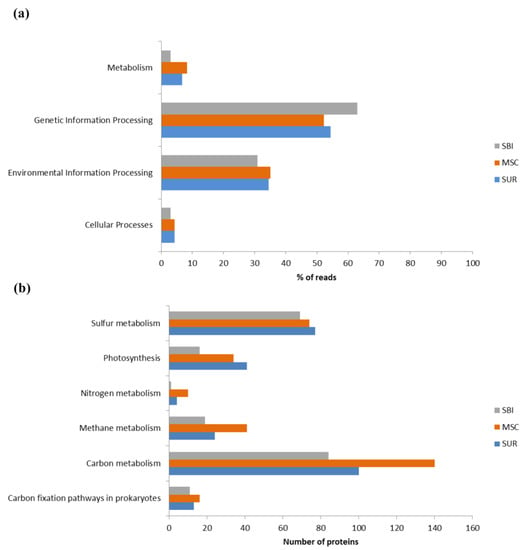
Figure 4.
General functional annotation of the analyzed Caspian viromes: (a)—the main functional categories (were indicated according to the KEGG Orthology); (b)—the involvement of viral proteins in global ecological processes (according to KEGG pathway enrichment analysis).
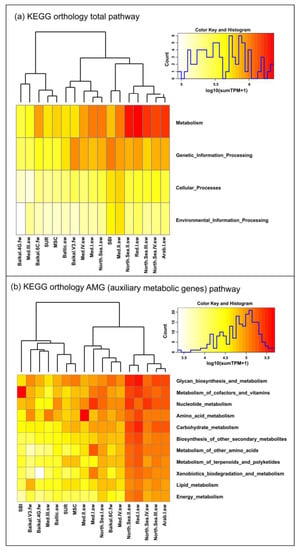
Figure 5.
Clustering of virome datasets based on functional analysis. (a) The main functional groups of identified genes according to the KEGG database; (b) main functional metabolic categories of auxiliary genes (the number of reads of the functional groups is shown at log10 scale).
The highest percentage of reads in all three Caspian Sea samples belonged to the “Genetic Information Processing” category, followed by the “Environmental Information Processing category. However, the percentage of reads in the “Genetic Information Processing” category was higher in the SBI sample, whereas the percentage of reads belonging to the “Environmental Information Processing” category was higher in the SUR and MSC samples. The percentage of reads related to cellular processes was the lowest in all the samples and had various levels under 4%. The percentage of reads in the “Metabolism” category was the highest in the MSC sample (8%), followed by the SUR sample (6.8%) and the SBI sample (4.3%).
Analysis of the profile of auxiliary metabolic genes (Figure 5b) showed that, overall, proteins involved in the metabolism of glycans, cofactors and vitamins, nucleotides, carbohydrates and terpenoids and polyketides dominated the northern region of the Caspian Sea, although the percentage of individual auxiliary genes in each sample differed. In the SUR and MSC samples, the greatest number of reads corresponded to the scaffolds with genes encoding proteins of glycan biosynthesis and metabolism, carbohydrate metabolism, nucleotides, terpenoids and polyketides, whereas in the SBI sample, proteins involved in cofactor and vitamin synthesis significantly prevailed.
A more detailed analysis of the metabolism category showed that the SBI sample had a high content of the hemX gene involved in porphyrin metabolism (cofactor and vitamin metabolism category), with the maximum number of reads attributed to the scaffold k141_3133183 (Sulfitobacter phage pCB2047-A, Podoviridae) that contains this gene.
The SUR and MSC samples mainly contained auxiliary genes from the “Glycan Biosynthesis and Metabolism” category encoding enzymes involved in the biosynthesis of lipopolysaccharides, lipoarabinomannan and teichoic acids. In the “Carbohydrate Metabolism” category, the largest number of reads represented the scaffold of the Synechococcus phage S-SKS1 (myovirus, k141_886840) and Synechococcus phage S-SSM7 (myovirus, k141_1343683) virotypes, in which auxiliary genes encoding amino- and nucleotide sugar metabolism proteins were identified.
The enzymes from the “Metabolism of terpenoids and polyketides” category were related to the biosynthesis of secondary metabolites: novU, C-methyltransferase; tylC3, NDP-4-keto-2,6-dideoxyhexose 3-C-methyltransferase; mtmC, D-mycarose 3-C-methyltransferase; and elmMII, 8-demethyl-8-(2-methoxy-alpha-L-rhamnosyl)tetracenomycin-C 3’-O-methyltransferase, which were mainly identified in the scaffolds of the following virotypes: the Synechococcus phage S-CBS4 (siphovirus, k141_1383679) and Paramecium bursaria Chlorella virus 1 (Phycodnaviridae, k141_268211). The Lipid Metabolism category was represented by genes encoding bacterial membrane biogenesis enzymes (bgsB; 1,2-diacylglycerol 3-alpha-glucosyltransferase) in the SUR and MSC samples (Supplementary Table S4).
The KEGG pathway enrichment analysis revealed 259, 315 and 200 proteins of energy metabolism in the SUR, MSC and SBI samples, respectively. The detected proteins were involved in the metabolism of carbon, nitrogen, methane and sulfur, as well as in photosynthesis (Figure 4b). The greatest number of proteins detected pertained to carbon and sulfur metabolism, while nitrogen metabolism proteins were the least represented.
The largest number of proteins involved in carbon, methane and nitrogen metabolism was recorded in the MSC sample. The number of proteins involved in photosynthesis was dominant in the SUR sample. Proteins involved in carbon fixation pathways in prokaryotes and sulfur metabolism were found in relatively equal amounts in all three studied samples from the Caspian Sea.
3.8. Comparative Analysis of Viromes from Various Marine and Freshwater Ecosystems
A comparative analysis of the virome of the Northern Caspian with viromes of freshwater (Lake Baikal) and marine (Baltic Sea, North Sea, Mediterranean Sea, Red Sea, Arabian Sea) aquatic ecosystems (Table 1) was carried out.
The composition of viral families determined in the samples of the North Caspian Sea (Table 4) differed from the composition of viral families in comparison samples from other reservoirs (Table 1). In the comparison samples, in addition to the main families of viruses indicated in Table 4, viruses belonging to the Monodnaviria (families Circoviridae, Microviridae, Pleolipoviridae), Adnaviria (Lipothrixviridae) and Varidnaviria (Tectiviridae, Adenoviridae, Poxviridae, Ascoviridae families) realms, and also to the Baculoviridae family of the Naldaviricetes class, were identified (Supplementary Table S5). Viruses of the Circoviridae family were detected only in one of the studied samples of the Mediterranean Sea (Med.III.sw), while representatives of the Microviridae family were found in two other samples (Med.I.sw and Med.II.sw). The archaeal viruses of the family Pleolipoviridae were identified in insignificant amounts only in a sample from the Baltic Sea (Baltic.sw). Representatives of the Tectiviridae family were present only in the virome from Lake Baikal with quantitative predominance in the sample Baikal.V3.fw. Sequences related to adenoviruses were also found in small amounts in Lake Baikal (Baikal.6C.fw). A small number of reads close to the viruses of the Poxviridae and Ascoviridae families were identified in samples from the North Sea (North.Sea.II.sw and North.Sea.III.sw) and the Mediterranean Sea (Med.III.sw and Med.IV.sw), respectively. Viruses of the Lipothrixviridae family that infect archaea were present only in Red Sea samples. Insect baculoviruses were identified in two Baikal samples, with quantitative dominance in the Baikal.6C.fw sample.
According to the hierarchical clustering, when comparing the taxonomic composition of viral communities of the North Caspian samples with other freshwater and marine samples, two main clusters were identified: the first and most numerous cluster included samples from the Mediterranean, Arabian, Red and North seas; the second cluster included samples from the North Caspian Sea, Baltic Sea and Lake Baikal (Figure 6). The Caspian Sea samples SUR and MSC were similar to each other but differed from SBI.
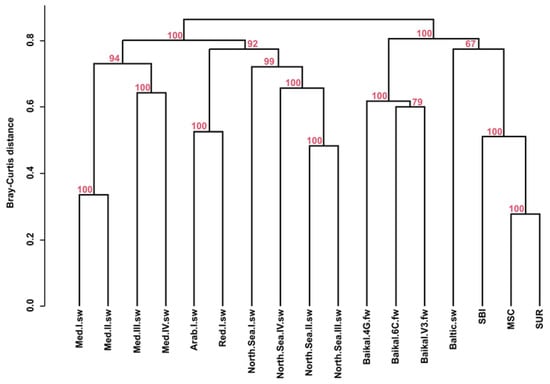
Figure 6.
UPGMA (unweighted pair group method with arithmetic mean) cluster dendrogram, (average clustering method with Bray–Curtis distances).
The differences in viral communities were investigated using nonmetric multidimensional scaling (NMDS) analysis (Figure 7). In general, according to the analysis, the gradient vector in the samples of the Caspian Sea, Baltic Sea and Lake Baikal shifted towards the dominance of siphoviruses. The samples from Lake Baikal (Baikal.6C.fw and Baikal.V3.fw) were more similar to each other and differed from Baikal.4G.fw by the predominance of the virotypes of the family Tectiviridae and the presence of baculoviruses. The sample from the Baltic Sea was placed into the main cluster with the samples from the Caspian Sea and Lake Baikal based on the taxonomic composition of the identified virotypes and families, but differed in their percentage. Virotypes related to siphoviruses also predominated in the samples from the Mediterranean Sea, but the Med.III.sw and Med.IV.sw samples differed from the Med.I.sw and Med.II.sw samples by the predominance of Haloviruses in them, the presence of representatives of Ascoviridae and the absence of viruses of the Microviridae family. In all the samples from the North Sea, the gradient vector shifted towards the predominance of myoviruses, but samples North.Sea.II.sw and North.Sea.III.sw differed in the presence of reads similar to poxviruses in them.
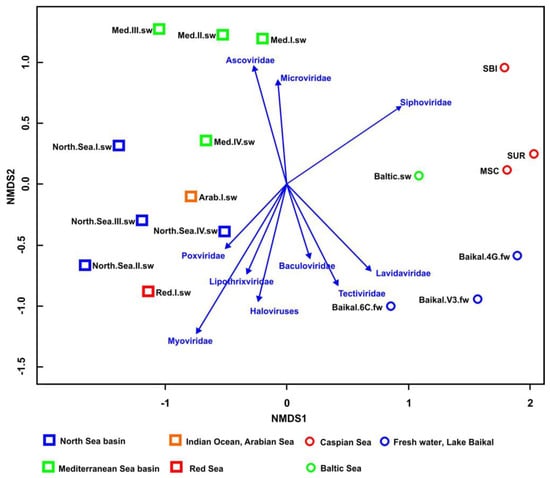
Figure 7.
NMDS (nonmetric multidimensional scaling) biplot based on Bray–Curtis distances showing the similarity of the samples based on virotype counts (similarity of the taxonomic composition of viral communities).
As a result of the comparative functional analysis of the viral communities of the North Caspian viromes and those of other freshwater and marine ecosystems, based on the assembled datasets and the KEGG database, the genes of the four highest hierarchical categories were identified in each of the samples (Figure 5a, Supplementary Table S6). A dendrogram based on the distribution of the highest functional categories grouped the analyzed samples into four clusters and showed the similarity of the virus communities of two Caspian samples (SUR and MSC) with freshwater samples Baikal_4G.fw and Baikal.6C.fw and with a sample from the Mediterranean Sea Med.III.sw (1 cluster). The second cluster combined samples from the Baltic Sea (Baltic.sw), Lake Baikal (Baikal.V3.fw), Mediterranean Sea (Med.IV.sw and Med,I,sw) and North Sea (North.Sea.I.sw). The smallest number of samples represented the third cluster: the Caspian Sea (SBI) and Mediterranean Sea (Med.II.sw) samples. The fourth cluster (4) consisted of five samples and included samples from the North Sea (North.Sea.II.sw; North.Sea.III.sw; North.Sea.IV.sw), Red Sea (Red.I.sw) and Arabian Sea (Arab.I.sw) (Figure 5a).
The analysis of the metavirome reads using the KEGG Orthology (KO) AMG pathway database showed the presence of auxiliary metabolic genes belonging to 11 secondary KO categories (Figure 5b). According to the heat map distribution, the highest proportion of reads of auxiliary metabolic genes in all the secondary categories was determined in samples from the North Sea (North.Sea.II.sw, North.Sea.III.sw, North.Sea.IV.sw), Red Sea (Red.I.sw) and Arabian Sea (Arab.I.sw), and the distribution of reads among secondary categories in these samples, as well as in sample North.Sea.I.sw, was relatively even. In the remaining samples, identified proteins predominantly represented categories “Glycan Biosynthesis and metabolism”, “Metabolism of Cofactors and Vitamins”, “Nucleotide metabolism” and “Metabolism of amino acids”, with the exception of the Med.I.sw sample, where proteins of the “Nucleotide metabolism”, “Metabolism of other amino acids” and “Xenobiotics biodegradation and metabolism” categories dominated, as well as samples Med.II.sw, Med.III.sw, Baikal.4G.fw and SBI, in which a significant dominance of auxiliary genes was identified in only one of the categories: “Amino Acids Metabolism” (Med.II.sw, Med.III.sw), “Glycan Biosynthesis and metabolism” (Baikal.4G.fw) or “Metabolism of Cofactors and Vitamins”(SBI). At the same time, Baikal.4G.fw was one of the studied samples where virotypes with genes encoding proteins involved in the process of metabolism and biodegradation of xenobiotics were least represented. Moreover, the heat map based on the distribution of the number of reads of auxiliary metabolic genes demonstrated the similarity of the viral communities of the Baika.6C.fw and Med.IV.sw samples, in which the largest number of encoded functions was related to glycan biosynthesis and metabolism, nucleotide, terpenoid and polyketide metabolism, and lipid metabolism. The most numerous proteins in terms of the number of reads were the categories “Glycan Biosynthesis and metabolism”, “Metabolism of cofactor and vitamin”, “Nucleotide Metabolism” and Amino Acid Metabolism in the Baikal.V3.fw and Baltic.sw samples, as well as the categories of lipid metabolism in the sample Baikal.V3.fw. Thus, the cluster analysis of the studied virotypes based on the profile of auxiliary metabolic genes showed that the samples of the Caspian Sea, Baltic Sea, Lake Baikal (Baikal.V3.fw, Baikal.4G.fw) and Mediterranean Sea (Med.III.sw) were in the same cluster, that SUR and MSC viromes of the Caspian Sea were similar to the virome of the Baltic Sea and that the SBI sample stood apart from the others (Figure 5b). Clustering of data from taxonomic analysis of virus communities grouped the studied samples mainly by bodies of water and revealed the highest similarity of all the samples of the Caspian Sea with the sample of the Baltic Sea (Figure 6a).
4. Discussion
Although viruses are recognized as the most widespread marine organisms and play a major role in nutrient cycling and genetic material transfer in marine ecosystems [8,14,67], our knowledge of the diversity, structure and functional features of marine virus communities is still far from being comprehensive.
This study, for the first time, investigated three viromes obtained from surface water samples from the Northern Caspian Sea and determined the genetic diversity of DNA viruses in the viromes. Taxonomic classification using the k-mers analysis based on the Kraken2 search using the NCBI nr (nonredundant) database revealed that viral sequences made up 11.96–43.19% of all reads. The taxonomic analysis showed that among the annotated viral sequences, the predominant group in the viromes of the SUR, MSC and SBI samples were tailed phages with double-stranded DNA along with other representatives of Phycodnaviridae that infect algae. These results are consistent with the fact that podo-, sipho- and myoviruses are known to be the main viral communities in aquatic viromes, and they greatly exceed eukaryotic DNA viruses in number [68].
These results are similar to the findings of other investigations of marine viromes, such as Monterey Bay (65% of reads belong to tailed phages), the Indian Ocean (95.3%), the East China Sea, the Baltic Sea, the Southern Ocean near the Antarctic Peninsula and the Pacific Ocean (~80%), where this group of bacteriophages also predominated [23,24,25,69,70,71]. The predominance of dsDNA phages in marine viral communities is related both to the abundance of their hosts, the bacterial microflora and to their capacity for lysogenic and lytic replication [26]. It is known that viruses of the Caudoviricetes class infect a wide range of host bacteria, including Proteobacteria and Bacteroidetes types, whose representatives are a dominant and integral part of marine bacterial communities [68,72] and are directly involved in the degradation of biopolymers, phytoplankton lysis and processing of organic matter obtained after the death of living organisms [73]. In general, phages belonging to the Myoviridae and Siphoviridae groups were the most numerous in the SUR and MSC samples but not in the SBI sample where up to 55% of the viral reads belonged to the Podoviridae group. In addition, eukaryotic DNA viruses of the Lavidaviridae family, commonly known as virophages, whose reproduction depends on a co-infecting giant virus of the Mimiviridae family, were detected in all the investigated viromes. Reads related to virophages predominated in the MSC sample, as did those related to mimiviruses. Other studies of viromes [46,74,75,76] showed that virophages are widely spread throughout the world in a variety of environments, both marine and freshwater. In addition to virophages and tailed phages, viruses of seven other different families were quantitatively predominant in the MSC compared to the SUR and SBI samples. This diversity of viruses is most likely due to the high species diversity of the biota in the wetland ecosystem of the Ural River delta and along the Ural–Caspian navigation channel in the location where this sample was collected [77].
The analysis of the viral community composition revealed that there were up to 631 different virotypes in the Northern Caspian Sea, while the composition of the viral communities of the SUR and MSC samples was similar to each other and significantly differed from that of SBI, which is explained by the fact that the collection areas of the first two samples are geographically connected by the influence of the inflowing Ural River.
SUR and MSC samples were dominated by viruses closely related to known cyanophages of unicellular cyanobacteria, mainly Synechococcus sp. The dominance of cyanophages in these samples may be a consequence of increased cyanobacterial content because of the introduction of freshwater phytoplankton by the Ural River and their more active growth in the shallow, and hence warmer, water of the Northern Caspian Sea [78].
In the SBI sample, the viral sequences predominantly belonged to the virotype that infects bacteria of the Sulfitobacter genus (Sulfitobacter phage pCB2047-A). Bacteria of this genus belong to the Alphaproteobacteria class and are gram-negative and sulfur-oxidizing chemolithoheterotrophs [79]. It was reported that representatives of this genus, on the one hand, stimulate the growth of some algae through the production of a hormone (indole-3-acetic acid) using the secreted and endogenous tryptophan of diatom algae [80]. On the other hand, they can also have an algicidal effect in microalgae in response to the presence of dimethylsulfoniopropionate (DMSP) [81] that is produced in large quantities by certain microalgae species involved in the sulfur cycle [82]. Sulfitobacter species, being in an environment with a high content of phytoplankton, are able to degrade DMSP and use it as a source of sulfur [80]. Thus, the presence of an active phytoplankton community is probably responsible for the presence of a dominant number of sequences similar to the Sulfitobacter phage virus in the SBI water sample. Unfortunately, it is not yet possible to confirm this assumption since we did not analyze bacterio- and phytoplankton in the SBI area at the time of sampling. To determine the cause of the dominance of the virus related to Sulfitobacter phage pCB2047-A, more extensive and targeted studies of the SBI area are needed, including the analysis of hydrochemical parameters and microbiota diversity. Such a difference in the SBI sample may be due to the formation of special conditions of environmental factors at a given sampling point (local pollution or other anthropogenic factor, nesting place of birds or habitat of a certain species of animals, etc.). It should be noted that strong fluctuations in the abundance and species composition of phytoplankton and bacterioplankton with a clear dominance of individual taxa were found in Lake Baikal, at sites less than 20 km away from each other, studied at the same time [83]. It is possible that such a heterogeneous development of bacteria and algae under similar environmental conditions is typical for large lakes, given that the Caspian Sea is essentially a lake.
In addition, the detection of a large number of reads similar to the Psychrobacter phage Psymv2 in the SUR sample was unexpected. Host bacterial strains of this phage include representatives of the Psychrobacter genus, which are found primarily in Antarctic latitudes from surface and deep water, sediments and soils [84,85].
The prevalence of phages specific to the widespread groups of the Candidatus Pelagibacter bacteria was detected in all the Caspian Sea samples studied. Pelagiphages HTVC010P morphologically belong to the group of short-tailed podoviruses, and are widely distributed in marine ecosystems. Pelagibacter phage HTVC010P was found in large quantities in surface ocean waters and is one of the most common members of the Caudoviricetes class in the biosphere [86]. Moreover, the viral communities were dominated by Puniceispirillum phages belonging to podoviruses. Another dominant virotype in the northern region of the Caspian Sea was Cellulophaga phage phi38:1, and the number of reads per virotype prevailed in the SUR and MSC samples. This virus infects marine heterotrophs belonging to the phylum Bacteroidetes. The number of representatives of the phylum Bacteroidetes increases during algal blooms because they are responsible for the degradation of polysaccharides and participate in the processing of organic matter from algal blooms [87]. To date, most Bacteroidetes infecting phage isolates affect bacteria of the Cellulophaga genus, which inhabit the surface of macroalgae [88]. The wide distribution of this phage in the environment is partially due to the structure of its genome, which contains a large number of tRNAs, allowing it to have a wider range of hosts [88].
In the studied viromes, sequences related to phages of pathogenic bacteria were found. The presence of such virotypes as Shigella phage Sf13, Bordetella virus BPP1 and Burkholderia phage KS9, specific for bacteria of the Shigella, Bordetella and Burkholderia genera, shows the presence in the studied water area of pathogens of bacterial infections, which may have ended up in the water with wastewater discharge.
Sequences of the Sphingobium phage were also identified in the Northern Caspian Sea samples. Bacteria of the species Sphingobium are their hosts. The detection of the Sphingobium bacteria indicates the presence of various oil products in this reservoir since these bacteria can degrade polycyclic aromatic hydrocarbons that constitute an integral part of crude oil. This fact is confirmed by active oil production on the Caspian shelf that causes contamination of seawater during transportation and pumping on oil tankers and in terminals [89]. The presented data confirm the fact that the population growth in the cities of the Caspian basin, increasing discharge of various wastewaters (industrial, agricultural and urban wastewater), development of oil production in the area, growing river navigation and eventual connection of the Caspian Sea with the World Ocean resulted in a significant increase in anthropogenic pressure on the unique ecosystem of the Caspian Sea and disturbance of its balance [90].
After assembling the reads, no full-genome viral sequences were found that showed a significant similarity to the reference sequences. Long scaffolds (greater than 25 kb) accounted for 1.5%, 1.4% and 1.7% of the total number of scaffolds with identified virotypes in the SUR, MSC and SBI samples, respectively. The k141_540989 scaffold was the longest, and was similar to the reference sequence, having the maximum number of predicted ORFs with a similarity of 71.8% according to the RefSeq protein database. Bacillus phage BCD7 was the closest relative of this scaffold. The k141_1441527 scaffold with the Synechococcus phage S-CBS4 virotype identified it was next in terms of the number of predicted ORFs with the maximum similarity of 83.8% in the RefSeq protein database.
One of the key aspects of virus evolution is the acquisition of host genes and their fixation in viral genomes [91]. It was suggested that such genes could improve the efficiency of viral reproduction and adaptation [92,93] by supporting key stages of host metabolism, due to which they are called “auxiliary metabolic genes”. Expression of auxiliary metabolic genes accelerates cellular processes of the host cell during viral infection [94]. Such auxiliary genes play crucial roles in successful viral proliferation. Thus, in energy metabolism, virotypes carrying genes involved in sulfur metabolism were identified in all the examined samples. Among them, the most frequent AMG was the cysH gene. The enzyme encoded by this gene is involved in the synthesis of sulfite from phosphoadenosine-5’-phosphosulfate (PAPS) and is thus part of the sulfate reduction pathway [95]. This enzyme is normally repressed during photoautotrophic growth using hydrogen sulfide as an electron donor and is used to incorporate sulfate into amino acids. Expression of cysH, encoded by the phage, can increase the intake of sulfite consumed by Mo-containing enzymes, which leads to an increase in cysteine synthesis and, presumably, a decrease in the difference of sulfur isotope fractionation between sulfate and sulfide [96,97]. The cysH gene has been identified in phages affecting members of the SAR11 clade that lack phosphoadenosine-5’-phosphosulfate reductase and other genes required for assimilative sulfate reduction, but has recently been found to be widely distributed among marine phages [98]. Additionally, in the Caspian Sea SBI sample, the genes responsible for porphyrin metabolism dominated: hemX, ahbD and CobS. These genes encode proteins that catalyze two sequential methylation reactions involved in the conversion of uroporphyrinogen III to precorrin-2 through the intermediate formation of precorrin-1, which are steps in the biosynthesis of both cobalamin (vitamin B12) and siroheme. The CobS gene encodes a protein that catalyzes the last stage of bacterial cobalamin (vitamin B12) biosynthesis, which promotes a more active proliferation of bacterial cells [99]. Assumptions about the participation of viruses in the biosynthesis of cobalamin in the marine ecosystem are very tempting but require more targeted research and experimental evidence to confirm this fact. The ahbD gene encodes an AdoMet-dependent hemosintase, which participates in protoheme biosynthesis by catalyzing the conversion of Fe-coproporphyrin III to heme [100]. This phenomenon was discovered and studied in sulfate-reducing bacteria of the Desulfivibrio genus and in methanogenic archaea [101]. Heme is an important prosthetic group involved in fundamental biological processes such as respiration, photosynthesis, metabolism and oxygen transport [102]. Similar accessory genes were also previously found in the Baltic Sea virome. Presumably, the genes cysH, ahbD and CobS are some of the conservative AMGs that are present independently of hosts and the environment and are found in various viral assemblies of different origins, such as the human gut, marine sediments and deep sea subsoil [103,104].
An analysis of the Global Ocean Survey metagenomic data based on the global metabolic network suggests that many auxiliary metabolic genes of viruses from various categories are closely related to purine and pyrimidine metabolic pathways. Therefore, there are suggestions that metabolic genes contained in marine virus genomes expand the nucleotide pool in infected hosts using two combined strategies: (1) recycling the building blocks of the cellular genome and transcriptome; and (2) altering host metabolism to form a substrate for de novo synthesis of purines and pyrimidines [105]. Thus, viral transfer into the host cell and subsequent expression of auxiliary metabolic genes result in increased energy metabolism and the accelerated assembly of viral particles by reducing the latent period, which increases the rate of the infection process and the accumulation of subsequent virus generations.
The functional analysis of viral scaffolds revealed the predominant accessory genes (pimA, pimC) in the SUR and MSC samples. These genes encode enzymes belonging to a new group of membrane-associated glycosyltransferase B (GT-B) [106,107]. There is evidence that a significant number of members of this group of enzymes are involved in the biosynthesis of major glycoconjugates in bacteria, including major human pathogens such as Neisseria meningitidis, Pseudomonas aeruginosa, Staphylococcus aureus and Streptococcus pneumoniae [108,109]. The predominance of such genes in these samples may indicate the presence of human pathogenic microorganisms in the environment, and as a consequence, a significant anthropogenic load.
It is known that the Caspian Sea is a relict basin from the ancient oceans Tethys and Paratethys that existed in the Mesozoic and early Cenozoic eras. Thus, the Caspian Sea was part of the World Ocean and finally separated from it about 14–10 million years ago as part of the Sarmatian Sea [110]. Therefore, probably, the general profile of the Caspian Sea virome retained some features of and similarity to the viromes of the open seas. Therefore, it was interesting for us to compare the viromes of the studied samples of the Caspian Sea with the viromes of both the open sea (North Sea, Baltic Sea, Mediterranean Sea, Red Sea, Arabian Sea) and lake ecosystems (Lake Baikal) uploaded to public databases.
Nonmetric multidimensional scaling of the identified viral families in the North Caspian Sea and comparison samples resulted in the formation of two main clusters: the first one containing samples from the Mediterranean, Arabian, Red and North Seas; the second cluster consisting of samples from the Caspian Sea, Baltic Sea and Lake Baikal. Such a distribution of the investigated samples in these two clusters on the NMDS scale indicated significant differences in the structure of viral communities. The comparison showed the similarity of the taxonomic diversity of the North Caspian Sea viromes with those of the Baltic Sea and Lake Baikal. The similarity of viromes in these reservoirs, located at a considerable distance from each other, can be explained by several reasons. Firstly, it is known that during the glacial maximum of the Quaternary period, ice sheets formed in the north of the Eurasian continent, and this prevented the flow of rivers into the open seas and oceans. As a result, vast ice-dammed lakes were formed at the base of glaciers. The flow of these lakes often occurred in the southern reservoirs, the path to which was not blocked by ice sheets. About 90,000 to 80,000 years ago, the Barents and Kara Ice Sheet blocked the flow of Siberian rivers, including the Yenissei and the Ob into the Arctic Ocean, rerouting them to the Aral and the Caspian Seas and thus creating a direct connection between Lake Baikal and these seas at that time. About 18–17 thousand years ago, due to the Scandinavian Ice Sheet in the north of Eastern Europe, a network of ice-dammed lakes was formed, directly connecting the Baltic Sea with the Volga river and the Caspian Sea [111]. Secondly, presumably since the second half of the 4th century, there has been the Volga–Baltic trade route (from the Varangians to the Persians), along which portage river transport has moved a lot of people and goods for 17 centuries, including the present, through a system of rivers and lakes. Moreover, the Volga–Baltic Waterway was created, thanks to whose canals, the Baltic Sea and the Caspian Sea became directly connected, which led to a significant intensification of river navigation and a multifold increase in the movement of ships, goods and people along waterways, and as a consequence, the transfer of microflora between the Baltic and Caspian Seas [112,113].
5. Conclusions
The study of the diversity of viruses of water samples of the Northern Caspian Sea showed that the formation of the main features of the virome is a long historical process affected by global abiogenic and biogenic factors. Thus, the movement of large volumes of water from the North to the South has caused some commonality in the viromes of the Northern Caspian Sea, the Baltic Sea and Lake Baikal. The development of oil fields in the Caspian Sea has led to the appearance of viruses that infect microorganisms feeding on oil products, while the identification of viruses similar to those that infect pathogenic human microflora indicates the presence of wastewater from human settlements. Functional analysis of the virome revealed auxiliary metabolic genes that are part of nutrient cycling pathways such as sulfur, carbon, nitrogen and others. These and other identified auxiliary genes are likely to enhance the adaptability of the viruses in the respective ecosystem by increasing the viability of their hosts, including under conditions of global climate change [27,114,115].
To comprehensively assess the viral diversity and determine its impact on the Caspian Sea ecosystem, the study of viruses from different regions of the Caspian Sea will be continued, and studies on the vertical distribution of marine viral communities of the Caspian basin will be made.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15070813/s1, Table S1: The list and taxonomy of virotypes revealed in analyzed viromes; Table S2: Obtained viral scaffolds after metagenomic assembly of all analyzed datasets; Table S3: Taxonomic identification of the scaffolds, the similarity of detected ORFs and RefSeq proteins; Table S4: General functional description of the proteins identified in specific scaffold with metabolic function; Table S5: List of identified viral families; Table S6: Main KO (KEGG Orthology) functional categories of predicted viral proteins and the number of reads related to these functions in the samples.
Author Contributions
Conceptualization, M.S.A., A.P.B. and T.V.B.; methodology, M.S.A. and A.P.B.; software, Y.S.B.; validation, T.V.B., Y.S.B. and M.S.A.; formal analysis, Y.S.B., T.V.B., M.S.A. and P.G.A.; investigation, M.S.A., Y.S.B. and P.G.A.; resources, M.S.A.; data curation, Y.S.B.; writing—original draft preparation, M.S.A.; writing—review and editing, all authors; visualization, Y.S.B., P.G.A. and M.S.A.; supervision, V.E.B.; project administration, M.S.A.; funding acquisition, M.S.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Ministry of Education and Science of the Republic of Kazakhstan, grant number: AP09058580.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Unprocessed virome reads for samples SUR, MSC and SBI of the Northern Caspian Sea were submitted to the NCBI SRA database (BioProject PRJNA961337, BioSamples SAMN34358971, SAMN34358972, SAMN34358973). The count tables on the taxonomic and functional analyses (Tables S1–S6) and R script (Supplementary File S1) are presented in this study as Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dimitrakopoulos, P.G.; Troumbis, A.Y. Biotopes. In Encyclopedia of Ecology, 2nd ed.; Fath, B., Ed.; Elsevier: Oxford, UK, 2019; pp. 359–365. ISBN 978-0-444-64130-4. [Google Scholar]
- Munang’andu, H.M.; Mugimba, K.K.; Byarugaba, D.K.; Mutoloki, S.; Evensen, Ø. Current Advances on Virus Discovery and Diagnostic Role of Viral Metagenomics in Aquatic Organisms. Front. Microbiol. 2017, 8, 406. [Google Scholar] [CrossRef]
- Zhang, Y.-Z.; Shi, M.; Holmes, E.C. Using Metagenomics to Characterize an Expanding Virosphere. Cell 2018, 172, 1168–1172. [Google Scholar] [CrossRef] [PubMed]
- Rumbou, A.; Vainio, E.J.; Büttner, C. Towards the Forest Virome: High-Throughput Sequencing Drastically Expands Our Understanding on Virosphere in Temperate Forest Ecosystems. Microorganisms 2021, 9, 1730. [Google Scholar] [CrossRef] [PubMed]
- Turmagambetova, A.S.; Alexyuk, M.S.; Bogoyavlenskiy, A.P.; Linster, M.; Alexyuk, P.G.; Zaitceva, I.A.; Smith, G.J.D.; Berezin, V.E. Monitoring of Newcastle Disease Virus in Environmental Samples. Arch. Virol. 2017, 162, 2843–2846. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Rodriguez, T.M.; Hollister, E.B. Human Virome and Disease: High-Throughput Sequencing for Virus Discovery, Identification of Phage-Bacteria Dysbiosis and Development of Therapeutic Approaches with Emphasis on the Human Gut. Viruses 2019, 11, 656. [Google Scholar] [CrossRef]
- Maranger, R.; Bird, D. Viral Abundance in Aquatic Systems:A Comparison between Marine and Fresh Waters. Mar. Ecol. Prog. Ser. 1995, 121, 217–226. [Google Scholar] [CrossRef]
- Middelboe, M.; Brussaard, C.P.D. Marine Viruses: Key Players in Marine Ecosystems. Viruses 2017, 9, 302. [Google Scholar] [CrossRef]
- Bergh, O.; Børsheim, K.Y.; Bratbak, G.; Heldal, M. High Abundance of Viruses Found in Aquatic Environments. Nature 1989, 340, 467–468. [Google Scholar] [CrossRef]
- Coutinho, F.H.; Silveira, C.B.; Gregoracci, G.B.; Thompson, C.C.; Edwards, R.A.; Brussaard, C.P.D.; Dutilh, B.E.; Thompson, F.L. Marine Viruses Discovered via Metagenomics Shed Light on Viral Strategies throughout the Oceans. Nat. Commun. 2017, 8, 15955. [Google Scholar] [CrossRef]
- Gregory, A.C.; Zayed, A.A.; Conceição-Neto, N.; Temperton, B.; Bolduc, B.; Alberti, A.; Ardyna, M.; Arkhipova, K.; Carmichael, M.; Cruaud, C.; et al. Marine DNA Viral Macro- and Microdiversity from Pole to Pole. Cell 2019, 177, 1109–1123.e14. [Google Scholar] [CrossRef]
- Alarcón-Schumacher, T.; Guajardo-Leiva, S.; Antón, J.; Díez, B. Elucidating Viral Communities During a Phytoplankton Bloom on the West Antarctic Peninsula. Front. Microbiol. 2019, 10, 1014. [Google Scholar] [CrossRef]
- Yuan, S.; Friman, V.-P.; Balcazar, J.L.; Zheng, X.; Ye, M.; Sun, M.; Hu, F. Viral and Bacterial Communities Collaborate through Complementary Assembly Processes in Soil to Survive Organochlorine Contamination. Appl. Environ. Microbiol. 2023, 89, e01810-22. [Google Scholar] [CrossRef]
- Suttle, C.A. Viruses in the Sea. Nature 2005, 437, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M. Marine Viruses: Truth or Dare. Annu. Rev. Mar. Sci. 2012, 4, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Marine Viruses–Major Players in the Global Ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, S.W.; Suttle, C.A. Viruses and Nutrient Cycles in the Sea: Viruses Play Critical Roles in the Structure and Function of Aquatic Food Webs. BioScience 1999, 49, 781–788. [Google Scholar] [CrossRef]
- Thingstad, T.F. Elements of a Theory for the Mechanisms Controlling Abundance, Diversity, and Biogeochemical Role of Lytic Bacterial Viruses in Aquatic Systems. Limnol. Oceanogr. 2000, 45, 1320–1328. [Google Scholar] [CrossRef]
- Thingstad, T.F.; Lignell, R. Theoretical Models for the Control of Bacterial Growth Rate, Abundance, Diversity and Carbon Demand. Aquat. Microb. Ecol. 1997, 13, 19–27. [Google Scholar] [CrossRef]
- Knowles, B.; Silveira, C.B.; Bailey, B.A.; Barott, K.; Cantu, V.A.; Cobián-Güemes, A.G.; Coutinho, F.H.; Dinsdale, E.A.; Felts, B.; Furby, K.A.; et al. Lytic to Temperate Switching of Viral Communities. Nature 2016, 531, 466–470. [Google Scholar] [CrossRef]
- Silveira, C.B.; Rohwer, F.L. Piggyback-the-Winner in Host-Associated Microbial Communities. Npj Biofilms Microbiomes 2016, 2, 16010. [Google Scholar] [CrossRef]
- Angly, F.E.; Felts, B.; Breitbart, M.; Salamon, P.; Edwards, R.A.; Carlson, C.; Chan, A.M.; Haynes, M.; Kelley, S.; Liu, H.; et al. The Marine Viromes of Four Oceanic Regions. PLoS Biol. 2006, 4, e368. [Google Scholar] [CrossRef]
- Steward, G.F.; Preston, C.M. Analysis of a Viral Metagenomic Library from 200 m Depth in Monterey Bay, California Constructed by Direct Shotgun Cloning. Virol. J. 2011, 8, 287. [Google Scholar] [CrossRef] [PubMed]
- Williamson, S.J.; Allen, L.Z.; Lorenzi, H.A.; Fadrosh, D.W.; Brami, D.; Thiagarajan, M.; McCrow, J.P.; Tovchigrechko, A.; Yooseph, S.; Venter, J.C. Metagenomic Exploration of Viruses throughout the Indian Ocean. PLoS ONE 2012, 7, e42047. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, B.L.; Sullivan, M.B. The Pacific Ocean Virome (POV): A Marine Viral Metagenomic Dataset and Associated Protein Clusters for Quantitative Viral Ecology. PLoS ONE 2013, 8, e57355. [Google Scholar] [CrossRef] [PubMed]
- Brum, J.R.; Ignacio-Espinoza, J.C.; Roux, S.; Doulcier, G.; Acinas, S.G.; Alberti, A.; Chaffron, S.; Cruaud, C.; de Vargas, C.; Gasol, J.M.; et al. Patterns and Ecological Drivers of Ocean Viral Communities. Science 2015, 348, 1261498. [Google Scholar] [CrossRef]
- Roux, S.; Brum, J.R.; Dutilh, B.E.; Sunagawa, S.; Duhaime, M.B.; Loy, A.; Poulos, B.T.; Solonenko, N.; Lara, E.; Poulain, J.; et al. Ecogenomics and Potential Biogeochemical Impacts of Globally Abundant Ocean Viruses. Nature 2016, 537, 689–693. [Google Scholar] [CrossRef]
- Rohwer, F.; Thurber, R.V. Viruses Manipulate the Marine Environment. Nature 2009, 459, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.H.; Sullivan, M.B. Marine Phage Genomics: What Have We Learned? Curr. Opin. Biotechnol. 2005, 16, 299–307. [Google Scholar] [CrossRef]
- Coutinho, F.H.; Gregoracci, G.B.; Walter, J.M.; Thompson, C.C.; Thompson, F.L. Metagenomics Sheds Light on the Ecology of Marine Microbes and Their Viruses. Trends Microbiol. 2018, 26, 955–965. [Google Scholar] [CrossRef]
- Perez Sepulveda, B.; Redgwell, T.; Rihtman, B.; Pitt, F.; Scanlan, D.J.; Millard, A. Marine Phage Genomics: The Tip of the Iceberg. FEMS Microbiol. Lett. 2016, 363, fnw158. [Google Scholar] [CrossRef]
- Warwick-Dugdale, J.; Buchholz, H.H.; Allen, M.J.; Temperton, B. Host-Hijacking and Planktonic Piracy: How Phages Command the Microbial High Seas. Virol. J. 2019, 16, 15. [Google Scholar] [CrossRef]
- Garin-Fernandez, A.; Pereira-Flores, E.; Glöckner, F.O.; Wichels, A. The North Sea Goes Viral: Occurrence and Distribution of North Sea Bacteriophages. Mar. Genomics 2018, 41, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Dumont, H.J. The Caspian Lake: History, Biota, Structure, and Function. Limnol. Oceanogr. 1998, 43, 44–52. [Google Scholar] [CrossRef]
- Van der Boon, A. From Peri-Tethys to Paratethys: Basin Restriction and Anoxia in Central Eurasia Linked to Volcanic Belts in Iran. Available online: https://dspace.library.uu.nl/handle/1874/356088 (accessed on 5 June 2023).
- Kosarev, A.N.; Yablonskaya, E.A. The Caspian Sea; SPB Academic Publishing: The Hague, The Netherlands, 1994; ISBN 978-90-5103-088-4. [Google Scholar]
- Kosarev, A.N. Physico-Geographical Conditions of the Caspian Sea. In The Caspian Sea Environment; Kostianoy, A.G., Kosarev, A.N., Eds.; The Handbook of Environmental Chemistry; Springer: Berlin/Heidelberg, Germany, 2005; pp. 5–31. ISBN 978-3-540-31505-6. [Google Scholar]
- Leroy, S.A.G.; Marret, F.; Gibert, E.; Chalié, F.; Reyss, J.-L.; Arpe, K. River Inflow and Salinity Changes in the Caspian Sea during the Last 5500 Years. Quat. Sci. Rev. 2007, 26, 3359–3383. [Google Scholar] [CrossRef]
- Van de Velde, S.; Wesselingh, F.P.; Yanina, T.A.; Anistratenko, V.V.; Neubauer, T.A.; ter Poorten, J.J.; Vonhof, H.B.; Kroonenberg, S.B. Mollusc Biodiversity in Late Holocene Nearshore Environments of the Caspian Sea: A Baseline for the Current Biodiversity Crisis. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2019, 535, 109364. [Google Scholar] [CrossRef]
- Mahmoudi, N.; Robeson, M.S.; Castro, H.F.; Fortney, J.L.; Techtmann, S.M.; Joyner, D.C.; Paradis, C.J.; Pfiffner, S.M.; Hazen, T.C. Microbial Community Composition and Diversity in Caspian Sea Sediments. FEMS Microbiol. Ecol. 2015, 91, 1–11. [Google Scholar] [CrossRef]
- Miller, J.I.; Techtmann, S.; Fortney, J.; Mahmoudi, N.; Joyner, D.; Liu, J.; Olesen, S.; Alm, E.; Fernandez, A.; Gardinali, P.; et al. Oil Hydrocarbon Degradation by Caspian Sea Microbial Communities. Front. Microbiol. 2019, 10, 995. [Google Scholar] [CrossRef]
- Prata, C.; Ribeiro, A.; Cunha, Â.; Gomes, N.C.M.; Almeida, A. Ultracentrifugation as a Direct Method to Concentrate Viruses in Environmental Waters: Virus-like Particle Enumeration as a New Approach to Determine the Efficiency of Recovery. J. Environ. Monit. 2012, 14, 64–70. [Google Scholar] [CrossRef]
- López-Pérez, M.; Haro-Moreno, J.M.; Gonzalez-Serrano, R.; Parras-Moltó, M.; Rodriguez-Valera, F. Genome Diversity of Marine Phages Recovered from Mediterranean Metagenomes: Size Matters. PLoS Genet. 2017, 13, e1007018. [Google Scholar] [CrossRef]
- Butina, T.V.; Bukin, Y.S.; Krasnopeev, A.S.; Belykh, O.I.; Tupikin, A.E.; Kabilov, M.R.; Sakirko, M.V.; Belikov, S.I. Estimate of the Diversity of Viral and Bacterial Assemblage in the Coastal Water of Lake Baikal. FEMS Microbiol. Lett. 2019, 366, fnz094. [Google Scholar] [CrossRef] [PubMed]
- Butina, T.V.; Bukin, Y.S.; Petrushin, I.S.; Tupikin, A.E.; Kabilov, M.R.; Belikov, S.I. Extended Evaluation of Viral Diversity in Lake Baikal through Metagenomics. Microorganisms 2021, 9, 760. [Google Scholar] [CrossRef]
- Butina, T.V.; Petrushin, I.S.; Khanaev, I.V.; Bukin, Y.S. Metagenomic Assessment of DNA Viral Diversity in Freshwater Sponges, Baikalospongia Bacillifera. Microorganisms 2022, 10, 480. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining Viral Signal from Microbial Genomic Data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Ye, Z.; Stanton, R. Misuse of RPKM or TPM Normalization When Comparing across Samples and Sequencing Protocols. RNA 2020, 26, 903–909. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog Assignment Based on Profile HMM and Adaptive Score Threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated Genome Annotation and Pathway Identification Using the KEGG Orthology (KO) as a Controlled Vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Tenenbaum, D.; Volkening, J.; Maintainer, B.P. KEGGREST: Client-Side REST Access to the Kyoto Encyclopedia of Genes and Genomes (KEGG). 2023. Available online: https://bioconductor.org/packages/release/bioc/html/KEGGREST.html (accessed on 27 April 2023).
- O’hara, R.B. Species Richness Estimators: How Many Species Can Dance on the Head of a Pin? J. Anim. Ecol. 2005, 74, 375–386. [Google Scholar] [CrossRef]
- Colwell, R.K.; Coddington, J.A. Estimating Terrestrial Biodiversity through Extrapolation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1994, 345, 101–118. [Google Scholar] [CrossRef]
- Hill, M.O. Diversity and Evenness: A Unifying Notation and Its Consequences. Ecology 1973, 54, 427–432. [Google Scholar] [CrossRef]
- Suzuki, R.; Shimodaira, H. Pvclust: An R Package for Assessing the Uncertainty in Hierarchical Clustering. Bioinformatics 2006, 22, 1540–1542. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P.; Stevenes, M.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.0-2. 2012. Available online: https://www.researchgate.net/publication/282247686_Vegan_Community_Ecology_Package_R_package_version_20-2 (accessed on 27 April 2023).
- Gplots: Various R Programming Tools for Plotting Data—ScienceOpen. Available online: https://www.scienceopen.com/document?vid=0e5d8e31-1fe4-492f-a3d8-8cd71b2b8ad9 (accessed on 2 April 2023).
- La Scola, B.; Desnues, C.; Pagnier, I.; Robert, C.; Barrassi, L.; Fournous, G.; Merchat, M.; Suzan-Monti, M.; Forterre, P.; Koonin, E.; et al. The Virophage as a Unique Parasite of the Giant Mimivirus. Nature 2008, 455, 100–104. [Google Scholar] [CrossRef]
- Weynberg, K.D. Viruses in Marine Ecosystems: From Open Waters to Coral Reefs. Adv. Virus Res. 2018, 101, 1–38. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M. Origins and Evolution of Viruses of Eukaryotes: The Ultimate Modularity. Virology 2015, 479, 2–25. [Google Scholar] [CrossRef]
- Wu, S.; Zhou, L.; Zhou, Y.; Wang, H.; Xiao, J.; Yan, S.; Wang, Y. Diverse and Unique Viruses Discovered in the Surface Water of the East China Sea. BMC Genom. 2020, 21, 441. [Google Scholar] [CrossRef] [PubMed]
- Brum, J.R.; Hurwitz, B.L.; Schofield, O.; Ducklow, H.W.; Sullivan, M.B. Seasonal Time Bombs: Dominant Temperate Viruses Affect Southern Ocean Microbial Dynamics. ISME J. 2016, 10, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Zeigler Allen, L.; McCrow, J.P.; Ininbergs, K.; Dupont, C.L.; Badger, J.H.; Hoffman, J.M.; Ekman, M.; Allen, A.E.; Bergman, B.; Venter, J.C. The Baltic Sea Virome: Diversity and Transcriptional Activity of DNA and RNA Viruses. mSystems 2017, 2, e00125-16. [Google Scholar] [CrossRef] [PubMed]
- Alexyuk, M.; Bogoyavlenskiy, A.; Alexyuk, P.; Moldakhanov, Y.; Berezin, V.; Digel, I. Epipelagic Microbiome of the Small Aral Sea: Metagenomic Structure and Ecological Diversity. MicrobiologyOpen 2020, 10, e1142. [Google Scholar] [CrossRef]
- Fernández-Gómez, B.; Richter, M.; Schüler, M.; Pinhassi, J.; Acinas, S.G.; González, J.M.; Pedrós-Alió, C. Ecology of Marine Bacteroidetes: A Comparative Genomics Approach. ISME J. 2013, 7, 1026–1037. [Google Scholar] [CrossRef]
- Gong, C.; Zhang, W.; Zhou, X.; Wang, H.; Sun, G.; Xiao, J.; Pan, Y.; Yan, S.; Wang, Y. Novel Virophages Discovered in a Freshwater Lake in China. Front. Microbiol. 2016, 7, 5. [Google Scholar] [CrossRef]
- Zhou, J.; Sun, D.; Childers, A.; McDermott, T.R.; Wang, Y.; Liles, M.R. Three Novel Virophage Genomes Discovered from Yellowstone Lake Metagenomes. J. Virol. 2014, 89, 1278–1285. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, W.; Yan, S.; Xiao, J.; Zhang, Y.; Li, B.; Pan, Y.; Wang, Y. Diversity of Virophages in Metagenomic Data Sets. J. Virol. 2013, 87, 4225–4236. [Google Scholar] [CrossRef]
- Ural River Delta, Kazakhstan. Available online: https://earthobservatory.nasa.gov/images/5551/ural-river-delta-kazakhstan (accessed on 8 June 2023).
- Heydari, N.; Fatemi, S.M.R.; Mashinchian, A.; Nadushan, R.M.; Raeisi, B. Seasonal Species Diversity and Abundance of Phytoplankton from the Southwestern Caspian Sea. Int. Aquat. Res. 2018, 10, 375–390. [Google Scholar] [CrossRef]
- Sorokin, D. Sulfitobacter Pontiacus Gen. Nov., Sp. Nov.—A New Heterotrophic Bacterium from the Black Sea, Specialized on Sulfite Oxidation. Microbiology 1995, 64, 295–305. [Google Scholar]
- Amin, S.A.; Hmelo, L.R.; van Tol, H.M.; Durham, B.P.; Carlson, L.T.; Heal, K.R.; Morales, R.L.; Berthiaume, C.T.; Parker, M.S.; Djunaedi, B.; et al. Interaction and Signalling between a Cosmopolitan Phytoplankton and Associated Bacteria. Nature 2015, 522, 98–101. [Google Scholar] [CrossRef]
- Barak-Gavish, N.; Frada, M.J.; Ku, C.; Lee, P.A.; DiTullio, G.R.; Malitsky, S.; Aharoni, A.; Green, S.J.; Rotkopf, R.; Kartvelishvily, E.; et al. Bacterial Virulence against an Oceanic Bloom-Forming Phytoplankter Is Mediated by Algal DMSP. Sci. Adv. 2018, 4, eaau5716. [Google Scholar] [CrossRef]
- Holligan, P.M.; Fernández, E.; Aiken, J.; Balch, W.M.; Boyd, P.; Burkill, P.H.; Finch, M.; Groom, S.B.; Malin, G.; Muller, K.; et al. A Biogeochemical Study of the Coccolithophore, Emiliania Huxleyi, in the North Atlantic. Glob. Biogeochem. Cycles 1993, 7, 879–900. [Google Scholar] [CrossRef]
- Mikhailov, I.S.; Zakharova, Y.R.; Bukin, Y.S.; Galachyants, Y.P.; Petrova, D.P.; Sakirko, M.V.; Likhoshway, Y.V. Co-Occurrence Networks Among Bacteria and Microbial Eukaryotes of Lake Baikal During a Spring Phytoplankton Bloom. Microb. Ecol. 2019, 77, 96–109. [Google Scholar] [CrossRef]
- Meiring, T.L.; Marla Tuffin, I.; Cary, C.; Cowan, D.A. Genome Sequence of Temperate Bacteriophage Psymv2 from Antarctic Dry Valley Soil Isolate Psychrobacter Sp. MV2. Extremophiles 2012, 16, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Romanenko, L.A.; Schumann, P.; Rohde, M.; Lysenko, A.M.; Mikhailov, V.V.; Stackebrandt, E. Psychrobacter submarinus Sp. Nov. and Psychrobacter marincola Sp. Nov., Psychrophilic Halophiles from Marine Environments. Int. J. Syst. Evol. Microbiol. 2002, 52, 1291–1297. [Google Scholar] [CrossRef]
- Zhao, Y.; Temperton, B.; Thrash, J.C.; Schwalbach, M.S.; Vergin, K.L.; Landry, Z.C.; Ellisman, M.; Deerinck, T.; Sullivan, M.B.; Giovannoni, S.J. Abundant SAR11 Viruses in the Ocean. Nature 2013, 494, 357–360. [Google Scholar] [CrossRef]
- Krüger, K.; Chafee, M.; Ben Francis, T.; Glavina del Rio, T.; Becher, D.; Schweder, T.; Amann, R.I.; Teeling, H. In Marine Bacteroidetes the Bulk of Glycan Degradation during Algae Blooms Is Mediated by Few Clades Using a Restricted Set of Genes. ISME J. 2019, 13, 2800–2816. [Google Scholar] [CrossRef]
- Bischoff, V.; Zucker, F.; Moraru, C. Marine Bacteriophages. In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: Oxford, UK, 2021; pp. 322–341. ISBN 978-0-12-814516-6. [Google Scholar]
- Madsen, E.L. 6.10—Biodegradability of Recalcitrant Aromatic Compounds. In Comprehensive Biotechnology, 2nd ed.; Moo-Young, M., Ed.; Academic Press: Burlington, ON, Canada, 2011; pp. 95–103. ISBN 978-0-08-088504-9. [Google Scholar]
- Bagheri, S.; Turkoglu, M.; Abedini, A. Phytoplankton and Nutrient Variations in the Iranian Waters of the Caspian Sea (Guilan Region) during 2003–2004. Turk. J. Fish. Aquat. Sci. 2014, 14, 231–245. [Google Scholar] [CrossRef]
- Irwin, N.A.T.; Pittis, A.A.; Richards, T.A.; Keeling, P.J. Systematic Evaluation of Horizontal Gene Transfer between Eukaryotes and Viruses. Nat. Microbiol. 2022, 7, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Lindell, D.; Jaffe, J.D.; Johnson, Z.I.; Church, G.M.; Chisholm, S.W. Photosynthesis Genes in Marine Viruses Yield Proteins during Host Infection. Nature 2005, 438, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Dammeyer, T.; Bagby, S.C.; Sullivan, M.B.; Chisholm, S.W.; Frankenberg-Dinkel, N. Efficient Phage-Mediated Pigment Biosynthesis in Oceanic Cyanobacteria. Curr. Biol. 2008, 18, 442–448. [Google Scholar] [CrossRef]
- Sullivan, M.B.; Lindell, D.; Lee, J.A.; Thompson, L.R.; Bielawski, J.P.; Chisholm, S.W. Prevalence and Evolution of Core Photosystem II Genes in Marine Cyanobacterial Viruses and Their Hosts. PLoS Biol. 2006, 4, e234. [Google Scholar] [CrossRef]
- Bick, J.A.; Dennis, J.J.; Zylstra, G.J.; Nowack, J.; Leustek, T. Identification of a New Class of 5’-Adenylylsulfate (APS) Reductases from Sulfate-Assimilating Bacteria. J. Bacteriol. 2000, 182, 135–142. [Google Scholar] [CrossRef]
- Haverkamp, T.; Schwenn, J.D. Structure and Function of a CysBJIH Gene Cluster in the Purple Sulphur Bacterium Thiocapsa Roseopersicina. Microbiology 1999, 145 Pt 1, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Hesketh-Best, P.J.; Bosco-Santos, A.; Garcia, S.L.; O’Beirne, M.D.; Werne, J.P.; Gilhooly, W.P.; Silveira, C.B. Viruses of Sulfur Oxidizing Phototrophs Encode Genes for Pigment, Carbon, and Sulfur Metabolisms. Commun. Earth Environ. 2023, 4, 126. [Google Scholar] [CrossRef]
- Kieft, K.; Zhou, Z.; Anderson, R.E.; Buchan, A.; Campbell, B.J.; Hallam, S.J.; Hess, M.; Sullivan, M.B.; Walsh, D.A.; Roux, S.; et al. Ecology of Inorganic Sulfur Auxiliary Metabolism in Widespread Bacteriophages. Nat. Commun. 2021, 12, 3503. [Google Scholar] [CrossRef]
- Magnúsdóttir, S.; Ravcheev, D.; de Crécy-Lagard, V.; Thiele, I. Systematic Genome Assessment of B-Vitamin Biosynthesis Suggests Co-Operation among Gut Microbes. Front. Genet. 2015, 6, 148. [Google Scholar] [CrossRef]
- Bali, S.; Lawrence, A.D.; Lobo, S.A.; Saraiva, L.M.; Golding, B.T.; Palmer, D.J.; Howard, M.J.; Ferguson, S.J.; Warren, M.J. Molecular Hijacking of Siroheme for the Synthesis of Heme and D1 Heme. Proc. Natl. Acad. Sci. USA 2011, 108, 18260–18265. [Google Scholar] [CrossRef] [PubMed]
- Buchenau, B.; Kahnt, J.; Heinemann, I.U.; Jahn, D.; Thauer, R.K. Heme Biosynthesis in Methanosarcina Barkeri via a Pathway Involving Two Methylation Reactions. J. Bacteriol. 2006, 188, 8666–8668. [Google Scholar] [CrossRef] [PubMed]
- Layer, G.; Reichelt, J.; Jahn, D.; Heinz, D.W. Structure and Function of Enzymes in Heme Biosynthesis. Protein Sci. Publ. Protein Soc. 2010, 19, 1137–1161. [Google Scholar] [CrossRef] [PubMed]
- Heyerhoff, B.; Engelen, B.; Bunse, C. Auxiliary Metabolic Gene Functions in Pelagic and Benthic Viruses of the Baltic Sea. Front. Microbiol. 2022, 13, 863620. [Google Scholar] [CrossRef] [PubMed]
- Kieft, K.; Zhou, Z.; Anantharaman, K. VIBRANT: Automated Recovery, Annotation and Curation of Microbial Viruses, and Evaluation of Viral Community Function from Genomic Sequences. Microbiome 2020, 8, 90. [Google Scholar] [CrossRef]
- Enav, H.; Mandel-Gutfreund, Y.; Béjà, O. Comparative Metagenomic Analyses Reveal Viral-Induced Shifts of Host Metabolism towards Nucleotide Biosynthesis. Microbiome 2014, 2, 9. [Google Scholar] [CrossRef]
- Albesa-Jové, D.; Giganti, D.; Jackson, M.; Alzari, P.M.; Guerin, M.E. Structure–Function Relationships of Membrane-Associated GT-B Glycosyltransferases. Glycobiology 2014, 24, 108. [Google Scholar] [CrossRef]
- Berg, S.; Kaur, D.; Jackson, M.; Brennan, P.J. The Glycosyltransferases of Mycobacterium Tuberculosis—Roles in the Synthesis of Arabinogalactan, Lipoarabinomannan, and Other Glycoconjugates. Glycobiology 2007, 17, 35R–56R. [Google Scholar] [CrossRef]
- Berg, S.; Edman, M.; Li, L.; Wikström, M.; Wieslander, Å. Sequence Properties of the 1,2-Diacylglycerol 3-Glucosyltransferase from Acholeplasma LaidlawiiMembranes: Recognition of a Large Group of Lipid Glycosyltransferases in Eubacteria and Archaea. J. Biol. Chem. 2001, 276, 22056–22063. [Google Scholar] [CrossRef]
- Lind, J.; Rämö, T.; Klement, M.L.R.; Bárány-Wallje, E.; Epand, R.M.; Epand, R.F.; Mäler, L.; Wieslander, A. High Cationic Charge and Bilayer Interface-Binding Helices in a Regulatory Lipid Glycosyltransferase. Biochemistry 2007, 46, 5664–5677. [Google Scholar] [CrossRef]
- Esin, N.V.; Yanko-Hombach, V.V.; Esin, N.I. Evolutionary Mechanisms of the Paratethys Sea and Its Separation into the Black Sea and Caspian Sea. Quat. Int. 2018, 465, 46–53. [Google Scholar] [CrossRef]
- Mangerud, J.; Jakobsson, M.; Alexanderson, H.; Astakhov, V.; Clarke, G.K.C.; Henriksen, M.; Hjort, C.; Krinner, G.; Lunkka, J.-P.; Möller, P.; et al. Ice-Dammed Lakes and Rerouting of the Drainage of Northern Eurasia during the Last Glaciation. Quat. Sci. Rev. 2004, 23, 1313–1332. [Google Scholar] [CrossRef]
- Zimnitskaya, H.; von Geldern, J. Is the Caspian Sea a Sea; and Why Does It Matter? J. Eurasian Stud. 2011, 2, 1–14. [Google Scholar] [CrossRef]
- Dmitrievich, F.O. Rus: The Way From the Varangians to the Persians. Humanitarian Paradigm 2018, 2, 27–36. (In Russian) [Google Scholar]
- Danovaro, R.; Corinaldesi, C.; Dell’anno, A.; Fuhrman, J.A.; Middelburg, J.J.; Noble, R.T.; Suttle, C.A. Marine Viruses and Global Climate Change. FEMS Microbiol. Rev. 2011, 35, 993–1034. [Google Scholar] [CrossRef] [PubMed]
- Ettinger, C.L.; Saunders, M.; Selbmann, L.; Delgado-Baquerizo, M.; Donati, C.; Albanese, D.; Roux, S.; Tringe, S.; Pennacchio, C.; del Rio, T.G.; et al. Highly Diverse and Unknown Viruses May Enhance Antarctic Endoliths’ Adaptability. Microbiome 2022, 11, 103. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).