Abstract
The basic knowledge of freshwater bivalves in the Unionida in some regions of the world is still limited, hindering potential conservation efforts, including in Vietnam. A subset of these mussels, the freshwater bivalve tribe Anodontini, is especially difficult to properly identify morphologically due to intraspecific shell similarity. This study aims to define the species of Anodontini in Vietnam and describe their evolutionary relationships and distributions by estimating phylogenies and analyzing collected specimens. The Anodontini are represented in Vietnam by five species divided among three genera: Sinanodonta, Cristaria, and Pletholophus. Sinanodonta woodiana, a large species complex, is represented in Vietnam by Sinanodonta jourdyi. Cristaria is confirmed to include the widespread Cristaria plicata and substantiates the validity of Cristaria truncata. Finally, Pletholophus is here recognized as distinct from Cristaria, containing two species in Vietnam, Pletholophus tenuis, and a species new to science. Our study is an important baseline for future studies on Vietnamese freshwater mussels and highlights the importance of surveys, molecular work, and taxonomic expertise to describe the biodiversity of understudied regions.
1. Introduction
Freshwater bivalves in the order Unionida, also recognized as freshwater mussels, are known for their unique life cycle, with a parasitic phase primarily on fish [1] and an uncommon doubly mitochondrial inheritance [2]. These animals are notorious for providing vital ecosystem functions and services [3]. They are useful for exploring past geological and hydrological events due to their stable biogeography [4]. Unfortunately, this group is among the most imperiled worldwide, with many species considered extinct or on the brink of extinction [5]. This highlights the urgency of describing their diversity, distribution, and evolution, which is reflected in the growing research and conservation attention paid to this group [6,7]. However, in many regions of the world, the necessary baseline knowledge to guide conservation efforts is still lacking [5]. For instance, in Asia, our understanding of the biology and systematics of these animals’ ranges from poor to absent in many areas [8]. Many Asian River basins have never been surveyed and may be home to numerous undescribed genera and species (e.g., [9,10]) that may even go extinct before being described or studied [11,12,13,14]. Despite being poorly known, the freshwater mussel fauna of Asia has recently attracted intense research interest, especially in taxonomy, phylogeny, and biogeography (e.g., [8,10,15,16,17,18,19,20,21,22,23,24,25]). However, these efforts have been mostly regional or national, with several countries or regions being overlooked, such as Vietnam. In this country, research on freshwater mussels is limited, and only recently has a wide study on the Vietnamese freshwater bivalve fauna been published to document its diversity and distribution [26]. However, it relied solely on shell characters, which often fail to properly identify and separate species.
Species belonging to the freshwater tribe Anodontini generally present high cryptic (species that can only be distinguished molecularly) diversity and exhibit high habitat and host-fish plasticity. However, virtually nothing is reported about the habitat requirements and main life history traits of these species in Vietnam. Furthermore, until this study, the number and identity of species belonging to this tribe in Vietnam were still contentious, with previous morphological identifications not being verified molecularly. Therefore, the present study aims to overcome this issue and update the systematics, taxonomy, phylogeny, and distribution of Anodontini freshwater mussels from Vietnam based upon extensive surveys by (i) examining and defining species boundaries and distributions; (ii) estimating phylogenies with new data from additional specimens; (iii) revising species taxonomy and systematics; and (iv) discussing conservation implications based on these results.
2. Materials and Methods
2.1. Field and Tissue Sampling
Representative specimens of Anodontini were collected during extensive surveys across Vietnam from 2010 to 2016, also covering local markets where freshwater mussels are typically sold as food (Figure 1) [26]. Specimens and shells were collected as vouchers and deposited in the IEBR—Institute of Ecology and Biological Resources, Hanoi, Vietnam, and the NCSM—North Carolina Museum of Natural Sciences, U.S.A. Selected specimens were sampled for molecular analyses (Figure 2) (Supplementary Tables S1–S3). For this, a small sample of foot tissue was collected (see [27]) and placed in 95% ethanol.

Figure 1.
Freshwater mussels for sale as food in local market in Hanoi, Vietnam. Photograph by Arthur Bogan, 2014.
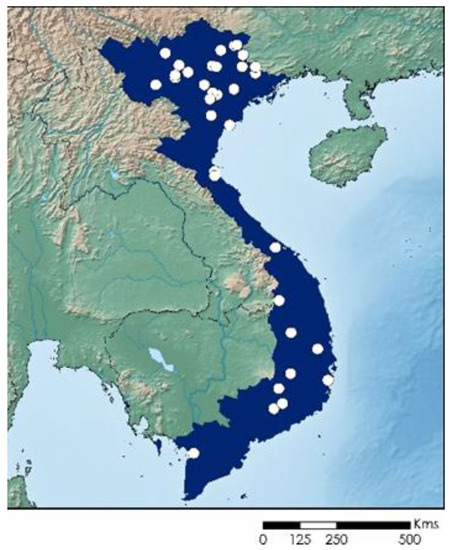
Figure 2.
Map of Vietnam, with the study area shaded in blue. White dots mark the location of collected specimens or shells.
2.2. DNA Extraction, PCR, and Sequencing
Genomic DNA was extracted from the tissue samples using a standard high-salt protocol [28]. PCRs were done for the female lineages of mitochondrial cytochrome c oxidase subunit 1, COI (annealing temperature of 48 °C), and 28S ribosomal RNA (annealing temperature of 50 °C) as described in [29]. Amplified DNA templates were purified and sequenced using the same primers by Macrogen.
2.3. Phylogenetic Analyses
In order to estimate the phylogenetic position of all the newly sequenced individuals, a preliminary COI alignment was produced, including all available Anodontini. A Bayesian Inference phylogenetic tree was produced using MrBayes 3.2.7a [30] with 20 × 106 generations sampled at intervals of 1000 generations on a single partition with the model GTR + I + G.
Based on this preliminary analysis, three genera were revealed to be present in the data set, i.e., Cristaria, Pletholophus, and Sinanodonta (Supplementary Tables S1–S3). Therefore, a concatenated COI + 28S dataset was constructed using one individual of each clade inside each genus (Table 1). Sequences from species belonging to the same three genera as well as a single representative of all previously published sequences of congeneric taxa and four other taxa as an outgroup were also included (Table 1). This data set was aligned with GUIDANCE2 [31], following [32], and analyzed using Maximum Likelihood (ML) in IQ-TREE v. 1.6.12 [33] and Bayesian inference (BI) using MrBayes 3.2.7a [30]. For the BI analyses, the best-fit models of nucleotide substitution and partition schemes were selected using PartitionFinder 2 [34] under the Bayesian information criterion. BI analyses were initiated with program-generated trees and four Markov chains with default incremental heating. Two independent runs of 20 × 106 generations were sampled at intervals of 1000 generations, producing a total of 20,000 trees. Burn-in was determined upon the convergence of log-likelihood and parameter values using Tracer 1.7.1 [35]. The best-fit models of nucleotide substitution and partition schemes were selected using ModelFinder [36] for the ML, and then the analyses were conducted with initial tree searches, followed by 10 independent runs and 10,000 ultrafast bootstrap replicates. Substitution saturation tests for all codon positions were performed for COI as implemented in DAMBE 6 [37].

Table 1.
List of sequences used for the phylogenies of the Cristariini with respective species, countries, voucher numbers, and GenBank references selected from the combined dataset (COI + 28S rRNA). Acronyms for the voucher deposit institutions are as follows: IEBR—Institute of Ecology and Biological Resources, Hanoi, Vietnam; MHS—Matsuyama High School; NCSM—North Carolina Museum of Natural Sciences, U.S.A.; NNIBR—Nakdonggang National Institute of Biological Resources, South Korea; NCU—Nanchang University, China; OMNH—Osaka Museum of Natural History, Japan; RMBH—Russian Museum of Biodiversity Hotspots, Arkhangelsk, Russia.
Three individual COI data sets, corresponding to the three genera found, i.e., Cristaria, Pletholophus, and Sinanodonta, were also produced with all newly sequenced individuals, including different outgroup taxa (Supplementary Tables S1–S3). Alignments were produced in GUIDANCE2 and analyzed using ML and BI methods, following the same steps applied to the concatenated data set (above).
2.4. Species Delimitation
Three distinct methods were applied to each COI dataset, without outgroup taxa, to determine the number of molecular operational taxonomic units (MOTUs). Two are distance-based: the BIN system implemented in BOLD [40] and the automatic barcode gap discovery (ABGD) [41]; and another that uses haplotype network reconstructions in TCS 1.21 [42] with a 95% statistical parsimony connection limit. Sequence divergences (uncorrected p-distance) were estimated using MEGA X [43].
For the BINs system, each COI dataset was analyzed with the cluster sequences tool implemented in BOLD 4 (http://v4.boldsystems.org) (accessed 28 March 2020) [40]. The ABGD was applied to each dataset using its online version (http://wwwabi.snv.jussieu.fr/public/abgd/abgdweb.html) (accessed 28 March 2020) with the default settings and the Kimura-2-parameter (K2P) distance matrix [41].
2.5. Distribution Maps
The range of each species in the study area was mapped using the current results and previous reference works [18,26,44,45,46]. The distribution data were then integrated and represented as colored potential distribution maps using a level 8 HydroBASINS [47] shapefile. For pictorial reasons, Earth topography layers made by Natural Earth http://naturalearthdata.com (accessed 26 March 2020) were also included.
3. Results and Discussion
Here we present the diversity of the Anodontini of Vietnam, recognizing five valid Anodontini species, including the newly described Pletholophus honglinhensis sp. nov. We also describe their distribution, revise their taxonomy, and give guidance for future studies and conservation action.
The preliminary Bayesian inference phylogeny retrieved three clades that include Vietnamese Anodontini taxa, which were considered for further phylogenetic and species delimitation analyses. The composition, size, and parameters of the four datasets (3 COI and 1 COI + 28S) and the partition schemes and nucleotide substitution models for all analyses are presented in Table 2. No indels or stop codons were found in any of the COI datasets. The saturation test showed significantly lower values of ISS than ISS.C (a critical value determined from the computer simulation), indicating that the evaluated (COI) data set is not site saturated and is useful for phylogenetic comparisons. Both (COI + 28S) BI and ML phylogenies presented the same topology (Figure 3). All the generic-level clades are well supported, but the relationships among some of them still lack resolution (Figure 3). In both phylogenies, the newly described species Pletholophus honglinhensis sp. nov. clusters with Pletholophus tenuis, Sinanodonta jourdyi within the Sinanodonta clade, and Cristaria truncata within the Cristaria clade (Figure 3).

Table 2.
Number of sequences, haplotypes, and sizes of all datasets used, as well as substitution models for each partition for all phylogenetic analyses.
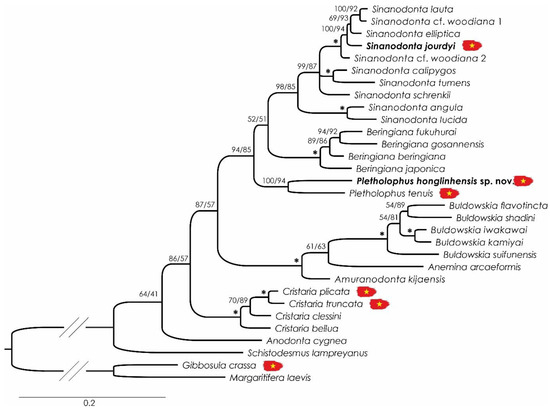
Figure 3.
Bayesian inference phylogenetic tree inferred from the combined cytochrome c oxidase subunit I + 28S ribosomal RNA gene fragments. The values above the branches indicate Bayesian posterior probability percentage/maximum likelihood ultrafast bootstrap values. Values over 95% for both analyses are represented by an asterisk. Sequences marked with the Vietnamese flag represent specimens collected in Vietnam.
Molecular species delimitation for the COI individual datasets revealed the existence of five MOTUs present in the study area, here recognized as species (Figure 4, Figure 5 and Figure 6).
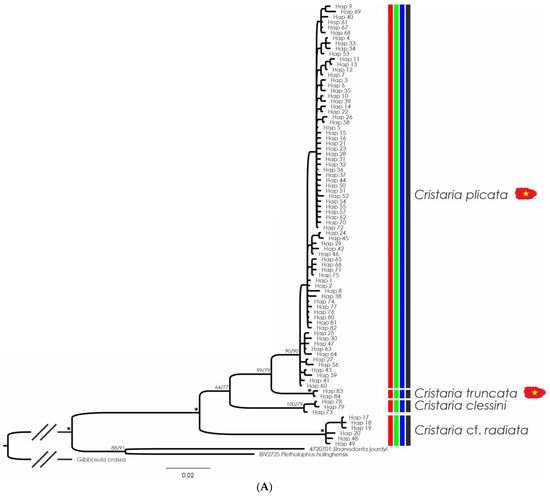
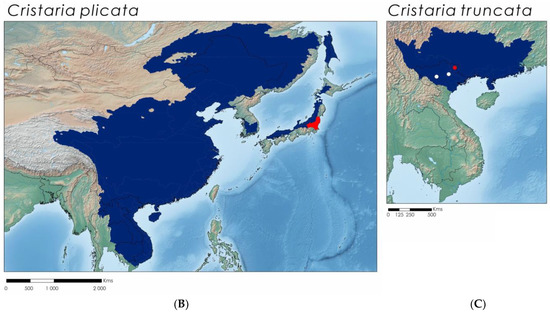
Figure 4.
(A) Bayesian Inference phylogenetic tree inferred from the cytochrome c oxidase subunit I (COI) gene fragment, species delimitation of Cristaria species, and distribution maps of the species present in the study area (marked with Vietnamese flags). Support values above the branches are percent posterior probabilities/ultrafast bootstraps. Vertical bars correspond to molecular operational taxonomic units by various species delimitation methods: red—TCS (95%); green—ABGD; blue—BINS of BOLD; and black—consensus. Support values >95% for both phylogenetic analyses are represented by an asterisk; support values within each recognized MOTU were erased for clarity. Distribution maps of each species (B,C) depicting the potential distribution across the main river basins in shaded colors, the native range in blue, and the introduced range in red. White dots represent collected specimens or shells, red dots represent sequenced individuals. Sequences from the current study are represented in bold.
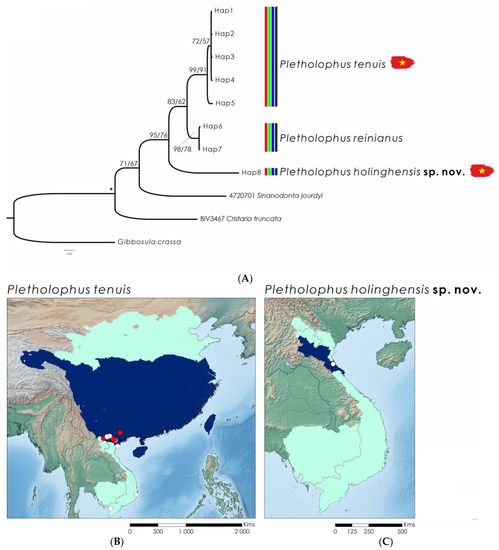
Figure 5.
(A) Bayesian Inference phylogenetic tree inferred from the cytochrome c oxidase subunit I (COI) gene fragment, species delimitation of Pletholophus species, and distribution maps of species present in the study area (marked with Vietnamese flags). Support values above the branches are percent posterior probabilities/ultrafast bootstraps. Vertical bars correspond to molecular operational taxonomic units by various species delimitation methods: red—TCS (95%); green—ABGD; blue—BINS of BOLD; and black—consensus. Support values >95% for both phylogenetic analyses are represented by an asterisk. Distribution maps for each species (B,C) depicting the potential distribution across the main river basins are in shaded colors; the known native range is in dark blue, and the questionable range is in light blue. White dots represent collected specimens or shells; red dots represent sequenced individuals. Sequences from the current study are represented in bold.
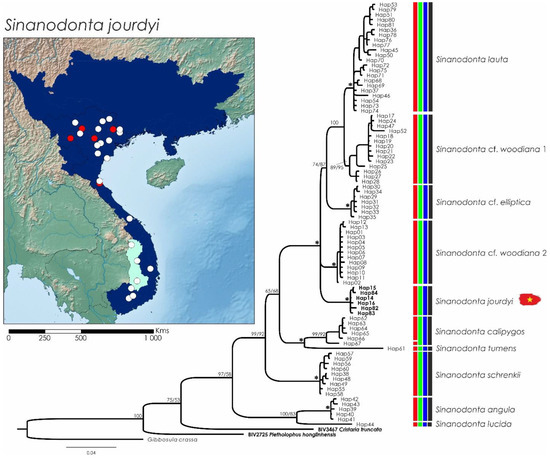
Figure 6.
Bayesian Inference phylogenetic tree inferred from the cytochrome c oxidase subunit I (COI) gene fragment, species delimitation of Sinanodonta species, and distribution maps of S. jourdyi present in the study area (marked with a Vietnamese flag). Support values above the branches are percent posterior probabilities/ultrafast bootstraps. Vertical bars correspond to molecular operational taxonomic units by various species delimitation methods: red—TCS (95%); green—ABGD; blue—BINS of BOLD; and black—consensus. Support values >95% for both phylogenetic analyses are represented by an asterisk; support values within each recognized MOTU were erased for clarity. Distribution maps of each species depict the potential distribution across the main river basins in shaded colors; white dots represent newly sequenced individuals. Distribution map of S. jourdyi depicting the potential distribution across the main river basins in shaded colors, the known native range in dark blue, and the questionable range in light blue. White dots represent collected specimens or shells; red dots represent sequenced individuals. Sequences from the current study are represented in bold.
From the Cristaria genus, 12 specimens were collected across Vietnam, but only a single Cristaria truncata lot was sequenced. Both COI (ML and BI) phylogenies for Cristaria exhibit similar topologies, retrieving five clades recovered as separate MOTUs by all species delimitation methods (Figure 4). Uncorrected p-distances among the delineated MOTUs (here recognized as species; see Taxonomic Account Section) ranged from 4.4% (between C. plicata and C. truncata) to 11.9% (between C. bellua and Cristaria sp.) (Table 3).

Table 3.
Pairwise genetic distance matrixes among species from the three Cristariini genera present in Vietnam. Mean uncorrected p-distance among species of cytochrome oxidase subunit I (COI) gene fragment (below the diagonal) and associated standard error (above the diagonal in blue). Taxa occurring in the study region are represented in bold. The lines are colored to make reading across the table easier.
From Pletholophus, 80 specimens were collected throughout Vietnam (Figure 5). Both COI ML and BI phylogenies for Pletholophus exhibit similar topologies, retrieving three clades recovered as separate MOTUs by all species delimitation methods (Figure 5). Uncorrected p-distances among the delineated MOTUs (here recognized as species; see Taxonomic Account Section) ranged from 5.0% (between P. tenuis and P. reinianus) to 10.9% (between P. honglinhensis sp. nov. and P. tenuis) (Table 3). Pletholophus tenuis has been recorded from the Mekong Basin in Vietnam north to the Yellow River Basin in China. Given that no molecular data is available from the distribution edges in Vietnam south of the city of Thanh Hoa and in the Yellow River Basin in China, the specific status of the Pletholophus specimens from these areas still needs to be confirmed. Pletholophus honglinhensis sp. nov. was found only in a coastal basin south of Hanoi.
From the Sinanodonta genus, 45 specimens were collected throughout Vietnam (Figure 6). Both COI (ML and BI) phylogenies for the Sinanodonta genus presented similar topologies, retrieving ten clades, defined as MOTUs by a consensus of all species delimitation methods. Only one of these MOTUs is present in Vietnam (Figure 6). Uncorrected p-distances among the delineated MOTUs (here recognized as species) ranged from 3.2% (between S. lauta (Martens, 1877) [48] and S. cf. woodiana 1) to 13.7% (between S. angula (Tchang et al., 1965) [49] and S. cf. elliptica (Heude, 1878) [50]) (Table 3). Sinanodonta jourdyi was detected throughout Vietnam, from the Pearl River Basin in the northeast to the southern coastal basins. The species has been detected in the lower Mekong Basin in Vietnam, but it is still contentious whether the species is native to this area (Figure 6).
3.1. Taxonomy of Sinanodonta, Cristaria, and Pletholophus
Ðặng et al. [44] recognized three separate species of Sinanodonta in Vietnam, while Haas [51] included all those names as synonyms of S. woodiana. Later, Graf and Cummings [52] followed Haas [51] and recognized only a single Sinanodonta species for Vietnam, i.e., S. woodiana. However, Sinanodonta woodiana (Lea, 1834) [53] in Asia has been recently shown to be a complex of at least seven mitochondrial lineages [54]. Here, we show that only one of these lineages of Sinanodonta woodiana occurs in Vietnam and was previously identified as Sinanodonta jourdyi (Morlet, 1886) [8,26,55,56,57,58,59].
Cristaria Schumacher, 1817 [60] was erected to separate the type species, Cristaria tuberculata Schumacher, 1817 [60] (type species by monotypy), from other thin-shelled taxa with wings but with a unique hinge. Simpson [61] erected Cristaria (Pletholophus) with the type species Symphynota discoidea Lea, 1834 [53] (original designation). Heard [62], in describing the comparative anatomy of C. plicata, made observations on Cristaria (Pletholophus) discoidea and mentioned anatomical distinctions between C. discoidea and C. plicata. A new Cristaria species restricted to northern Vietnam, C. truncata Ðặng in Ðặng et al. [44], was described based on shell characters (Figure 7). They also moved Cristaria discoidea out of Cristaria, elevating Pletholophus Simpson, 1900 [61] to the generic level. Graf & Cummings [52] recognized four species of Cristaria, including C. truncata, but did not recognize Pletholophus. He & Zhuang [63] recognized three species of Cristaria and placed another species of Cristaria erroneously in Middendorffinaia Moskvicheva & Starobogatov, 1973 [64] in China. Graf & Cummings [58] recognized five species in Cristaria, including C. truncata, and recognized Pletholophus, which included two species.
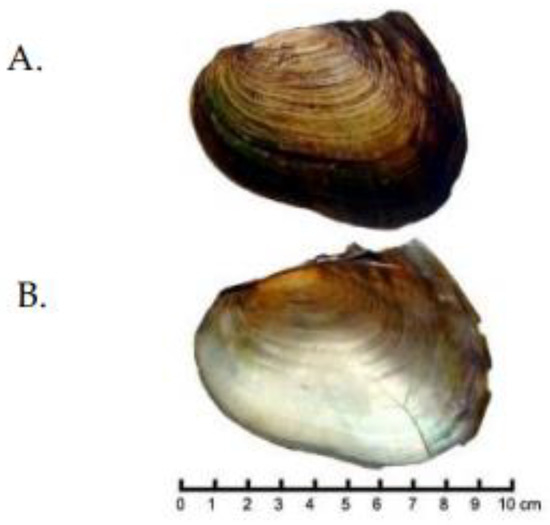
Figure 7.
Shell of Cristaria truncata. (A). Outside of left valve. (B). Inside of right valve. [From [45] Đặng & Hồ, 2017: 208, fig. 108] Reprinted with permission from Ref. [45]. Copyright 2017, Dr. Hồ, [personal communication, February 2023].
Cristaria plicata from eastern Russia was examined using conchological, anatomical, and molecular data and found to represent a single species [65,66]. Cristaria plicata is distributed from eastern Russia (Amur River basin), South Korea, Japan, China, Vietnam, Cambodia, the Lao People’s Republic, and Thailand [8,26,42,63,66,67,68]. Another unidentified species of Cristaria has been sequenced and was originally identified as C. plicata but is a separate species (Jia & Li, unpublished data, GenBank, [69]). Recently collected specimens of C. truncata Ðặng, 1980 [44] (Figure 7) were sequenced to test the validity and placement of this species. Graf & Cummings [56,57] and MolluscaBase [70] recognized four species in Cristaria, including two from Vietnam, C. plicata and C. truncata.
Cristaria discoidea (Lea, 1834) [53] has been reported in China, Taiwan, Hong Kong, northern Vietnam, Cambodia, and Thailand [8,26,62,63,67]. The species has been placed in Cristaria (Pletholophus) by Simpson [61,71] and Haas [53], and Pletholophus by Ðặng et al. [44]. However, both Cristaria plicata (Leach, 1814) [72] and Cristaria discoidea were overlooked by Liu et al. [73]. Historically, Anodon tenuis and Unio tenuis Gray in Griffith & Pidgeon have been dated as of 1834. It has been listed as a junior synonym of Unio discoidea Lea, 1834 [52] since the first edition of Lea’s Synopsis [74,75,76,77,78,79] and continued by Simpson [61,71], Preston [78], Modell [79], and Haas [51]. This was the state of the synonymy until Petit and Coan (2008) [80] reviewed all the taxa described by Gray in Griffiths and Pidgeon (1833–1834) [81]. Petit and Coan [80] (p. 229) determined the date of publication for the figure of Unio tenuis Gray in Griffith and Pidgeon to be 1833. This taxon was also mentioned as Anodon tenuis Gray in Griffith and Pidgeon [81] (p. 595) and again as Unio tenuis Gray in Griffith and Pidgeon 81] (p. 601), citing the same figure. Petit and Coan [80] noted that Unio tenuis has priority over the later name Unio discoidea Lea, 1834 [53]. This case does not meet the requirements of Code 23.9 [82] for usage, and the older name has date priority and must prevail. The correct name for Cristaria discoidea is Pletholophus tenuis (Gray in Griffith and Pidgeon, 1833) [81]. The distribution of Pletholophus tenuis was listed by Simpson [61] as only in China; Simpson [71] listed a subspecies from Taiwan. Haas [51] (p. 389) reported it only from China, Taiwan, Tonkin, and Cambodia, with no mention of Japan. Kondo [74] mentioned only Cristaria plicata as present in Japan. Imai [83] documented the first record of Cristaria discoidea from the Nagura River system on lshigakijima Island, Ryukyu Archipelago, Japan. The published range of Pletholophus tenuis included China, Taiwan, Japan, Vietnam, and Cambodia [59]. Do et al. [26] confirmed its occurrence in the northern half of Vietnam.
A subspecies, Cristaria discoidea reiniana Martens, 1875 [84], was recognized by [61,71]. Preston [78] listed Pletholophus reiniana from Lake Biwa, Japan; Modell [79] listed Pletholophus discoidea reiniana from central Japan; Haas [51] noted it was only known from Japan. However, discoidea, tenuis, and reiniana were not reported from Japan by Habe [85], Masuda and Uchiyam [86], or Kondo [68]. Habe [87] listed Cristaria reiniana as a junior synonym of Anodonta (Sinanodonta) woodiana, while Graf and Cummings [58] consider Cristaria reiniana a junior synonym of Pletholophus tenuis. Cristaria reiniana is considered a valid species [70]. The conservation status of this taxon is unknown currently, and the known distribution in Japan is very limited.
More recently, Cristaria bellua from the Mekong Basin was recognized as a valid species and the only species that crossed the strong biogeographical barrier of the Mekong River [38].
3.2. Systematics
Class: Bivalvia Linnaeus, 1758 [88]
Order: Unionida Gray, 1854 [89]
Superfamily: Unionoidea Rafinesque, 1820 [90]
Family: Unionidae Rafinesque, 1820 [90]
Subfamily: Unioninae Rafinesque, 1820 [90]
Tribe: Cristariini Lopes-Lima, Bogan, & Froufe, 2017 [39]
Genus: Pletholophus Simpson, 1900 [61]
Species: Pletholophus honglinhensis sp. nov.
New Taxa listed with Zoobank. urn:lsid:zoobank.org:pub:D224D8EB-5305-4458-8BB3-9C8F55E7606E
Common Name: Hồng Līnh Asianfloater, Vietnamese Name: Trai hồng lĩnh
Comparative Diagnosis:
Shells of Pletholophus honglinhensis sp. nov. are diagnosed by a nearly straight dorsal margin and an elongate oval shell with a marked posterior ridge and a minor secondary ridge, a lack of any surface sculpture, and a lack of anterior or posterior dorsal wings (Figure 8). The shells of Pletholophus tenuis are taller and rounder in outline than those of P. honglinhensis sp. nov. Pletholophus honglinhensis sp. nov. and P. tenuis both lack any evidence of the dorsal plications on the dorsum of Cristaria plicata. Pseudocardinal teeth in P. honglinhensis sp. nov. are elongate lamellar, and the lateral teeth are long and simple, often reduced as in P. tenuis. Pseudocardinal teeth are typically absent, and lateral teeth are single in specimens of Cristaria plicata. Cristaria truncata (Figure 7) is greatly inflated, lacks pseudocardinal teeth, and is truncated posteriorly as opposed to the more compressed and elongated shells and the presence of lamellate pseudocardinal teeth in P. honglinhensis sp. nov.
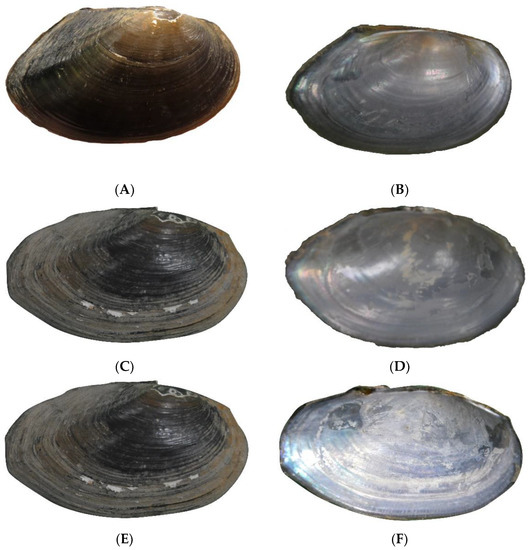
Figure 8.
Pletholophus honglinhensis sp. nov. Holotype NCSM 47211, total shell length 74.5 mm. (A). Outside of right valve. (B). Inside of left valve. Paratype NCSM 103062.2, total shell length 77.5 mm. (C). Outside of right valve. (D). Inside of left valve. Paratype NCSM 103062.3, total shell length 90.2 mm. (E). Outside of right valve. (F). Inside of left valve.
3.3. Description
Shells reach about 91 mm in total length (Figure 8 and Table 4); the shell shape is slightly rectangular to elongate oval in outline; and the shell is inflated. The anterior shell margin is evenly rounded, the dorsal shell margin is nearly straight, the ventral margin is broadly rounded, the posterior margin is rather straight and forms either a point posteriorly where it meets the upturned ventral margin or a more rounded end, the posterior ridge is prominent but not sharp, becoming obscured in older shells, and the posterior slope is smooth with a slight secondary posterior ridge. The umbo area is not elevated above the dorsal shell margin. Umbo sculpture appears double-looped with an indentation in the middle of parallel ridges (Figure 9). Periostracum is brown to black, lacking evidence of rays; the shell surface is smooth, lacking any sculpture, and marked by growth lines. Pseudocardinal teeth in the right valve: one long, thin lamellar tooth and a long, narrow lateral tooth. In the left valve, the pseudocardinal tooth is one long, thin lamellar tooth, and the lateral teeth are straight and well developed. Anterior adductor muscle scars are deep and smooth; pedal protractor muscle scars are separate; anterior pedal retractor muscle scars are united with anterior adductor muscle scars; and posterior adductor muscle scars are very faint. The pallial line impressed anteriorly before fading posteriorly. Umbo cavities are open and shallow. The nacre color is white, becoming bluish-iridescent toward the posterior margin.

Table 4.
Measurements of the type series of Pletholophus honglinhensis n. sp.
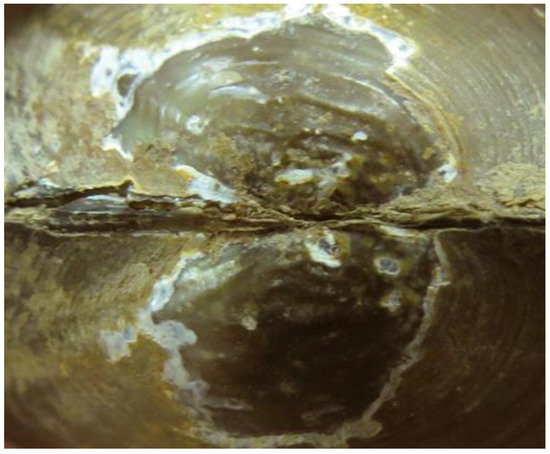
Figure 9.
Pletholophus honglinhensis sp. nov. umbo sculpture on the holotype NCSM 47211.
Type material:
Holotype: NCSM 47211
Paratypes: NCSM 103062 (2), all from the type locality.
Type Locality: Vietnam, Bắc Trung Bộ, Hà Tĩīnh Province, Market in Hồng Lĩnh. The specimens were collected from the Lam River, also known as the Cả River, in an area about eight kilometers north/northwest of Hồng Līnh. Latitude 18.573486 N, Longitude 105.673998 E. [Based on Google Maps. https://www.google.com/maps/place/H%E1%BB%93ng+L%C4%A9nh,+Ha+Tinh,+Vietnam/@18.533164,105.6719373,13z/data=!3m1!4b1!4m6!3m5!1s0×3139ca1076ed304b:0x3572bef7b81dcdef!8m2!3d18.5299746!4d105.7060872!16s%2Fm%2F03mct8b, accessed on 22 April 2023].
Collector: Dr. Phan Quoc Toan collected these specimens from the market in Hồng Lĩīnh, Hà Tĩnh Province, Vietnam, 12 July 2009.
Comparison: Pletholophus honglinhensis sp. nov. does not appear to have been collected before and may have been mistaken for P. tenuis. The shell shape of P. honglinhensis n. sp. is more elongated and not as tall as P. tenuis. The posterior ridge in P. tenuis is more rounded than in P. honglinhensis sp. nov. This new species will not be mistaken for Cristaria plicata because it lacks both anterior and posterior dorsal wings and the dorsal plications often seen in C. plicata. Pletholophus honglinhensis sp. nov. can be readily separated from Sinanodonta jourdyi (Morlet, 1886) [55] by being less inflated, typically having a more elongated oval shell, and lacking the heavy parallel bars of the umbo sculpture. Sinanodonta jourdyi lacks any evidence of hinge teeth compared with Pletholophus and Cristaria. Cristaria typically only has well-developed lateral teeth, while Pletholophus species both have weak lamellate pseudocardinal teeth and a single lateral tooth.
Distribution: Pletholophus honglinhensis sp. nov. is only known from the market in Hồng Lĩnh, Hà Tĩnh Province, Vietnam.
Conservation status: This species is known only from the type locality in Hà Tĩnh Province, Vietnam. The distribution of this species is currently unknown, so conservation status is not possible.
Etymology: The species is named after the Hồng Lĩnh Market, Hà Tĩnh Province, Vietnam, where it was collected.
Comparative shell material examined.
3.4. Pletholophus tenuis
NCSM 63599: Vietnam, Lang Son, Ky Cung River [Zhu River Basin, Pearl Basin], another stream flowing S, [0.24 air miles NNE center] Trung Thành, [Trang Dinh/Tràng Định] district]. Latitude 22.25553 N, Longitude 106.6068038 E.
NCSM 84943: Vietnam, Lang Son, Ky Cung River [Zhu River Basin, Pearl Basin], Trang Dinh, Trang Dinh District. Latitude 22.23858 N Longitude 106.5905303 E.
NCSM 85273: Vietnam, Ha Noi Province, Hong River, Phùng town, [Dan Phuomng district], [point estimated 0.21 air miles WNW Phượng Trì]. Latitude 21.087821 N, Longitude 105.6603546 E.
IEBR_QN_0001: Vietnam, Quang Ninh Province, Tien Yen River, Binh Lieu town, [Binh Lieu district]. Latitude 21.524945 N, Longitude 107.391095.
3.5. Cristaria plicata
NCSM 84956: Vietnam, Hanoi Province, Hong River, Ha Dong City, Viet Tri market Latitude 21.13333 N Longitude 105.5 E.
NCSM 100665: Vietnam, Đồng Nai Province, Cát Tiên market [point estimated 20.5 air miles SE of Đồng Xoài] Latitude 11.39935 N, Longitude 107.3306 E.
3.6. Sinanodonta jourdyi
NCSM 84916: Vietnam, Lang Son Province, Van Quan District, [Văn Quan], [0.7 air miles S center Vĩnh Lai]. Latitude 21.885499 N Longitude 106.5633499 E NCSM 84948: Vietnam, Nam Dinh Province, Day River, edge of Son la. Day River [Sông Đáy], Nam Dinh Province [Nam Định], S of Hanoi, second market on the E edge of Sơn La [point estimated center Sơn La]. Latitude 21.326944 N Longitude 103.918891 E NCSM 85282: Vietnam, Bắc Kạn Province, Nà Phặc, Ngân Sơn district, Northeast (Đông Bắc), [point estimated 1.01 air miles ENE center Na Young]. Latitude 22.385978 N, Longitude 105.9101409 E.
4. Conclusions
In this study, we unveiled the systematics, taxonomy, phylogeny, and distribution of Anodontini freshwater mussels from Vietnam, recognizing five valid species, including a species new to science, Pletholophus honglinhensis nov. sp. We also give guidance for future studies and conservation action.
The results reported here are an important baseline for further studies related to the conservation and biogeography of freshwater mussels at the regional and global levels. Additionally, the species delimitation provided here is key to planning and executing further ecological and physiological research on these species. Given that most of the basic life history traits and habitat requirements of freshwater mussels in Vietnam are still unknown, we advocate for investment in these fields to provide better conservation guidance. Of special concern is the complete lack of knowledge about the host fish range, which is crucial for mussels to complete their life cycle. Wider and more detailed surveys that not only provide information about the distribution but also the abundance and population structure of populations should be accomplished periodically to evaluate population trends and make more accurate conservation status assessments. The lack of such studies is hampering proper conservation status assessments, and an example is the single collection of the new Pletholophus species from the market of Hồng Lĩnh, Hà Tĩnh Province in the Cả or Lam River Basin, south of the Red River Basin.
Furthermore, a major threat that should be considered is the overexploitation of the five species here examined, which are sold in Vietnamese markets as human food items. This activity exerts constant pressure on mussel populations and should be regulated and managed by local authorities. Other threats such as dams, agricultural practices, over-harvesting, gravel/sand dredging, etc. are just some of the multiple impacts on these species that need to be addressed by developing a conservation plan for the freshwater mussels of Vietnam.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15060710/s1, Table S1: List of sequences used for the phylogenetic relationships within the genus Cristaria with respective haplotypes, localities, voucher numbers or specimen codes, and GenBank references. Table S2: List of sequences used for the phylogenetic relationships within the genus Pletholophus with respective haplotypes, localities, voucher numbers or specimen codes, and GenBank references. Table S3: List of sequences used for the phylogenetic relationships within the genus Sinanodonta with respective haplotypes, localities, voucher numbers or specimen codes, and GenBank references. References [91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110] are cited in Supplementary Materials.
Author Contributions
Conceptualization, A.E.B., E.F. and M.L.-L.; formal analysis, E.F. and M.L.-L.; funding acquisition, M.L.-L.; investigation, V.T.D.; methodology, E.F.; supervision, A.E.B.; writing—original draft, A.E.B., V.T.D., and M.L.-L.; writing—review and editing, A.E.B., V.T.D., E.F. and M.L.-L. All authors have read and agreed to the published version of the manuscript.
Funding
The Portuguese Foundation for Science and Technology (FCT), through national funds, funded MLL under contract 2020.03608. CEECIND and EF under contract CEECINST/00027/2021. This research of VTD was funded by the Institute of Ecology and Biological Resources (IEBR) under grant number IEBR.ĐT.2-23.
Institutional Review Board Statement
This was not applicable.
Data Availability Statement
The sequences used in this study are all available from GenBank. GenBank accession numbers for all sequences used in this work are listed in Supplementary Tables S1–S3. https://www.ncbi.nlm.nih.gov/genbank/about/ (accessed on 28 Marh 2023). Shell and body vouchers and tissue snips for the new species are available at the North Carolina Museum of Natural Sciences.
Acknowledgments
The late Richard Petit and Eugene Coan are thanked for their time and efforts to help clarify the unionid taxa of Gray and their date of publication. Ellen Strong is thanked for continuing the discussion on the intricacies of nomenclature. Jamie Smith is thanked for her patience and assistance with data and photographs. T.H. Hồ of Ha Noi, Vietnam, is thanked for allowing the reproduction of his figure of Cristaria truncata. Phan Quoc Toan, Director of the Center for Entomology and Parasitology Research, Institute of Research and Training of Medicine, Biology, and Pharmacy, Duy Tan University, Da Nang City, Vietnam, is thanked for his continued interest in freshwater mussels and for sending specimens of the new species. Cynthia M. Bogan endured reading many drafts of this paper and is thanked for her critical editing and suggestions. Jamie M. Smith aided with cleaning type photographs.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Modesto, V.; Ilarri, M.; Sopuza, A.T.; Lopes-Lima, M.; Douda, K.; Clavero, M.; Sousa, R. Fish and mussels: Importance of fish for freshwater mussel conservation. Fish Fish. 2018, 19, 244–259. [Google Scholar] [CrossRef]
- Guerra, D.; Lopes-Lima, M.; Froufe, E.; Gan, H.M.; Ondina, P.; Amaro, R.; Klunzinger, M.W.; Callil, C.; Prié, V.; Bogan, A.E.; et al. Variability of mitochondrial ORFans hints at possible differences in the system of doubly uniparental inheritance of mitochondria among families of freshwater mussels (Bivalvia: Unionida). BMC Evol. Biol. 2019, 19, 229. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, C.C. Ecosystem services provided by freshwater mussels. Hydrobiologia 2018, 810, 15–27. [Google Scholar] [CrossRef]
- Zieritz, A.; Froufe, E.; Bolotov, I.; Gonçalves, D.V.; Aldridge, D.C.; Bogan, A.E.; Gan, H.M.; Gomes-dos-Santos, A.; Sousa, R.; Teixeira, A.; et al. Mitogenomic phylogeny and fossil-calibrated mutation rates for all F- and M-type mtDNA genes of the largest freshwater mussel family (Bivalvia: Unionidae). Zool. J. Linn. Soc. 2021, 193, 1088–1107. [Google Scholar] [CrossRef]
- Lopes-Lima, M.; Bolotov, I.N.; Do, V.T.; Aldridge, D.C.; Fonseca, M.F.; Gan, H.M.; Gofarov, M.Y.; Kondakov, A.V.; Prié, V.; Sousa, R.; et al. Expansion and systematics redefinition of the most threatened freshwater mussel family, the Margaritiferidae. Mol. Phylogenet. Evol. 2018, 127, 98–118. [Google Scholar] [CrossRef]
- Lopes-Lima, M.; Teixeira, A.; Froufe, E.; Lopes, A.; Varandas, S.; Sousa, R. Biology and conservation of freshwater bivalves: Past, present and future perspectives. Hydrobiologia 2014, 735, 1–13. [Google Scholar] [CrossRef]
- Böhm, M.; Dewhurst-Richman, N.I.; Seddon, M.; Ledger, S.E.H.; Albrecht, C.; Allen, D.; Bogan, A.E.; Cordeiro, J.; Cummings, K.S.; Cuttelod, A.; et al. The conservation status of the world’s freshwater molluscs. Hydrobiologia 2021, 848, 3231–3254. [Google Scholar] [CrossRef]
- Zieritz, A.; Bogan, A.E.; Froufe, E.; Klishko, A.; Kondo, T.; Kovitvadhi, U.; Kovitvadhi, S.; Lee, J.H.; Lopes-Lima, M.; Pfeiffer, J.M.; et al. Diversity, biogeography, and conservation of freshwater mussels (Bivalvia: Unionida) in East and Southeast Asia. Hydrobiologia 2018, 810, 29–44. [Google Scholar] [CrossRef]
- Bolotov, I.N.; Vikhrev, I.V.; Kondakov, A.V.; Konopleva, E.S.; Gofarov, M.Y.; Aksenova, O.V.; Tumpeesuwan, S. New taxa of freshwater mussels (Unionidae) from a species-rich but overlooked evolutionary hotspot in Southeast Asia. Sci. Rep. 2017, 7, 11573. [Google Scholar] [CrossRef]
- Bolotov, I.N.; Pfeiffer, J.M.; Konopleva, E.S.; Vikhrev, I.V.; Kondakov, A.V.; Aksenova, O.V.; Gofarov, M.Y.; Tumpeesuwan, S.; Win, T. A new genus and tribe of freshwater mussel (Unionidae) from Southeast Asia. Sci. Rep. 2018, 8, 10030. [Google Scholar] [CrossRef]
- Régnier, C.; Fontaine, B.; Bouchet, P. Not knowing, not recording, not listing: Numerous unnoticed mollusk extinctions. Conser. Biol. 2009, 23, 1214–1221. [Google Scholar] [CrossRef] [PubMed]
- Régnier, C.; Achaz, G.; Lambert, A.; Cowie, R.H.; Bouchet, P.; Fontaine, B. Mass extinction in poorly known taxa. Proc. Nat. Acad. Sci. USA 2015, 112, 7761–7766. [Google Scholar] [CrossRef] [PubMed]
- Tedesco, P.A.; Bigorne, R.; Bogan, A.E.; Giam, X.; Jezequel, C.; Hugueny, B. Estimating how many undescribed species have gone extinct. Conser. Biol. 2014, 28, 1360–1370. [Google Scholar] [CrossRef] [PubMed]
- Bogan, A.E.; Do, V.T. An overlooked new species of freshwater bivalve from Northern Vietnam (Mollusca: Bivalvia: Unionidae). Raffles Bull. Zool. 2018, 66, 878–889. [Google Scholar]
- Bolotov, I.N.; Konopleva, E.S.; Vikhrev, I.V.; Gofarov, M.Y.; Lopes-Lima, M.; Bogan, A.E.; Lunn, Z.; Chan, N.; Win, T.; Aksenova, O.V.; et al. New freshwater mussel taxa discoveries clarify biogeographic division of Southeast Asia. Sci. Rep. 2020, 10, 6616. [Google Scholar] [CrossRef]
- Bolotov, I.N.; Kondakov, A.V.; Konopleva, E.S.; Vikhrev, I.V. A new genus of ultra-elongate freshwater mussels from Vietnam and eastern China (Bivalvia: Unionidae). Ecol. Montenegrina 2021, 39, 1–6. Available online: https://www.biotaxa.org/em/article/view/em.2021.39.1 (accessed on 20 March 2020). [CrossRef]
- Huang, X.C.; Su, J.H.; Ouyang, J.X.; Ouyang, S.; Zhou, S.H.; Wu, X.P. Towards a global phylogeny of freshwater mussels (Bivalvia: Unionida): Species delimitation of Chinese taxa, mitochondrial phylogenomics, and diversification patterns. Mol. Phylogenet. Evol. 2019, 130, 45–59. [Google Scholar] [CrossRef]
- Lopes-Lima, M.; Hattori, A.; Kondo, T.; Lee, J.H.; Kim, S.K.; Shirai, A.; Hayashi, H.; Usui, T.; Sakuma, K.; Toriya, T.; et al. Freshwater mussels (Bivalvia: Unionidae) from the Rising Sun (Far East Asia): Phylogeny, systematics, and distribution. Mol. Phylogenet. Evol. 2020, 146, 106755. [Google Scholar] [CrossRef]
- Ng, T.H.; Jeratthitikul, E.; Sutcharit, C.; Chhuoy, S.; Pin, K.; Pholyotha, A.; Siriwut, W.; Srisonchai, R.; Hogan, Z.S.; Ngor, P.B. Annotated checklist of freshwater molluscs from the largest freshwater lake in Southeast Asia. ZooKeys 2020, 958, 107–141. [Google Scholar] [CrossRef]
- Sano, I.; Saito, T.; Miyazaki, J.I.; Shirai, A.; Uechi, T.; Kondo, T.; Chiba, S. Evolutionary History and Diversity of Unionoid Mussels (Mollusca: Bivalvia) in the Japanese Archipelago. Plankton Benthos Res. 2020, 15, 97–111. [Google Scholar] [CrossRef]
- Konopleva, E.S.; Bolotov, I.N.; Pfeifer, J.M.; Vikhrev, I.V.; Kondakov, A.V.; Gofarov, M.Y.; Tomilova, A.A.; Tanmuangpak, K.; Tumpeesuwan, S. New freshwater mussels from two Southeast Asian genera Bineurus and Thaiconcha (Pseudodontini, Gonideinae, Unionidae). Sci. Rep. 2021, 11, 8244. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, J.M.; Graf, D.L.; Cummings, K.S.; Page, L.M. Taxonomic revision of a radiation of South-east Asian freshwater mussels (Unionidae: Gonideinae: Contradentini+Rectidentini). Invertebr. Systemat. 2021, 35, 394–470. [Google Scholar] [CrossRef]
- Sahidin, A.; Muhammad, G.; Hasan, Z.; Arief, M.C.W.; Marwoto, R.M.; Komaru, A. Indonesian freshwater bivalves: A meta-analysis of endemicity, ecoregion distributions, and conservation status. AACL Aquac. Aquar. Conserv. Legis. Bioflux 2021, 14, 3750–3775. [Google Scholar]
- Wu, R.W.; Liu, X.J.; Wang, S.; Roe, K.J.; Ouyang, S.; Wu, X.P. Analysis of mitochondrial genomes resolves the phylogenetic position of Chinese freshwater mussels (Bivalvia, Unionidae). ZooKeys 2019, 812, 23–46. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.P.; Dai, Y.T.; Yin, N.; Shu, F.Y.; Chen, Z.G.; Guo, L.; Zhou, C.H.; Ouyang, S.; Huang, X.C. Mitogenomic phylogeny resolves Cuneopsis (Bivalvia: Unionidae) as polyphyletic: The description of two new genera and a new species. Zool. Scr. 2022, 51, 173–184. [Google Scholar] [CrossRef]
- Do, V.T.; Le, Q.T.; Bogan, A.E. Freshwater mussels (Bivalvia: Unionoida) of Vietnam: Diversity, distribution, and conservation status. Freshw. Mollusk Biol. Conserv. 2018, 21, 1–18. [Google Scholar] [CrossRef]
- Naimo, T.J.; Damschen, E.D.; Rada, R.G.; Monroe, E.M. Nonlethal evaluation of the physiological health of unionid mussels: Methods for biopsy and glycogen analysis. J. N. Am. Benthol. Soc. 1998, 17, 121–128. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Harbor Spring Press: New York, NY, USA, 1989; p. 545. [Google Scholar]
- Froufe, E.; Sobral, C.; Teixeira, A.; Sousa, R.; Varandas, S.C.; Aldridge, D.; Lopes-Lima, M. Genetic diversity of the pan-European freshwater mussel Anodonta anatina (Bivalvia: Unionoida) based on CO1: New phylogenetic insights and implications for conservation. Aquat. Conserv. Mar. Freshw. Ecosyst. 2014, 24, 561–574. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Sela, I.; Ashkenazy, H.; Katoh, K.; Pupko, T. GUIDANCE2: Accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 2015, 43, W7–W14. [Google Scholar] [CrossRef]
- Fonseca, M.M.; Lopes-Lima, M.; Eackles, M.S.; King, T.L.; Froufe, E. The female and male mitochondrial genomes of Unio delphinus and the phylogeny of freshwater mussels (Bivalvia: Unionida). Mitochondrial DNA Part B 2016, 1, 954–957. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating Maximum-Likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A.; Susko, E. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. DAMBE6: New tools for microbial genomics, phylogenetics and molecular evolution. J. Hered. 2017, 108, 431–437. [Google Scholar] [CrossRef]
- Konopleva, E.S.; Bolotov, I.N.; Vikhrev, I.V.; Inkhavilay, K.; Gofarov, M.Y.; Kondakov, A.V.; Tomilova, A.A.; Chapurina, Y.E.; Do, V.T.; Pfeiffer, J.M., III; et al. A freshwater mussel species reflects a Miocene stream capture between the Mekong Basin and East Asian rivers. Zoosyst. Evol. 2023, 99, 29–42. [Google Scholar] [CrossRef]
- Lopes-Lima, M.; Froufe, E.; Do, V.T.; Ghamizi, M.; Mock, K.E.; Kebapçi, U.; Klishko, O.; Kovitvadhi, S.; Kovitvadhi, U.; Paul, O.S.; et al. Phylogeny of the most species rich freshwater bivalve family (Bivalvia: Unionida: Unionidae): Defining modern subfamilies and tribes. Mol. Phylogenet. Evol. 2017, 106, 174–191. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-based registry for all animal species: The Barcode Index Number (BIN) system. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef]
- Puillandre, N.; Modica, M.V.; Zhang, Y.; Sirovich, L.; Boisselier, M.-C.; Cruaud, C.; Holford, M.; Samadi, S. Large-scale species delimitation method for hyperdiverse groups. Mol. Ecol. 2012, 21, 2671–2691. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Đặng, N.T.; Thái, T.B.; Phạm, V.M. Identification of freshwater invertebrates of North Vietnam. In Định Loại Động Vật Không Xương Sống Nước Ngọt Bắc Việt Nam; Sciences and Technology Publishing Co.: Ha Noi, Vietnam, 1980; pp. 491–573. (In Vietnamese) [Google Scholar]
- Đặng, N.T.; Hồ, T.H. Trai Ốc Nước Ngọt Nội Địa Việt Nam (Mollusca: Gastropoda, Bivalvia); [Inland Freshwater Molluscs (Mollusca: Gastropoda, Bivalvia) of Vietnam] Ðộng Vật Chí Việt Nam [Fauna of Vietnam]; Science and Technology Publishing House: Hanoi, Vietnam, 2017; 360p, ISBN 978-604-913-677-1. (In Vietnamese, English Abstract). [Google Scholar]
- International Union for the Conservation of Nature (IUCN). The IUCN Red List of Threatened Species. 2023. Available online: https://www.iucnredlist.org/ (accessed on 1 January 2023).
- Lehner, B.; Grill, G. Global River hydrography and network routing: Baseline data and new approaches to study the world’s large river systems. Hydrol. Process. 2013, 27, 2171–2186. [Google Scholar] [CrossRef]
- von Martens, E. Herr von Martens gab im Anschluss an den vorhergehenden Vortrag eine Uebersicht über die von den Herren Dr. Fr. Hilgendorf und Dr. W. Dönitz in Japan gesammelten Binnenmollusken. Sitz. Ber. Ges. Nat. Freunde Berl. 1877, 1877, 97–123. [Google Scholar]
- Tchang, S.; Li, S.C.; Liu, Y.Y. Bivalves (Mollusca) of Tung-Ting Lake and its surrounding waters, Hunan Province, China. Acta Zool. Sin. 1965, 17, 197–213. [Google Scholar]
- Heude, P. Conchyliologie Fluviatile de la Province de Nanking et de la Chine Centrale; 10 Fascicules, plates, and un-numbered pages; Librairie F. Savy: Paris, France, 1875. [Google Scholar]
- Haas, F. Superfamilia Unionacea. Das. Tierreich. 1969, 88, 663. (In German) [Google Scholar]
- Graf, D.; Cummings, K. Review of the systematics and global diversity of freshwater mussel species (Bivalvia: Unionoida). J. Molluscan Stud. 2007, 73, 291–314. [Google Scholar] [CrossRef]
- Lea, I. Observations on the naïades; and descriptions of new species of that, and other families. Trans. Am. Philos. Soc. 1834, 5, 23–119. [Google Scholar] [CrossRef]
- Bolotov, I.N.; Bespalaya, Y.V.; Gofarov, M.Y.; Kondakov, A.V.; Konopleva, E.S.; Vikhrev, I.V. Spreading of the Chinese pond mussel, Sinanodonta woodiana, across Wallacea: One or more lineages invade tropical island and Europe. Biochem. Syst. Ecol. 2016, 67, 58–64. [Google Scholar] [CrossRef]
- Morlet, L. Diagnoses Molluscorum novorum Tonkini. J. Conchyliol. 1886, 34, 75–78. [Google Scholar]
- Bespalaya, Y.V.; Bolotov, I.N.; Aksenova, O.V.; Gofarov, M.Y.; Kondakov, A.V.; Vikhrev, I.V.; Vinarski, M.V. DNA barcoding reveals invasion of two cryptic Sinanodonta mussel species (Bivalvia: Unionidae) into the largest Siberian river. Limnologica 2018, 69, 94–102. [Google Scholar] [CrossRef]
- Kondakov, A.V.; Palatov, D.M.; Rajabov, Z.P.; Gofarov, M.Y.; Konopleva, E.S.; Tomilova, A.A.; Vikhrev, I.V.; Bolotov, I.N. DNA analysis of a non-native lineage of Sinanodonta woodiana species complex (Bivalvia: Unionidae) from Middle Asia supports the Chinese origin of the European invaders. Zootaxa 2018, 4462, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Graf, D.; Cummings, K. A ‘big data’ approach to global freshwater mussel diversity (Bivalvia: Unionoida), with an updated checklist of genera and species. J. Molluscan Stud. 2021, 87, 1–36. [Google Scholar] [CrossRef]
- Graf, D.; Cummings, K. Mussel-Project Webpage. 2023. Available online: http://mussel-project.uwsp.edu/ (accessed on 1 February 2023).
- Schumacher, C.F. Essai d’un Nouveau Système des Habitations des Vers Testacés Avec XXII Planches; De l’Imprimerie de Mr. le Directeur Schultz: Copenhague, Denmark, 1817; 287p. [Google Scholar]
- Simpson, C.T. Synopsis of the naiades, or pearly fresh-water mussels. Proc. U.S. Natl. Mus. 1900, 22, 501–1044. [Google Scholar] [CrossRef]
- Heard, W.H. Anatomical systematics of Cristaria plicata (Leach) (Pelecypoda: Unionidae, Unioninae). Malacol. Rev. 1977, 10, 59–70. [Google Scholar]
- He, J.; Zhuang, Z. The Freshwater Bivalves of China; Conchbooks: Harxheim, Germany, 2013; 197p. [Google Scholar]
- Moskvicheva, I.M.; Starobogatov, Y.I. About East-Asian Potomida similar Unionidae (Bivalvia). Bull. Mosc. Soc. Nat. Investig. Sect. Biol. 1973, 78, 21–37. (In Russian) [Google Scholar]
- Klishko, O.; Lima-Lopez, M.; Froufe, E.; Bogan, A.E. Are Cristaria herculea (Middendorff, 1847) and Cristaria plicata (Leach, 1815) (Bivalvia, Unionidae, Unioninae, Anodontini) separate species? Zookeys 2014, 438, 1–15. [Google Scholar] [CrossRef]
- Klishko, O.; Lopes-Lima, M.; Froufe, E.; Bogan, A.E.; Abakumova, V.Y. Systematics and distribution of Cristaria plicata (Bivalvia, Unionidae) from the Russian Far East. Zookeys 2016, 580, 13–27. [Google Scholar] [CrossRef]
- Brandt, R.A.M. The non-marine aquatic Mollusca of Thailand. Arch. Molluskenkd. 1974, 105, 1–423. [Google Scholar]
- Kondo, T. Monograph of Unionoida in Japan (Mollusca: Bivalvia); Special publication of the Malacological Society of Japan; Malacological Society of Japan: Ibaraki, Japan, 2008; 69p. [Google Scholar]
- GenBank. Nucleotide Sequences Database for the National Center for Biotechnology Information (NCBI). 2022. Available online: https://www.ncbi.nlm.nih.gov/nuccore/?term=Cristaria+plicata+mitochondrion (accessed on 1 January 2023).
- MolluscaBase. 2023. Available online: http://www.molluscabase.org/ (accessed on 1 January 2023).
- Simpson, C.T. A Descriptive Catalogue of the Naiades, or Pearly Fresh-Water Mussels; Parts I–III; Bryant Walker: Detroit, MI, USA, 1914; 1540p. [Google Scholar]
- Leach, W.E. The Zoological Miscellany; Being Descriptions of New, or Interesting Animals; McMillan: London, UK, 1814; Volume 1, 134p. [Google Scholar]
- Liu, Y.Y.; Zhang, W.; Wang, Y.; Wang, E. Economic Fauna of China. Freshwater Mollusks; Science Press: Beijing, China, 1979; 134p. (In Chinese) [Google Scholar]
- Lea, I. A Synopsis of the Family of Naïades; Cary, Lea and Blanchard, Philadelphia; John Miller: London, UK, 1836; 59p. [Google Scholar]
- Lea, I. A Synopsis of the Family of Naïades, 2nd ed.; Enlarged and Improved; Cary, Lea and Blanchard: Philadelphia, PA, USA, 1838; 44p. [Google Scholar]
- Lea, I. A Synopsis of the Family of Naïades, 3rd ed.; Greatly Enlarged and Improved; Blanchard and Lea: Philadelphia, PA, USA, 1852; 88p. [Google Scholar]
- Lea, I. A Synopsis of the Family Unionidae, 4th ed.; Very Greatly Enlarged and Improved; Henry C. Lea: Philadelphia, PA, USA, 1870; 184p. [Google Scholar]
- Preston, H.B. A catalogue of the Asiatic naiades in the collection of the Indian Museum, Calcutta, with descriptions of new species. Rec. Indian Mus. 1912, 7, 279–308. [Google Scholar] [CrossRef]
- Modell, H. Die Anodontinae, Ortm. Emend. (Najad., Mollusca). Eine studie über die Zusammenhänge von Klimazonen und Entwicklungsgeschichte. (Klimazonentheorie). Jena. Z. Med. Nat. 1945, 78, 58–100. [Google Scholar]
- Petit, R.E.; Coan, E.V. The molluscan taxa made available in the Griffith& Pidgeon (1833–1834) edition of Cuvier, with notes on the editions of Cuvier and on Wood’s Index Testaceologicus. Malacologia 2008, 50, 219–264. [Google Scholar]
- Griffith, E.; Pidgeon, E. The Mollusca and Radiata. In The Animal Kingdom Arranged in Conformity with Its Organization; Griffith, E., Ed.; Whittaker, and Co.: London, UK, 1833; Volume 12, pp. 8–601. [Google Scholar]
- International Commission on Zoological Nomenclature (ICZN). Incorporating Declaration 44, amendments of Article 74.7.3, with effect from 31 December 1999, and the Amendment on e-publication, amendments to Articles 8, 9, 10, 21 and 78, with effect from 1 January 2012. In International Code of Zoological Nomenclature, 4th ed.; International Trust for Zoological Nomenclature: Singapore, 1999; 306p, Available online: http://www.nhm.ac.uk/hosted-sites/iczn/code/ (accessed on 1 January 2023).
- Imai, H. The first record of Cristaria discoidea (Lea, 1834) (Bivalvia: Unionidae) from Ishigakijima Island, the Ryukyu Archipelago, Japan. Biol. Mag. Okinawa 2008, 46, 65–70. [Google Scholar]
- von Martens, E. Cristaria reiniana n. sp. Jahrbücher Dtsch. Malakozool. Ges. 1875, 2, 136. [Google Scholar]
- Habe, T. Shells of Japan; Color Books; Hoikusha Publishing Co. Ltd.: Osaka, Japan, 1971; 139p. [Google Scholar]
- Masuda, O.; Uchiyama, R. Freshwater Mollusks of Japan. Series 2. Freshwater Mollusks of Japan, Including Brackish Water Species; PICES, Ecological Field Guide Series; Pices Publishers, Co. Ltd.: Tokyo, Japan, 2004; 240p. (In Japanese) [Google Scholar]
- Habe, T. Bivalvia and Scaphopoda; Systematics of Mollusca in Japan; Hokuryukan Publisher: Tokyo, Japan, 1977; 372p. [Google Scholar]
- Linnaeus, C. Systema Naturae. Edition X. (Systema Naturae per Regna tria Naturae, Secundum Classes, Ordines, Genera, Species cum Characteribus, Differentiis, Synonymis, Locis. Tomus, I. Edtio Decima, Reformata); Laurentius Salvius: Stockholm, Sweden, 1758; Volume 1, pp. 1–824. [Google Scholar]
- Gray, J.E. A revision of the arrangement of the families of bivalve shells (Conchifera). Ann. Mag. Nat. Hist. 1854, 13, 408–418. [Google Scholar] [CrossRef]
- Rafinesque, C.S. Monographie des coquilles bivalves fluviatiles de la Rivière Ohio, contenant douze genres et soixante-huit espèces. Ann. Générales Des Sci. Phys. À Brux. 1820, 5, 287–322. [Google Scholar]
- Lee, J.H. Systematic Study of Korean Unionids (Bivalvia: Unionidae) Based on Morphological and Molecular Data. Ph.D. Thesis, School of Life Sciences, Kungpook National University, Daegu, Republic of Korea, 2017. [Google Scholar]
- Lee, J.H.; Choi, E.H.; Kim, S.K.; Ryu, S.H.; Hwang, U.W. Mitochondrial genome of the cockscomb pearl mussel Cristaria plicata (Bivalvia, Unionoida, Unionidae). Mitogenome Anoun. 2012, 23, 39–41. [Google Scholar]
- Jia, M.J.; Li, J.L.; Niu, D.H.; Bai, Z.Y. Sequence variation of COI gene in ten populations of Cristaria plicata from the Middle and Lower Yangtze River. Chin. J. Zool. 2009, 44, 1–8. [Google Scholar]
- Wang, X.F.; Liu, Z.M.; Wu, W.J. Transcriptome analysis of the freshwater pearl mussel (Cristaria plcaat) mantle unravels genes involved in the formation of shell and pearls. Mol. Genet. Genom. 2017, 292, 343–352. [Google Scholar] [CrossRef]
- Wu, R.W.; Liu, Y.T.; Wang, S.; Liu, X.J.; Zanatta, D.T.; Roe, K.J.; Song, X.L.; An, C.T.; Wu, X.P. Testing the utility of DNA barcodes and a preliminary phylogenetic framework for Chinese freshwater mussels (Bivalvia: Unionidae) from the middle and lower Yangtze River. PloS ONE 2018, 13, e0200956. [Google Scholar] [CrossRef]
- Song, X.L.; Ouyang, S.; Zhou, C.H.; Wu, X.P. Complete maternal mitochondrial genome of freshwater mussel Anodonta lucida (Bivalvia: Unionidae: Anodontinae). Mitochondrial DNA Part A 2016, 27, 549–550. [Google Scholar] [CrossRef] [PubMed]
- Sayenko, E.M.; Soroka, M.; Kholin, S.K. Comparison of the species Sinanodonta amurensis Moskvicheva, 1973 and Sinanodonta primorjensis Bogatov et Zatrawkin, 1988 (Bivalvia: Unionidae: Anodontinae) in view of variability of the mitochondrial DNA cox1 gene and conchological features. Biol. Bull. 2017, 44, 266–276. [Google Scholar] [CrossRef]
- Ng, T.H.; Tan, S.K.; Wong, W.H.; Meier, R.; Chan, S. Molluscs for Sale: Assessment of Freshwater Gastropods and Bivalves in the Ornamental Pet Trade. PLoS ONE 2016, 11, e0161130. [Google Scholar] [CrossRef] [PubMed]
- Stelbrink, B.; Rintelen, T.V.; Albrecht, C.; Clewing, C.; Naga, P.O. Forgotten for decades: Lake Lanao and the genetic assessment of its mollusc diversity. Hydrobiologi 2019, 843, 31–49. [Google Scholar] [CrossRef]
- Zieritz, A.; Lopes-Lima, M.; Bogan, A.E.; Sousa, R.; Walton, S.; Rahim, K.A.; Wilson, J.J.; Ng, P.Y.; Froufe, E.; McGowan, S. Factors driving changes in freshwater mussel (Bivalvia, Unionida) diversity and distribution in Peninsular Malaysia. Sci. Total Environ. 2016, 571, 1069–1078. [Google Scholar] [CrossRef]
- Clusa, L.; Miralles, L.; Basanta, A.; Escot, C.; Garcı, E. eDNA for detection of five highly invasive molluscs. A case study in urban rivers from the Iberian Peninsula. PloS ONE 2017, 12, e0188126. [Google Scholar] [CrossRef]
- Konečný, A.; Popa, O.P.; Bartáková, V.; Douda, K.; Bryja, J.; Smith, C.; Popa, L.O.; Reichard, M. Modelling the invasion history of Sinanodonta woodiana in Europe: Tracking the routes of a sedentary aquatic invader with mobile parasitic larvae. Evol. Appl. 2018, 11, 1975–1989. [Google Scholar] [CrossRef]
- Soroka, M. Genetic variability among freshwater mussel Anodonta woodiana (Lea, 1834) (Bivalvia: Unionidae) populations recently introduced in Poland. Zool. Sci. 2005, 22, 1137–1144. [Google Scholar] [CrossRef]
- Soroka, M. Characteristics of Mitochondrial DNA of Unionid Bivalves (Mollusca: Bivalvia: Unionidae). I. Detection andcharacteristics of doubly uniparental inheritance (DUI) of Unionid Mitochondrial DNA (1). Folia Malacol. 2010, 18, 147–188. [Google Scholar] [CrossRef]
- Soroka, M. Characteristics of mitochondrial DNA of unionid bivalves (Mollusca: Bivalvia: Unionidae). II. Detection and characteristics of doubly uniparental inheritance (DUI) of unionid mitochondrial DNA (2). Folia Malacol. 2010, 18, 189–209. [Google Scholar] [CrossRef]
- Soroka, M.; Urbańska, M.; Andrzejewsk, W. Chinese pond mussel Sinanodonta woodiana (Lea, 1834) (Bivalvia): Origin of the Polish population and GenBank data. J. Limnol. 2014, 73, 454–458. [Google Scholar] [CrossRef]
- Vikhrev, I.V.; Konopleva, E.S.; Gofarov, M.Y.; Kondakov, A.V.; Chapurina, Y.E.; Bolotov, I.N. A Tropical Biodiversity Hotspot Under the New Threat: Discovery and DNA Barcoding of the Invasive Chinese Pond Mussel Sinanodonta Woodiana in Myanmar. Trop. Conserv. Sci. 2017, 10, 1–11. [Google Scholar] [CrossRef]
- Guarneri, I.; Popa, O.P.; Gola, L.L.; Kamburska, L.; Lauceri, R.; Lopes-Lima, M.; Popa, L.O.; Riccardi, N. A morphometric and genetic comparison of Sinanodonta woodiana (Lea, 1834) populations: Does shape really matter? Aquat. Invasions 2014, 9, 183–194. [Google Scholar] [CrossRef]
- Froufe, E.; Lopes-Lima, M.; Riccardi, N.; Zaccara, S.; Vanetti, I.; Lajtner, J.; Teixeira, A.; Varandas, S.; Prié, V.; Zieritz, A.; et al. Lifting the curtain on the freshwater mussel diversity from the Italian Peninsula and Croatian Adriatic coast. Biodivers. Conserv. 2017, 26, 3255–3274. [Google Scholar] [CrossRef]
- Zhang, P.; Fang, H.Y.; Pan, W.J.; Pan, H.C. The complete mitochondrial genome of Chinese pond mussel Sinanodonta woodiana (Unionoida: Unionidae). Mitochondrial DNA Part A 2016, 27, 1620–1621. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).