The mtDNA D-Loop Legacy of Cattle: Fluctuations in Diversity from the Neolithic to Early Medieval Times in Switzerland
Abstract
1. Introduction
2. Materials and Methods
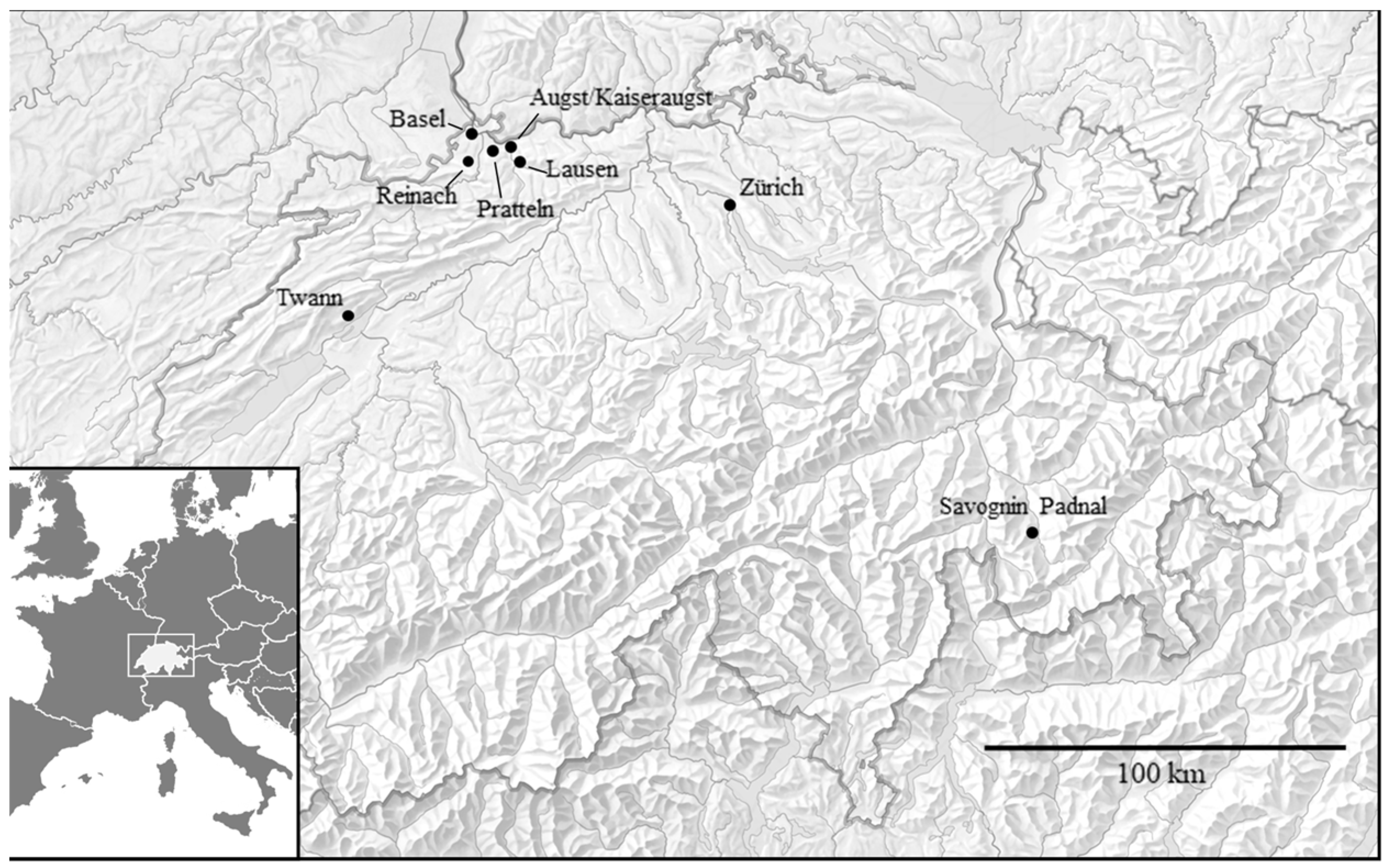
2.1. Archaeological Cattle
2.2. Methods
2.2.1. DNA Extraction and PCR Amplification
2.2.2. Sequence Analysis and Haplotype Identification
2.2.3. Population Parameters
2.3. Biometry
3. Results
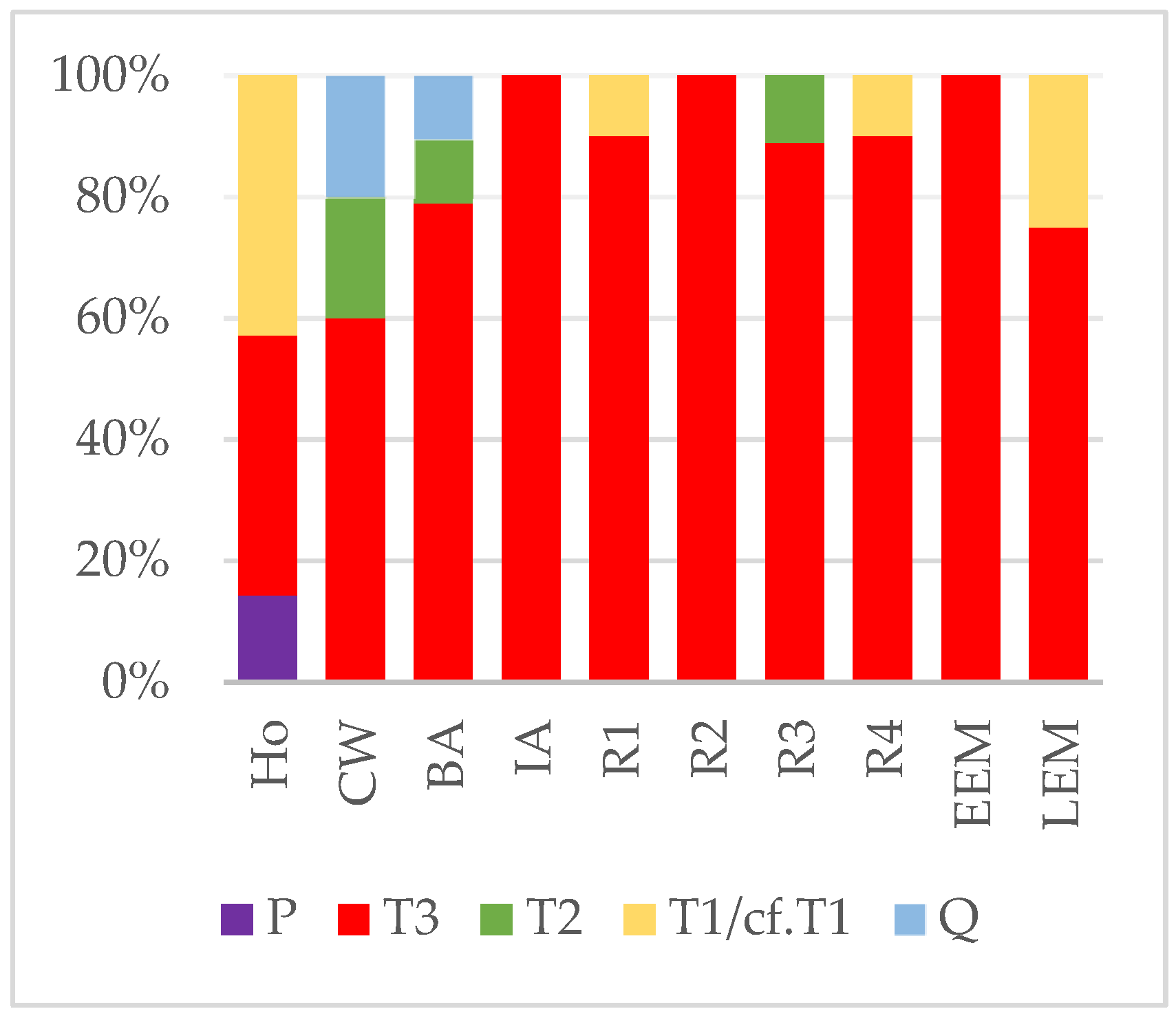
3.1. Haplogroup and Haplotype Identifications
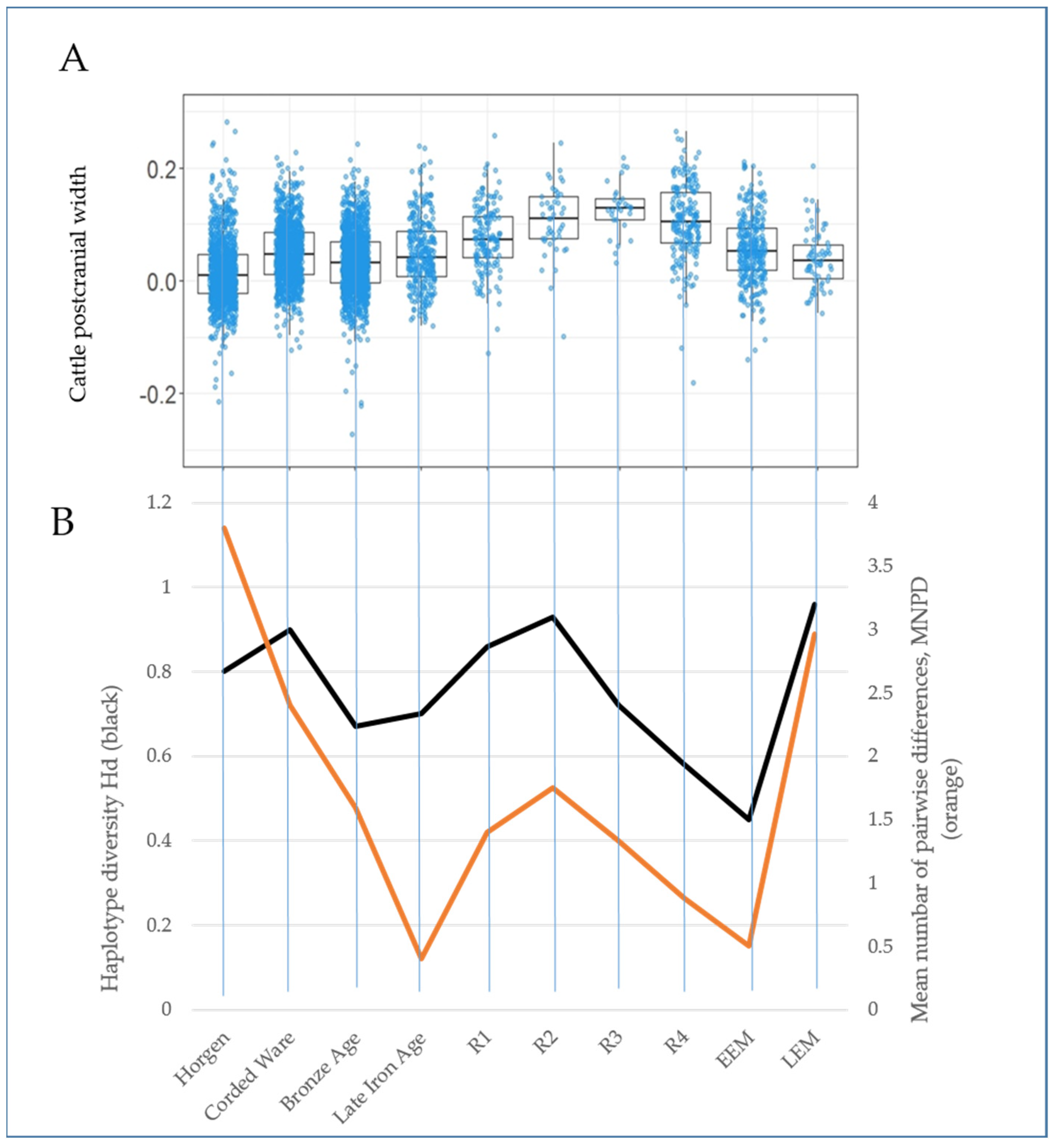
3.2. Genetic and Morphometric Diversity through Time
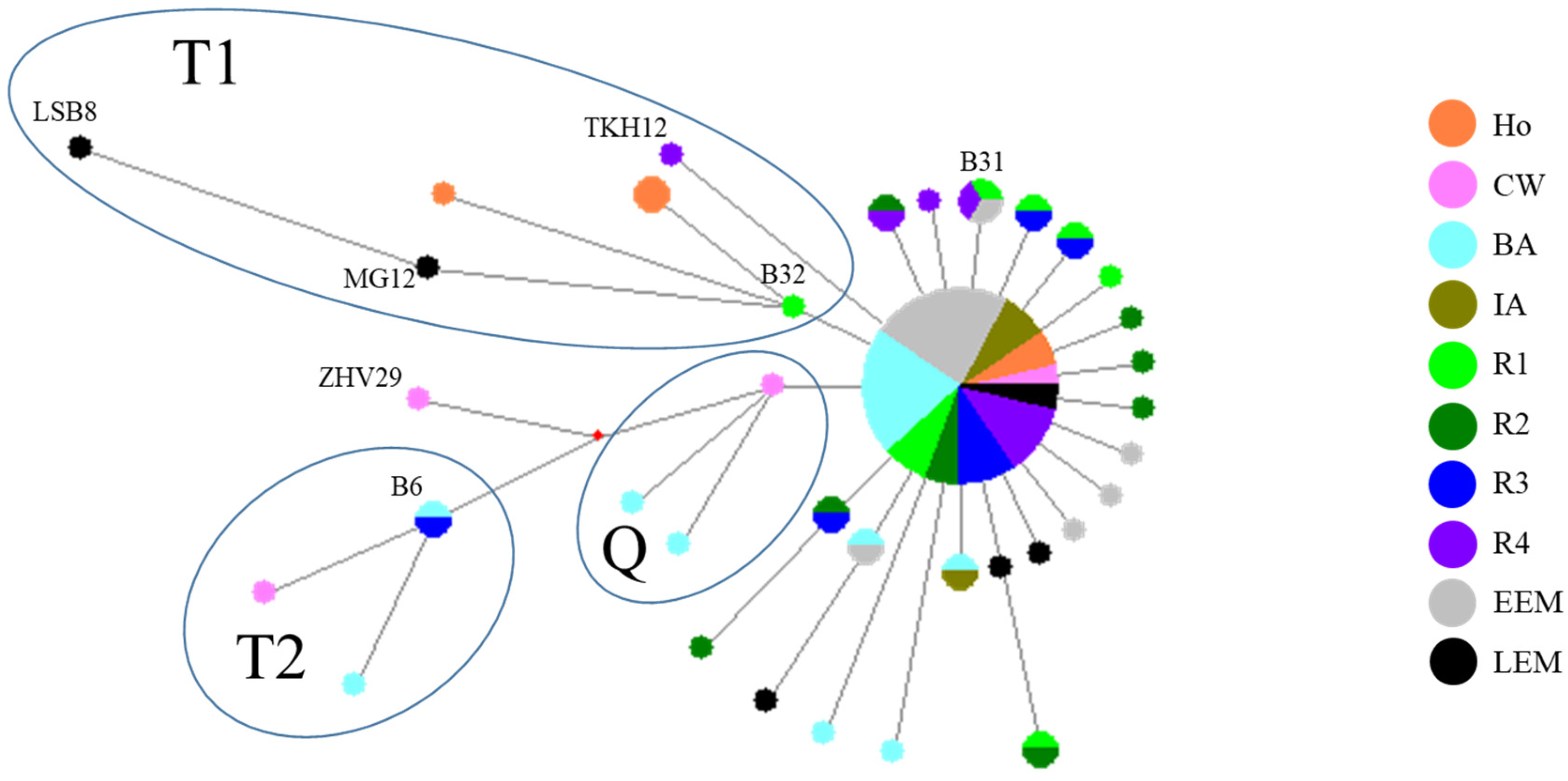
3.3. Median-Joining Network
3.4. Relationships between the Archaeological Cattle “Populations”
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schibler, J.; Schlumbaum, A. Geschichte und wirtschaftliche Bedeutung des Hausrindes (Bos taurus L.) in der Schweiz von der Jungsteinzeit bis ins frühe Mittelalter. Schweiz. Arch. Tierheilk. 2007, 149, 23–29. [Google Scholar] [CrossRef]
- Kantanen, J.; Olsaker, I.; Holm, L.E.; Lien, S.; Vilkki, J.; Brusgaard, K.; Eythorsdottir, E.; Danell, B.; Adalsteinsson, S. Genetic diversity and population structure of 20 north European cattle breeds. J. Hered. 2000, 91, 446–457. [Google Scholar] [CrossRef]
- Beja-Pereira, A.; Caramelli, D.; Lalueza-Fox, C.; Vernesi, C.; Ferrand, N.; Casoli, A.; Goyache, F.; Royo, L.J.; Conti, S.; Lari, M.; et al. The origin of European cattle: Evidence from modern and ancient DNA. Proc. Natl. Acad. Sci. USA 2006, 103, 8113–8118. [Google Scholar] [CrossRef]
- Cubric-Curik, V.; Novosel, D.; Brajkovic, V.; Stabelli, O.R.; Krebs, S.; Sölkner, J.; Šalamon, D.; Ristov, S.; Berger, B.; Trivizaki, S.; et al. Large-scale mitogenome sequencing reveals consecutive expansions of domestic taurine cattle and supports sporadic aurochs introgression. Evol. Appl. 2021, 15, 663–678. [Google Scholar] [CrossRef]
- Frantz, L.A.F.; Bradley, D.G.; Larson, G.; Orlando, L. Animal domestication in the era of ancient genomics. Nat. Rev. Genet. 2020, 21, 449–460. [Google Scholar] [CrossRef]
- Larson, G. Reconstructing Migration Trajectories Using Ancient DNA; Cambridge University Press: Cambridge, UK, 2017; pp. 237–260. [Google Scholar]
- Bonfiglio, S.; Ginja, C.; De Gaetano, A.; Achilli, A.; Olivieri, A.; Colli, L.; Tesfaye, K.; Agha, S.H.; Gama, L.T.; Cattonaro, F.; et al. Origin and Spread of Bos taurus: New Clues from Mitochondrial Genomes Belonging to Haplogroup T1. PLoS ONE 2012, 7, e38601. [Google Scholar] [CrossRef] [PubMed]
- Achilli, A.; Bonfiglio, S.; Olivieri, A.; Malusà, A.; Pala, M.; Kashani, B.H.; Perego, U.A.; Ajmone-Marsan, P.; Liotta, L.; Semino, O.; et al. The Multifaceted Origin of Taurine Cattle Reflected by the Mitochondrial Genome. PLoS ONE 2009, 4, e5753. [Google Scholar] [CrossRef] [PubMed]
- Achilli, A.; Olivieri, A.; Pellecchia, M.; Uboldi, C.; Colli, L.; Al-Zahery, N.; Accetturo, M.; Pala, M.; Kashani, B.H.; Perego, U.A.; et al. Mitochondrial genomes of extinct aurochs survive in domestic cattle. Curr. Biol. 2008, 18, R157–R158. [Google Scholar] [CrossRef]
- Pellecchia, M.; Negrini, R.; Colli, L.; Patrini, M.; Milanesi, E.; Achilli, A.; Bertorelle, G.; Cavalli-Sforza, L.L.; Piazza, A.; Torroni, A.; et al. The mystery of Etruscan origins: Novel clues from Bos taurus mitochondrial DNA. Proc. R. Soc. B Boil. Sci. 2007, 274, 1175–1179. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, A.; Gandini, F.; Achilli, A.; Fichera, A.; Rizzi, E.; Bonfiglio, S.; Battaglia, V.; Brandini, S.; De Gaetano, A.; El-Beltagi, A.; et al. Mitogenomes from Egyptian Cattle Breeds: New Clues on the Origin of Haplogroup Q and the Early Spread of Bos taurus from the Near East. PLoS ONE 2015, 10, e0141170. [Google Scholar] [CrossRef]
- Bonfiglio, S.; Achilli, A.; Olivieri, A.; Negrini, R.; Colli, L.; Liotta, L.; Marsan, P.A.; Torroni, A.; Ferretti, L. The Enigmatic Origin of Bovine mtDNA Haplogroup R: Sporadic Interbreeding or an Independent Event of Bos primigenius Domestication in Italy? PLoS ONE 2010, 5, e15760. [Google Scholar] [CrossRef]
- Scheu, A.; Powell, A.; Bollongino, R.; Vigne, J.-D.; Tresset, A.; Çakırlar, C.; Benecke, N.; Burger, J. The genetic prehistory of domesticated cattle from their origin to the spread across Europe. BMC Genet. 2015, 16, 54. [Google Scholar] [CrossRef] [PubMed]
- Ajmone-Marsan, P.; Garcia, J.F.; Lenstra, J.A. On the origin of cattle: How aurochs became cattle and colonized the world. Evol. Anthropol. 2010, 19, 148–157. [Google Scholar] [CrossRef]
- MacHugh, D.E.; Larson, G.; Orlando, L. Taming the Past: Ancient DNA and the Study of Animal Domestication. Annu. Rev. Anim. Biosci. 2017, 5, 329–351. [Google Scholar] [CrossRef]
- Gurke, M.; Vidal-Gorosquieta, A.; Pajimans, J.L.A.; Wȩcek, K.; Barlow, A.; González-Fortes, G.; Hartmann, S.; Grandal-D’anglade, A.; Hofreiter, M. Insight into the introduction of domestic cattle and the process of Neolithization to the Spanish region Galicia by genetic evidence. PLoS ONE 2021, 16, e0249537. [Google Scholar] [CrossRef]
- Colominas, L.; Edwards, C.J.; Beja-Pereira, A.; Vigne, J.-D.; Silva, R.M.; Castanyer, P.; Tremoleda, J.; Seguí, M.S.; Pérez-Ripoll, M.; Goyache, F.; et al. Detecting the T1 cattle haplogroup in the Iberian Peninsula from Neolithic to medieval times: New clues to continuous cattle migration through time. J. Archaeol. Sci. 2015, 59, 110–117. [Google Scholar] [CrossRef]
- Saliari, K.; Amory, C.; Draganits, E.; Ramsl, P.C.; Tobias, B.; Pucher, E.; Parson, W. Morphometric and genetic evidence for cattle imports from the Mediterranean into present-day Austria during the Iron Age. J. Archaeol. Sci. Rep. 2023, 48, 103842. [Google Scholar] [CrossRef]
- Svensson, E.; Häsler, S.; Nussbaumer, M.; Rehazek, A.; Omrak, A.; Götherström, A. Mediaeval cattle from Bern (Switzerland): An archaeozoological, genetic and historical approach. Schweiz. Arch. Tierheilkd. 2014, 156, 17–26. [Google Scholar] [CrossRef]
- Svensson, E.M.; Anderung, C.; Baubliene, J.; Persson, P.; Malmström, H.; Smith, C.; Vretemark, M.; Daugnora, L.; Götherström, A. Tracing genetic change over time using nuclear SNPs in ancient and modern cattle. Anim. Genet. 2007, 38, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Niemi, M.; Bläuer, A.; Iso-Touru, T.; Harjula, J.; Edmark, V.N.; Rannamäe, E.; Lõugas, L.; Sajantila, A.; Lidén, K.; Taavitsainen, J.-P. Temporal Fluctuation in North East Baltic Sea Region Cattle Population Revealed by Mitochondrial and Y-Chromosomal DNA Analyses. PLoS ONE 2015, 10, e0123821. [Google Scholar] [CrossRef]
- Niemi, M.; Sajantila, A.; Vilkki, J. Temporal variation in coat colour (genotypes) supports major changes in the Nordic cattle population after Iron Age. Anim. Genet. 2016, 47, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Meiri, M.; Stockhammer, P.W.; Morgenstern, P.; Maran, J. Mobility and trade in Mediterranean antiquity: Evidence for an ‘Italian connection’ in Mycenaean Greece revealed by ancient DNA of livestock. J. Archaeol. Sci. Rep. 2019, 23, 98–103. [Google Scholar] [CrossRef]
- Meiri, M.; Stockhammer, P.W.; Marom, N.; Bar-Oz, G.; Sapir-Hen, L.; Morgenstern, P.; Macheridis, S.; Rosen, B.; Huchon, D.; Maran, J.; et al. Eastern Mediterranean Mobility in the Bronze and Early Iron Ages: Inferences from Ancient DNA of Pigs and Cattle. Sci. Rep. 2017, 7, 701. [Google Scholar] [CrossRef]
- Fraser, S.; Elsner, J.; Hamilton, W.D.; Sayle, K.L.; Schlumbaum, A.; Bartosiewicz, L. Matrilines in Neolithic cattle from Orkney, Scotland reveals complex husbandry patterns of ancestry. J. Archaeol. Sci. Rep. 2017, 14, 46–54. [Google Scholar] [CrossRef]
- Schibler, J. The economy and environment of the 4th and 3rd millennia BC in the northern Alpine foreland based on studies of animal bones. Environ. Archaeol. 2006, 11, 49–64. [Google Scholar] [CrossRef]
- Schibler, J. Zooarchaeological results from Neolithic and Bronze Age wetland and dryland sites in the Central Alpine Foreland: Economic, ecologic and taphonomic relevance. In Oxford Handbook of Zooarchaeology; Albarella, U., Rizzetto, M., Russ, H., Vickers, K., Viner-Daniels, S., Eds.; Oxford University Press: Oxford, UK, 2017; pp. 83–98. [Google Scholar] [CrossRef]
- Hüster-Plogmann, H.; Schibler, J. Archäozoologie. In Ökonomie und Ökologie neolithischer und bronzezeitlicher Ufersiedlungen am Zürichsee; Schibler, J., Hüster-Plogmann, H., Jacomet, S., Brombacher, C., Gross-Klee, E., Rast-Eicher, A., Eds.; Direktion der Öffentlichen Bauten des Kantons Zürich, Hochbauamt, Abteilung Kantonsarchäologie: Zürich, Switzerland, 1997; pp. 40–121. [Google Scholar]
- Spangenberg, J.E.; Jacomet, S.; Schibler, J. Chemical analyses of organic residues in archaeological pottery from Arbon Bleiche 3, Switzerland—Evidence for dairying in the late Neolithic. J. Archaeol. Sci. 2006, 33, 1–13. [Google Scholar] [CrossRef]
- Deschler-Erb, S.; Marti-Grädel, E. Viehhaltung und Jagd. Ergebnisse der Untersuchungen der handaufgelesenen Tierknochen. In Die Jungsteinzeitliche Siedlung Arbon Bleiche 3. Umwelt und Wirtschaft; Jacomet, S., Leuzinger, U., Schibler, J., Eds.; Huber & Co AG: Frauenfeld, Switzerland, 2004; Volume 12, pp. 158–231. [Google Scholar]
- Arbogast, R.-M.; Jacomet, S.; Magny, M.; Schibler, J. The significance of climate fluctuations for lake level changes and shifts in subsistence economy during the late Neolithic (4300–2400 b.c.) in central Europe. Veg. Hist. Archaeobotany 2006, 15, 403–418. [Google Scholar] [CrossRef]
- Wright, E. Investigating cattle husbandry in the Swiss Late Neolithic using different scales of temporal precision: Potential early evidence for deliberate livestock “improvement” in Europe. Archaeol. Anthr. Sci. 2021, 13, 36. [Google Scholar] [CrossRef]
- Marti-Grädel, E. Archäozoologische untersuchungen zu viehhaltung, jagd und fischfang. In Um 2700 v. Chr.-Wandel und Kontinuität in den Ufersiedlungen am Bielersee; Suter, P.J., Ed.; Archäologischer Dienst des Kantons Bern: Bern, Switzerland, 2017; Volume 1, pp. 160–193. [Google Scholar]
- The End of the Lake-Dwellings in the Circum-Alpine Region; Menotti, F., Ed.; Oxbow Books: Oxford, UK; Philadelphia, PA, USA, 2015; p. 250. [Google Scholar]
- Bopp-Ito, M.; Deschler-Erb, S.; Vach, W.; Schibler, J. Size diversity in Swiss Bronze Age cattle. Int. J. Osteoarchaeol. 2018, 28, 294–304. [Google Scholar] [CrossRef]
- Stopp, B. Animal husbandry and hunting activities in the Late Bronze Age Circum-Alpine region. In The End of the Lake-Dwellings in the Circum-Alpine Region; Menotti, F., Ed.; Oxbow Books: Oxford, UK, 2015; pp. 179–210. [Google Scholar]
- Müller, F.; Kaenel, G.; Lüscher, G. (Eds.) SPM IV Eisenzeit; Schweizerische Gesellschaft für Ur- und Frühgeschichte: Basel, Switzerland, 1999; Volume SPM IV, p. 360. [Google Scholar]
- Stopp, B. Der Basler Münsterhügel am Übergang von spätkeltischer zu römischer Zeit. archäozoologische Auswertung der Grabungen FH 1978/13 und TEW 1978/26. Ph.D. Dissertation, University of Basel, Basel, Switzerland, 2009. Available online: https://edoc.unibas.ch/1043/ (accessed on 1 April 2023).
- Warnberg, O.; Knipper, C.; Röder, B.; Lassau, G.; Spichtig, N.; Ramsl, P.C.; Novotny, F.; Teschler-Nicola, M.; Marion, S.; Schönfelder, M.; et al. Missing lactase persistence in late Iron Age Central Europe. Archäologisches Korrespondenzblatt. 2022, 52, 225–248. [Google Scholar] [CrossRef]
- Deschler-Erb, S.; Akeret, E.Ö.; Vandorpe, P. Crop production and livestock breeding from the Late Iron Age to the Late Roman period in north western Switzerland. In Productions Agro-Pastorales, Pratiques Culturales et Élevage dans le Nord de la Gaule du Deuxième Siècle Avant J.-C. à la fin de la Période Romaine; Lepetz, S., Zech-Mattern, V., Eds.; Editions Mergoil: Quint-Fonsegrives, France, 2017; pp. 135–152. [Google Scholar]
- Valenzuela-Lamas, S.; Albarella, U. Animal Husbandry across the Western Roman Empire: Changes and Continuities. Eur. J. Archaeol. 2017, 20, 402–415. [Google Scholar] [CrossRef]
- Grau-Sologestoa, I.; Ginella, F.; Marti-Grädel, E.; Stopp, B.; Deschler-Erb, S. Animal husbandry between the Roman times and the High Middle Ages in central Europe: A biometrical analysis of cattle, sheep and pig. Archaeol. Anthr. Sci. 2021, 13, 176. [Google Scholar] [CrossRef]
- Columella. De re rustica. In Über Landwirtschaft; Akademie Verlag: Berlin, Germany, 1972; Volume 4. [Google Scholar]
- Cato. De agri cultura. In Vom Landbau; Tusculum-Bücherei, Heimeran Verlag: München, Germany, 1980. [Google Scholar]
- SPM V Römische Zeit; Flutsch, L., Niffeler, U., Rossi, F., Eds.; Schweizerische Gesellschaft für Ur- und Frühgeschichte: Basel, Switzerland, 2002; Volume SPM V, p. 432. [Google Scholar]
- MacKinnon, M. Cattle ‘breed’ variation and improvement in Roman Italy: Connecting the zooarchaeological and ancient textual evidence. World Archaeol. 2010, 42, 55–73. [Google Scholar] [CrossRef]
- Albarella, U.; Johnstone, C.; Vickers, K. The development of animal husbandry from the Late Iron Age to the end of the Roman period: A case study from South-East Britain. J. Archaeol. Sci. 2008, 35, 1828–1848. [Google Scholar] [CrossRef]
- Peters, J. Römische Tierhaltung und Tierzucht; Marie Leidorf GmbH: Rahden/Westf, Germany, 1998; Volume 5. [Google Scholar]
- Breuer, G.; Rehazek, A.; Stopp, B. Grössen- und Wuchsformveränderung beim Hausrind in römischer Zeit am Beispiel von Augst/Augusta Raurica. Jahresberichte aus Augst und Kaiseraugst 1999, 20, 207–228. [Google Scholar]
- Deschler-Erb, S. Archäozoologie—Leben am Rande der Welt. Die Tierknochen aus Brunnen und Gruben des römischen Vicus von Gross-Gerau. In Gross-Gerau, I. Der römische Vicus von Gross-Gerau, “Auf Esch”. Die Baubefunde des Kastellvicus und der Siedlung des 2–3, Jahrhunderts; Wenzel, C., Ed.; Verlag Dr. Rudolf Habelt GmbH: Bonn, Germany, 2009; pp. 255–300. [Google Scholar]
- Groot, M.; Deschler-Erb, S. Market strategies in the Roman provinces: Different animal husbandry systems explored by a comparative regional approach. J. Archaeol. Sci. Rep. 2015, 4, 447–460. [Google Scholar] [CrossRef]
- Lepetz, S. L’amélioration des espèces animales domestiques à la période romaine en France du Nord. Techniques et économie antiques et médiévales: Le temps de l’innovation. Aix-en Provence 21–23 mai 1996. In Colloque international (CNRS); Editions Errances: Paris, France, 1997. [Google Scholar]
- Trentacoste, A.; Nieto-Espinet, A.; Guimarães, S.; Wilkens, B.; Petrucci, G.; Valenzuela-Lamas, S. New trajectories or accelerating change? Zooarchaeological evidence for Roman transformation of animal husbandry in Northern Italy. Archaeol. Anthr. Sci. 2021, 13, 25. [Google Scholar] [CrossRef]
- Marti-Grädel, E. Archäozoologische Untersuchungen der Tierknochen aus der Burgstelle Altenberg, Kt. Basel-Landschaft (11. Jahrhundert): Im Kontext früh- bis hochmittelalterlicher Siedlung der Region (5.-12. Jahrhundert): Wirtschafts- und Umweltgeschichte des Früh- und Hochmittelalters in der Nordwestschweiz. Ph.D. Dissertation, University of Basel, Basel, Switzerland, 2012. Available online: https://edoc.unibas.ch/21278/ (accessed on 1 April 2023).
- Frosdick, R. Status and New Beginnings: Archaeozoological Research into the Early Medieval Rural Settlements of Northwest Switzerland; University of Basel: Basel, Switzerland, 2014. [Google Scholar] [CrossRef]
- Breuer, G.; Rehazek, A.; Stopp, B. Grössenveränderungen des Hausrindes. Osteometrische Untersuchungen grosser Fundserien aus der Nordschweiz von der Spätlatènezeit bis ins Frühmittelalter am Beispiel von Basel, Augst (Augusta Raurica) und Schleitheim-Brüel. Jahresberichte aus Augst und Kaiseraugst 1999, 20, 207–228. [Google Scholar]
- Granado, J.; Harmath, M.; Tecchiati, U.; Oeggl, K.; Schibler, J.; Schlumbaum, A. MtDNA D-Loop Diversity in Alpine Cattle during the Bronze Age. Diversity 2021, 13, 449. [Google Scholar] [CrossRef]
- Schibler, J.; Elsner, J.; Schlumbaum, A. Incorporation of aurochs into a cattle herd in Neolithic Europe: Single event or breeding? Sci. Rep. 2014, 4, 5798. [Google Scholar] [CrossRef]
- Grau-Sologestoa, I.; Groot, M.; Deschler-Erb, S. Innovation and Intensification: The Use of Cattle in the Roman Rhine Region. Environ. Archaeol. 2022, 1–19. [Google Scholar] [CrossRef]
- Schlumbaum, A.; Turgay, M.; Schibler, J. Near East mtDNA haplotype variants in Roman cattle from Augusta Raurica, Switzerland, and in the Swiss Evolene breed. Anim. Genet. 2006, 37, 373–375. [Google Scholar] [CrossRef] [PubMed]
- Schlumbaum, A.; Stopp, B.; Breuer, G.; Rehazek, A.; Turgay, M.; Blatter, R.; Schibler, J. Combining archaeozoology and molecular genetics: The reason behind the changes in cattle size between 150BC and 700AD in Switzerland. Antiquity 2003, 77. [Google Scholar]
- Elsner, J.; Schibler, J.; Hofreiter, M.; Schlumbaum, A. Burial condition is the most important factor for mtDNA PCR amplification success in Palaeolithic equid remains from the Alpine foreland. Archaeol. Anthr. Sci. 2014, 7, 505–515. [Google Scholar] [CrossRef]
- Warnberg, O.; Alt, K.W. Molekulargenetische Analysen an den Bestattungen aus dem endneolithischen Kollektivgrab von Spreitenbach. In Spreitenbach-Moosweg (Aargau, Schweiz): Ein Kollektivgrab um 2500 v. Chr.; Antiqua 51; Doppler, T., Ed.; Veröffentlichung der Archäologie Schweiz: Basel, Switzerland, 2012; pp. 158–169. [Google Scholar]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.; de Bruijn, M.; Coulson, A.; Eperon, I.; Sanger, F.; Young, I. Complete sequence of bovine mitochondrial DNA conserved features of the mammalian mitochondrial genome. J. Mol. Biol. 1982, 156, 683–717. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D.G.; MacHugh, D.E.; Cunningham, P.; Loftus, R.T. Mitochondrial diversity and the origins of African and European cattle. Proc. Natl. Acad. Sci. USA 1996, 93, 5131–5135. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.-J.; Macaulayb, V.; Richards, M. Median Networks: Speedy Construction and Greedy Reduction, One Simulation, and Two Case Studies from Human mtDNA. Mol. Phylogenetics Evol. 2000, 16, 8–28. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Pozo, J.M.; Trentacoste, A.; Nieto-Espinet, A.; Guimarães, S.; Valenzuela-Lamas, S. Zoolog R package: Zooarchaeological analysis with log-ratios. Quat. Int. 2022. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M. A measure of population subdivision based on microsatellite allele frequencies. Genetics 1995, 139, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Lenstra, J.A.; Ajmone-Marsan, P.; Beja-Pereira, A.; Bollongino, R.; Bradley, D.G.; Colli, L.; De Gaetano, A.; Edwards, C.J.; Felius, M.; Ferretti, L.; et al. Meta-Analysis of Mitochondrial DNA Reveals Several Population Bottlenecks during Worldwide Migrations of Cattle. Diversity 2014, 6, 178–187. [Google Scholar] [CrossRef]
- Schlumbaum, A.; Granado, J.; Hans-Peter, G.; Schibler, J. MtDNA d-loop variation in Prespa dwarf cattle (Bos taurus) from Albania. bioRxiv 2023. [Google Scholar] [CrossRef]
- Hristov, P.; Sirakova, D.; Mitkov, I.; Spassov, N.; Radoslavov, G. Balkan brachicerous cattle—The first domesticated cattle in Europe. Mitochondrial DNA Part A 2016, 29, 56–61. [Google Scholar] [CrossRef]
- Hristov, P.; Spassov, N.; Iliev, N.; Radoslavov, G. An independent event of Neolithic cattle domestication on the South-eastern Balkans: Evidence from prehistoric aurochs and cattle populations. Mitochondrial DNA Part A 2015, 28, 383–391. [Google Scholar] [CrossRef]
- Troy, C.S.; MacHugh, D.E.; Bailey, J.F.; Magee, D.A.; Loftus, R.T.; Cunningham, P.; Chamberlain, A.T.; Sykes, B.C.; Bradley, D.G. Genetic evidence for Near-Eastern origins of European cattle. Nature 2001, 410, 1088–1091. [Google Scholar] [CrossRef]
- Colominas, L.; Edwards, C.J. Livestock Trade during the Early Roman Period: First Clues from the Trading Post of Empúries (Catalonia). Int. J. Osteoarchaeol. 2016, 27, 167–179. [Google Scholar] [CrossRef]
- Furtwängler, A.; Rohrlach, A.B.; Lamnidis, T.C.; Papac, L.; Neumann, G.U.; Siebke, I.; Reiter, E.; Steuri, N.; Hald, J.; Denaire, A.; et al. Ancient genomes reveal social and genetic structure of Late Neolithic Switzerland. Nat. Commun. 2020, 11, 1915. [Google Scholar] [CrossRef] [PubMed]
- Knipper, C.; Mittnik, A.; Massy, K.; Kociumaka, C.; Kucukkalipci, I.; Maus, M.; Wittenborn, F.; Metz, S.E.; Staskiewicz, A.; Krause, J.; et al. Female exogamy and gene pool diversification at the transition from the Final Neolithic to the Early Bronze Age in central Europe. Proc. Natl. Acad. Sci. USA 2017, 114, 10083–10088. [Google Scholar] [CrossRef] [PubMed]
- Bopp-Ito, M. Archaeozoological Study on the Bronze Age Alpine Settlement Savognin-Padnal in the Canton of Grisons, Switzerland. Ph.D. Thesis, University of Basel, Basel, Switzerland, 2019. Available online: https://edoc.unibas.ch/70524/1/Bopp-Ito_Dissertation%202019_ohne_CV.pdf (accessed on 1 January 2023).
- Jennings, B. Exploring Late Bronze Age systems of bronzework production in Switzerland through Network Science. STAR Sci. Technol. Archaeol. Res. 2016, 2, 90–112. [Google Scholar] [CrossRef]
- Riedel, A.; Tecchiati, U. Settlements and economy in the Bronze and Iron Age in Trentino-South Tyrol: Notes for an archaeozoological model. Preistoria Alpina 2001, 35, 105–113. [Google Scholar]
- Moghaddam, N.; Müller, F.; Lösch, S. A bioarchaeological approach to the Iron Age in Switzerland: Stable isotope analyses (δ13C, δ15N, δ34S) of human remains. Archaeol. Anthropol. Sci. 2016, 10, 1067–1085. [Google Scholar] [CrossRef]
- Schibler, J.; Stopp, B.; Studer, J. Haustierhaltung und Jagd; Eisenzeit, Müller, F., Kaenel, G., Lüscher, G., Eds.; Schweizerische Gesellschaft für Ur- und Frühgeschichte: Basel, Switzerland, 1999; Volume 4, pp. 116–136. [Google Scholar]
- Brönnimann, D.; Rissanen, H.; Spichtig, N.; Wimmer, J. Die jüngerlatènezeitliche Zentralsiedlung Basel-Gasfabrik im Fokus. Ausgewählte Ergebnisse der interdisziplinären Forschung. Jahresbericht der Archäologischen Bodenforschung Basel-Stadt 2021, 2022, 116–151. [Google Scholar]
- Groot, M.; Deschler-Erb, S. Carnem et circenses—Consumption of animals and their products in Roman urban and military sites in two regions in the northwestern provinces. Environ. Archaeol. 2016, 22, 96–112. [Google Scholar] [CrossRef]
- Deschler-Erb, S.; Stopp, B.; Vandorpe, P. Big Data—65 Jahre archäobiologische Forschungen in Augusta Raurica. Jahresberichte aus Augst und Kaiseraugst 2021, 42, 293–368. [Google Scholar]
- Schucany, C. Die römische Villa von Biberist-Spitalhof/SO (Grabungen 1982, 1983, 1986–1989). Untersuchungen im Wirtschaftsteil und Überlegungen zum Umland. Ausgrabungen und Forschungen 4; BAG Verlag: Remshalden, Germany, 2006. [Google Scholar]
- Sütterlin, H. Kastelen 2: Die Aelteren Steinbauten in den Insulae 1 Und 2 von Augusta Raurica; Forsch. Augst: Neuenstadt, Germany, 1999; p. 281. [Google Scholar]
- Deschler-Erb, S. Die Tierknochen. In Die römische Villa von Biberist-Spitalhof/SO (Grabungen 1982, 1983, 1986–1989) Untersuchungen zum Wirtschaftsteil und Überlegungen zum Umland; Schucany, C., Ed.; B.A.G. Verlag: Remshalden, Germany, 2006; Volume 2, pp. 635–665. [Google Scholar]
- Schwarz, P.-A. Kastelen 4—Die Nordmauer und die Überreste der Innenbebauung der spätrömischen Befestigung auf Kastelen. Forsch. Augst 2002, 24, 511. [Google Scholar]
- Marti, R. Landwirtschaft lohnt sich—Goldfunde aus der römischen Villa rustica von Pratteln, Kästeli. In 50 Jahre—50 Funde; Marti, R., Fischer, A., Eds.; Archäologie im Kanton Baselland: Basel, Switzerland, 2018; pp. 74–75. [Google Scholar]
- Fischer, A. Pratteln, Kästeli: Ein römischer Gutshof wie kein anderer? Jahresbericht Archäologie Baselland 2009, 2010, 35–43. [Google Scholar]
- Marti, R.; Fischer, A. Pratteln Kästeliweg: Grossgrabung im römischen Gutshof. Jahresbericht Archäologie Baselland 2016, 2017, 31–38. [Google Scholar]
- Der Untergang des Römischen Reiches; Henrich, P., Ed.; WBG Theiss: Darmstadt, Germany, 2022; Volume 44. [Google Scholar]
- Hüster-Plogmann, H.; Kühn, M. Landwirtschaft: Das tägliche Brot. In SPM VI: Frühmittelalter. Die Schweiz vom Paläolithikum bis zum frühen Mittelalter; Windler, R., Marti, R., Niffeler, U., Steiner, L., Eds.; Schweizerische Gesellschaft für Ur- und Frühgeschichte: Basel, Switzerland, 2005; Volume 6, pp. 340–342. [Google Scholar]
- Windler, R.; Marti, R.; Niffeler, U.; Steiner, L. (Eds.) SPM VI Frühmittelalter; Schweizerische Gesellschaft für Ur- und Frühgeschichte: Basel, Switzerland, 2005; Volume 6, p. 443. [Google Scholar]
- Brombacher, C.; Kühn, M. Vegetationsentwicklung und Umland der Siedlungen—Waldentwicklung, Ackerland und Grünland. In SPM VI: Frühmittelalter. Die Schweiz vom Paläolithikum bis zum frühen Mittelalter; Windler, R., Marti, R., Niffeler, U., Steiner, L., Eds.; Schweizerische Gesellschaft für Ur- und Frühgeschichte: Basel, Switzerland, 2005; Volume 6, pp. 88–91. [Google Scholar]
- Bachrach, B. Charlemagne’s Early Campaigns (768–777). A Diplomatic and Military Analysis; Brill: Leiden/Boston, The Netherlands, 2013; Volume 82. [Google Scholar]
- Marti, R. Gesellschaft, Kultur, Wirtschaft: Aspekte des Frühmittelalters; Nordwestschweiz. In SPM VI: Frühmittelalter. Die Schweiz vom Paläolithikum bis zum frühen Mittelalter; Windler, R., Marti, R., Niffeler, U., Steiner, L., Eds.; Schweizerische Ge-sellschaft für Ur- und Frühgeschichte: Basel, Switzerland, 2005; Volume 6, pp. 247–249. [Google Scholar]
- Arango, J.A.; Cundiff, L.V.; Van Vleck, L.D. Genetic parameters for weight, weight adjusted for body condition score, height, and body condition score in beef cows. J. Anim. Sci. 2002, 80, 3112–3122. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, A.C.; Daetwyler, H.D.; Chamberlain, A.J.; Ponce, C.H.; Sargolzaei, M.; Schenkel, F.S.; Sahana, G.; Govignon-Gion, A.; Boitard, S.; Dolezal, M.; et al. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat. Genet. 2018, 50, 362–367. [Google Scholar] [CrossRef]
- Herre, W.; Röhrs, M. Haustiere—Zoologisch Gesehen; Gustav Fischer: Stuttgart, NY, USA, 1990. [Google Scholar]
- Funston, R.N.; Larson, D.M.; Vonnahme, K.A. Effects of maternal nutrition on conceptus growth and offspring performance: Implications for beef cattle production1. J. Anim. Sci. 2010, 88, E205–E215. [Google Scholar] [CrossRef]
- Yonova-Doing, E.; Calabrese, C.; Gomez-Duran, A.; Schon, K.; Wei, W.; Karthikeyan, S.; Chinnery, P.F.; Howson, J.M.M. An atlas of mitochondrial DNA genotype–phenotype associations in the UK Biobank. Nat. Genet. 2021, 53, 982–993. [Google Scholar] [CrossRef]
- Schutz, M.; Freeman, A.; Lindberg, G.; Koehler, C.; Beitz, D. The effect of mitochondrial DNA on milk production and health of dairy cattle. Livest. Prod. Sci. 1994, 37, 283–295. [Google Scholar] [CrossRef]
- Liu, H.; Zhai, J.; Wu, H.; Wang, J.; Zhang, S.; Li, J.; Niu, Z.; Shen, C.; Zhang, K.; Liu, Z.; et al. Diversity of Mitochondrial DNA Haplogroups and Their Association with Bovine Antral Follicle Count. Animals 2022, 12, 2350. [Google Scholar] [CrossRef]
- Dorji, J.; Jagt, C.J.V.; Chamberlain, A.J.; Cocks, B.G.; MacLeod, I.M.; Daetwyler, H.D. Recovery of mitogenomes from whole genome sequences to infer maternal diversity in 1883 modern taurine and indicine cattle. Sci. Rep. 2022, 12, 5582. [Google Scholar] [CrossRef]
- Schucany, C.; Mattmann, T. Die Keramik von Augusta Raurica: Typologie und Chronologie; Forsch. Augst: Neuenstadt, Germany, 2019; Volume 42, 481p. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Granado, J.; Wright, E.; Blatter, R.; Lange, J.; Turgay, M.; Bañuelos, L.; Deschler-Erb, S.; Stopp, B.; Marti-Grädel, E.; Schäfer, M.; et al. The mtDNA D-Loop Legacy of Cattle: Fluctuations in Diversity from the Neolithic to Early Medieval Times in Switzerland. Diversity 2023, 15, 687. https://doi.org/10.3390/d15050687
Granado J, Wright E, Blatter R, Lange J, Turgay M, Bañuelos L, Deschler-Erb S, Stopp B, Marti-Grädel E, Schäfer M, et al. The mtDNA D-Loop Legacy of Cattle: Fluctuations in Diversity from the Neolithic to Early Medieval Times in Switzerland. Diversity. 2023; 15(5):687. https://doi.org/10.3390/d15050687
Chicago/Turabian StyleGranado, José, Elizabeth Wright, Robert Blatter, Jürg Lange, Meral Turgay, Laura Bañuelos, Sabine Deschler-Erb, Barbara Stopp, Elisabeth Marti-Grädel, Marguerita Schäfer, and et al. 2023. "The mtDNA D-Loop Legacy of Cattle: Fluctuations in Diversity from the Neolithic to Early Medieval Times in Switzerland" Diversity 15, no. 5: 687. https://doi.org/10.3390/d15050687
APA StyleGranado, J., Wright, E., Blatter, R., Lange, J., Turgay, M., Bañuelos, L., Deschler-Erb, S., Stopp, B., Marti-Grädel, E., Schäfer, M., Grau-Sologestoa, I., Ammann, S., Schmid, D., Furger, A. R., Marti, R., Schibler, J., & Schlumbaum, A. (2023). The mtDNA D-Loop Legacy of Cattle: Fluctuations in Diversity from the Neolithic to Early Medieval Times in Switzerland. Diversity, 15(5), 687. https://doi.org/10.3390/d15050687







