Similar Ones Are Not Related and Vice Versa—New Dendronotus Taxa (Nudibranchia: Dendronotidae) from the North Atlantic Ocean Provide a Platform for Discussion of Global Marine Biodiversity Patterns †
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Morphological Analysis
2.3. Molecular Analysis
3. Results
3.1. Systematics
- Diagnosis
- Etymology
- Material examined
- Diagnosis
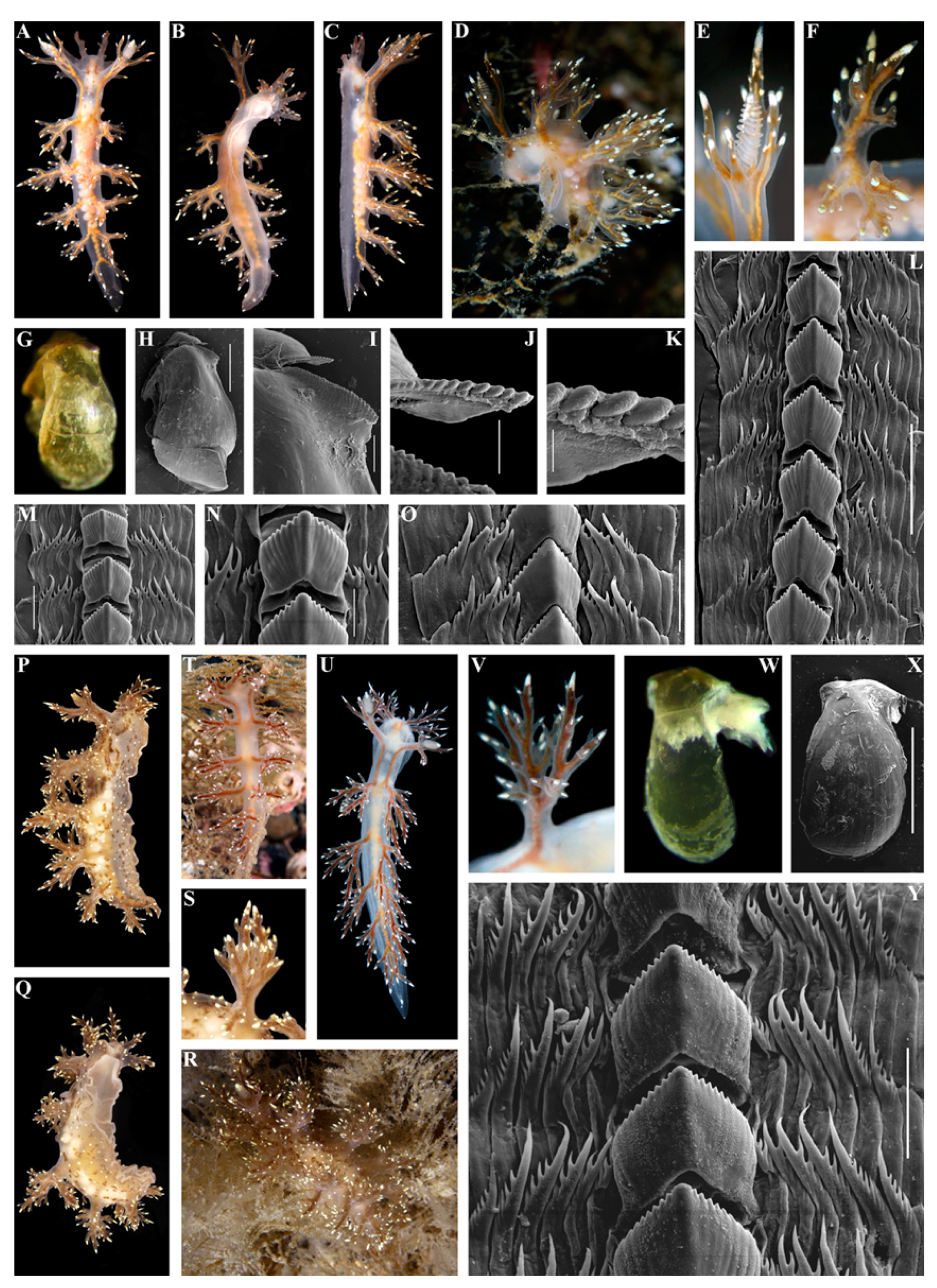
- Description
- Distribution and ecological observations
- Comparative remarks
| Colouration | Body Length | Dorsolateral Appendages (Pairs) | Appendages of Oral Veil | Appendages of Rhinophoral Stalks | Rhinophoral Lamellae | Radula | Ampulla | Bursa Copulatrix | Prostate | Copulatory Organ (Penis) | Distribution | References | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dendronotus frondosus | Brownish to reddish-brown, often with small white and yellow specks, but usually without opaque white stripes between dorsolateral processes, to completely white translucent specimens | Up to 50 mm (live) | Up to 6, with relatively long primary stalk; moderately branched secondary branches bear elongate tertiary branches | Up to 7 | Up to 5 | Up to 12 | Up to 42 rows of teeth. Central tooth with deep furrows and with up to 14 (common range 8–12) distinct denticles. Up to 10 (usually up to 8) lateral teeth, bearing up to seven denticles | Voluminous, folded | Large, oval to rounded | Up to 30 alveolar glands | Relatively long, curved | Boreal waters of North Atlantic; do not penetrate to the true Arctic further than easternmost Barents Sea | Korshunova et al., 2020 [23] |
| Dendronotus arcticus arcticus | Brownish, highly consistent in all observed specimens, with scattered distinct opaque white dots on dorsal side and appendages; upper part of the dorsolateral appendages not covered with golden yellow pigment | Up to 19 mm (preserved) | Up to 6, with a moderate primary stalk; moderately branched secondary branches, and short tertiary branches | Up to 8 | Up to 6 | Up to 18 | Up to 39 rows of teeth. Central tooth with up to 14 small denticles and reduced furrows. Up to nine lateral teeth, bearing up to 9 denticles | Folded | Large and rounded | Up to 30 alveolar glands | Long and twisted | Restricted to the true Arctic waters: Laptev Sea, Kara Sea, easternmost part of Barents Sea adjacent to Kara Sea | Korshunova et al., 2016 [22]; present study |
| Dendronotus arcticus gartensis n. subsp. | Semi-translucent, highly consistent in all observed specimens, with bright yellow spots scattered dorsally and laterally, supplemented with dark brownish to deep violet markings, and similarly coloured cores of the dorsolateral appendages; upper part of the dorsolateral appendages covered with bright golden yellow to yellowish pigment | Up to 40 mm (live) | Up to 6, with elongate and pointed primary stalk, secondary branches reduced towards the tip of primary stalk and short tertiary branches, general spinose appearance | Up to 8 | Up to 7 | Up to 13 | Up to 46 rows of teeth. Central tooth moderately denticulated and bearing up to 20 distinct denticles from each side. Denticles with relatively deep furrows. Up to eight lateral teeth, bearing up to eight denticles | Large, folded | Large, rounded | Up to 40 and more alveolar glands | Long, strongly convoluted | Restricted to the boreal waters of middle Norway | Present study |
| Dendronotus keatleyae n. sp. | Semi-transclucent white to pale greyish-brown, in some specimens almost without any additional spots and lines (only very rare small white spots) or with more distinct brownish spots and broken line-like patterns, including small yellowish spots and pointed tubercles; upper part of dorsolateral appendages not covered with bright golden-yellow pigment; tips of appendages covered with white to yellowish bright contrasting pigment | Up to 35 mm (live) | Up to 7, with elongate primary stalk, moderately branched elongate secondary branches and short tertiary branches | Up to 6 | Up to 6 | Up to 14 | Up to 34 rows of teeth. Central tooth moderately denticulated and bearing up to 11 distinct to reduced denticles from each side. Denticles with relatively deep to reduced furrows. Up to nine lateral teeth, bearing up to eight denticles | Massiv, large | Large, rounded | About 10–20 alveolar glands | Moderately long, partially curved | Middle to southern Norway, North Sea, the UK | Present study |
| D. yrjargul | Semi-translucent white to greyish, with characteristic two thin wavy subparallel brownish-orange to brownish lines running from the head nearly to the tail. There are no specimens with brownish basal colour; upper part of the dorsolateral appendages covered with bright golden- yellow to yellowish pigment. Foot bordered with thin brownish orange line | Up to 95 mm (live) | Up to 8, with relatively short primary stalk, strongly branched secondary branches and elongate tertiary branches | Up to 6 | Up to 5 | Up to 30 | Up to 50 rows of teeth. Central (rachidian) tooth moderately denticulated and bearing up to 16distinct denticles. Denticles with deep furrows. Up to 9 lateral teeth, bearing up to 7 denticles | Kidney-shaped, thickened | Large, rounded | Up to 50 alveolar glands | Long, thick, curved | Middle Norway to Kara Sea | Korshunova et al., 2020 [23]; present study |
| Dendronotus kalikal | Brownish, with creamy to whitish areas, dorsum, dorsolateral appendages, and upper sides of foot bear brownish orange lines and dots, partly arranged in blurred subparallel pattern, no golden- yellow pigment on the upper part of dorso-lateral appendages | Up to 15 mm (preserved) | Up to 5 (plus 2–3 smaller), with relatively long primary stalk, moderately branched thin secondary branches | Up to 5 | Up to 5 | Up to 14 mm | Up to 31 rows of teeth. Central tooth with deep furrows and with up to 18 distinct denticles. Up to eight lateral teeth, bearingup to 8 denticles | Moderately voluminous, folded | Large, considerably elongate, narrow ly-oval | Up to circa 20 alveolar glands | Relatively long, slightly curved | Kurile Islands to Bering Strait | Korshunova et al., 2019 [27], 2020 [23] |
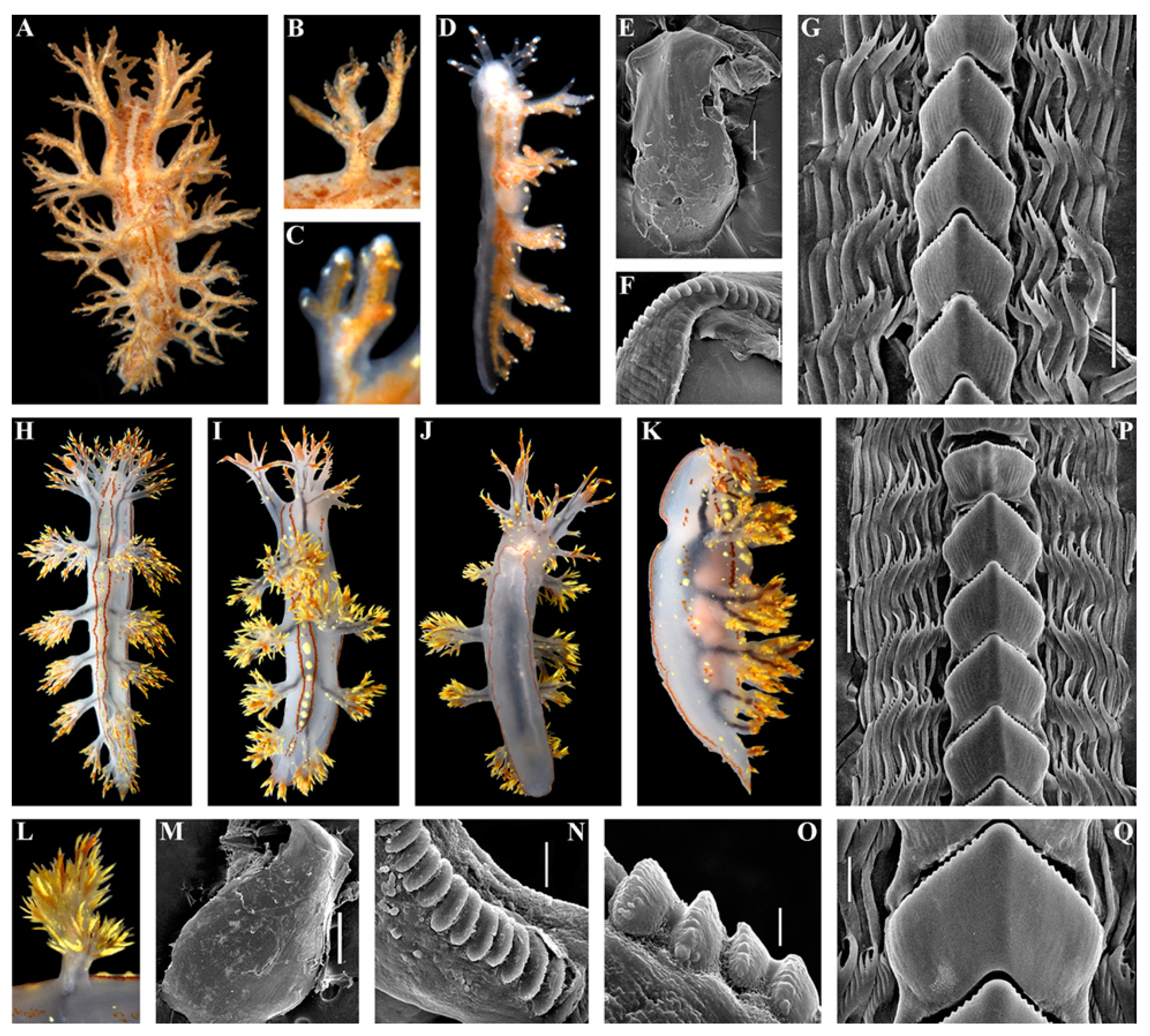
- Material examined
- Diagnosis
- Description
- Distribution and ecological observations
- Comparative remarks
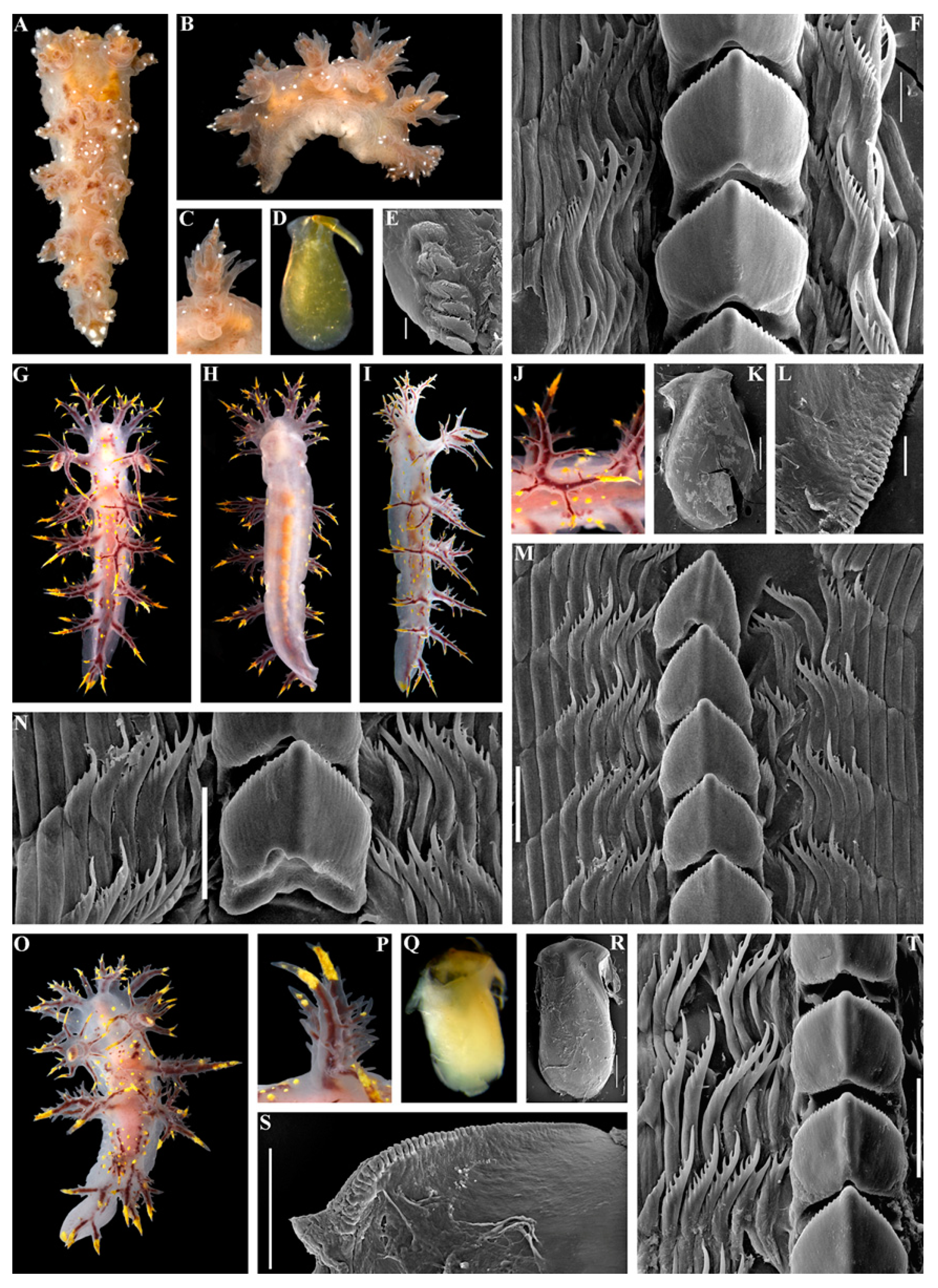
- Etymology
- Material examined
- Diagnosis
- Description
- Distribution and ecological observations
- Comparative remarks
3.2. Molecular Phylogeny

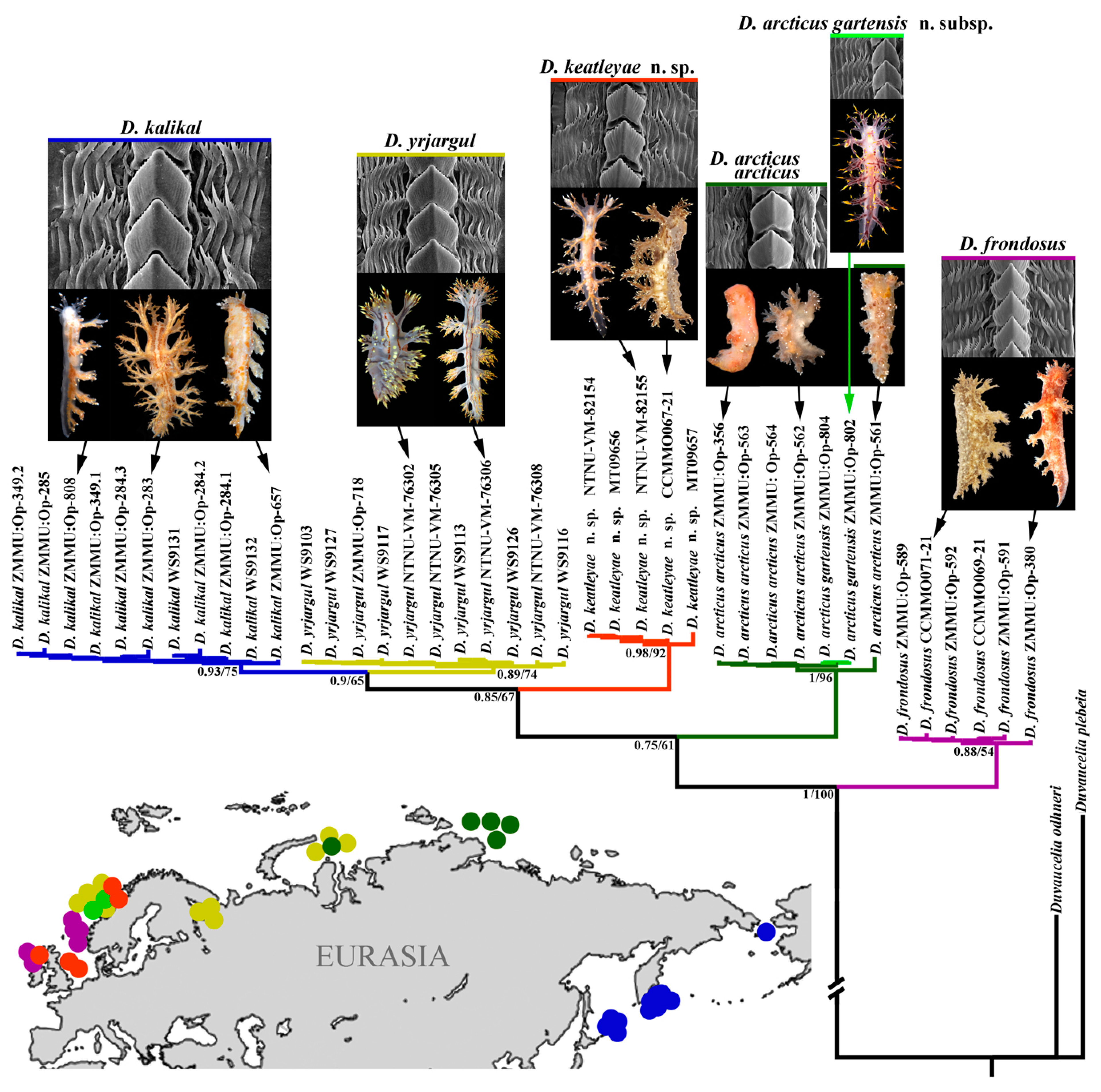
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blaxter, M. Molecular systematics: Counting angels with DNA. Nature 2003, 421, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Le Bagousse-Pinguet, Y.; Soliveres, S.; Gross, N.; Torices, R.; Berdugo, M.; Maestre, F.T. Phylogenetic, functional, and taxonomic richness have both positive and negative effects on ecosystem multifunctionality. Proc. Natl. Acad. Sci. USA 2019, 116, 8419–8424. [Google Scholar] [CrossRef] [PubMed]
- Pascual, U.; Adams, W.M.; Díaz, S.; Lele, S.; Mace, G.M.; Turnhout, E. Biodiversity and the challenge of pluralism. Nat. Sustain. 2021, 4, 567–572. [Google Scholar] [CrossRef]
- Velasco, D.; García-Llorente, M.; Alonsoa, B.; Dolera, A.; Palomo, I.; Iniesta-Arandia, I.; Martín-Lópeza, B. Biodiversity conservation research challenges in the 21st century: A review of publishing trends in 2000 and 2011. Environ. Sci. Policy 2015, 54, 90–96. [Google Scholar] [CrossRef]
- Johnson, C.N.; Balmford, A.; Brook, B.W.; Buettel, J.C.; Galetti, M.; Guangchun, L.; Wilmshurst, J.M. Biodiversity losses and conservation responses in the Anthropocene. Science 2017, 356, 270–275. [Google Scholar] [CrossRef]
- Proença, V.; Martin, L.J.; Pereira, H.M.; Fernandez, M.; McRae, L.; Belnap, J.; Böhm, M.; Brummitt, N.; García-Moreno, J.; Gregory, R.D.; et al. Global biodiversity monitoring: From data sources to essential biodiversity variables. Biol. Conserv. 2017, 213, 256–263. [Google Scholar] [CrossRef]
- Heberling, J.M.; Miller, J.T.; Noesgaard, D.; Weingart, S.B.; Schigel, D. Data integration enables global biodiversity synthesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2018093118. [Google Scholar] [CrossRef]
- Alder, J.; Hancock, A. A Monograph of the British Nudibranchiate Mollusca; Parts I–VII; The Ray Society: London, UK, 1845–1855.
- Odhner, N. Opisthobranchiate Mollusca from the western and northern coasts of Norway. Det Kong. Nor. Vidensk. Selsk. Skrift. 1939, 1, 1–93. [Google Scholar]
- Thompson, T.E.; Brown, G.H. Biology of Opisthobranch Molluscs; The Ray Society Publishing: London, UK, 1984; Volume 2. [Google Scholar]
- Picton, B.E.; Morrow, C. A Field Guide to the Nudibranchs of the British Isles; Immel Publishing: London, UK, 1994. [Google Scholar]
- Evertsen, J.; Bakken, T. Nudibranch diversity (Heterobranchia, Gastropoda) along the coast of Norway. Fauna Nor. 2005, 25, 1–37. [Google Scholar]
- Kienberger, K.; Carmona, L.; Pola, M.; Padula, V.; Gosliner, T.M.; Cervera, J.L. Aeolidia papillosa (Linnaeus, 1761) (Mollusca: Heterobranchia: Nudibranchia), single species or a cryptic species complex? A morphological and molecular study. Zool. J. Linn. Soc. 2016, 177, 481–506. [Google Scholar] [CrossRef]
- Korshunova, T.A.; Martynov, A.V.; Bakken, T.; Evertsen, J.; Fletcher, K.; Mudianta, I.W.; Saito, H.; Lundin, K.; Schrödl, M.; Picton, B. Polyphyly of the traditional family Flabellinidae affects a major group of Nudibranchia: Aeolidacean taxonomic reassessment with descriptions of several new families, genera, and species (Mollusca, Gastropoda). ZooKeys 2017, 717, 1–139. [Google Scholar] [CrossRef]
- Korshunova, T.A.; Lundin, K.; Malmberg, K.; Picton, B.; Martynov, A.V. First true brackish water nudibranch mollusc provides new insights for phylogeny and biogeography. PLoS ONE 2018, 13, e0192177. [Google Scholar] [CrossRef]
- Korshunova, T.; Picton, B.; Furfaro, G.; Mariottini, P.; Pontes, M.; Prkić, J.; Fletcher, K.; Malmberg, K.; Lundin, K.; Martynov, A. Multilevel fine-scale diversity challenges the ‘cryptic species’ concept. Sci. Rep. 2019, 9, 6732. [Google Scholar] [CrossRef] [PubMed]
- Korshunova, T.A.; Sanamyan, N.P.; Sanamyan, K.E.; Torkild, B.; Lundin, K.; Fletcher, K.; Martynov, A.V. Biodiversity hotspot in cold waters: A review of the genus Cuthonella with descriptions of seven new species (Mollusca, Nudibranchia). Contrib. Zool. 2021, 90, 216–283. [Google Scholar] [CrossRef]
- Korshunova, T.; Driessen, F.; Picton, B.; Martynov, A.V. The multilevel organismal diversity approach deciphers difficult to distinguish nudibranch species complex. Sci. Rep. 2021, 11, 18323. [Google Scholar] [CrossRef] [PubMed]
- Thollesson, M. Discrimination of two Dendronotus species by allozyme electrophoresis and the reinstatement of Dendronotus lacteus (Thompson, 1840) (Nudibranchia, Dendronotoidea). Zool. Scr. 1998, 27, 189–195. [Google Scholar] [CrossRef]
- Stout, C.; Pola, M.; Valdés, Á. Phylogenetic analysis of Dendronotus nudibranchs with emphasis on northeastern Pacific species. J. Moll. Stud. 2010, 76, 367–375. [Google Scholar] [CrossRef]
- Korshunova, T.A.; Sanamyan, N.; Martynov, A.V. Morphological and molecular evidence indicate Dendronotus primorjensis is a valid species that has priority over D. dudkai (Nudibranchia). ZooKeys 2016, 634, 15–28. [Google Scholar]
- Korshunova, T.A.; Sanamyan, N.; Zimina, O.; Fletcher, K.; Martynov, A.V. Two new species and a remarkable record of the genus Dendronotus from the North Pacific and Arctic oceans (Nudibranchia). ZooKeys 2016, 630, 19–42. [Google Scholar]
- Korshunova, T.A.; Bakken, T.; Grøtan, V.; Johnson, K.; Lundin, K.; Martynov, A.V. A synoptic review of the family Dendronotidae (Mollusca: Nudibranchia): A multilevel organismal diversity approach. Contr. Zool. 2020, 90, 93–153. [Google Scholar] [CrossRef]
- Martynov, A.V.; Fujiwara, Y.; Tsuchida, S.; Nakano, R.; Sanamyan, N.; Sanamyan, K.; Fletcher, K.; Korshunova, T.A. Three new species of the genus Dendronotus from Japan and Russia (Mollusca, Nudibranchia). Zootaxa 2020, 4747, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Korshunova, T.A.; Martynov, A.V.; Bakken, T.; Picton, B. External diversity is restrained by internal conservatism: New nudibranch mollusc contributes to the cryptic species problem. Zool. Scr. 2017, 46, 683–692. [Google Scholar] [CrossRef]
- Chandler, M.; See, L.; Copas, K.; Bonde, A.M.Z.; López, B.C.; Danielsen, F.; Legind, J.K.; Masinde, S.; Miller-Rushing, A.J.; Newman, G.; et al. Contribution of citizen science towards international biodiversity monitoring. Biol. Cons. 2017, 213, 280–294. [Google Scholar] [CrossRef]
- Korshunova, T.; Nakano, R.; Fletcher, K.; Sanamyan, N.; Martynov, A. First molecular confirmation of the presence of Dendronotus primorjensis Martynov, Sanamyan & Korshunova, 2015 in Japan and new distributional records of Dendronotus species in the North Pacific (Nudibranchia: Dendronotidae). Venus 2019, 77, 1–14. [Google Scholar]
- Díaz, S.; Malhi, Y. Biodiversity: Concepts, patterns, trends, and perspectives. Annu. Rev. Environ. Resour. 2022, 47, 31–63. [Google Scholar] [CrossRef]
- Ehrich, S.; Adlerstein, S.; Brockmann, U.; Floeter, J.; Garthe, S.; Hinz, H.; Kröncke, I.; Neumann, H.; Reiss, H.; Sell, A.F.; et al. 20 years of the German small-scale bottom trawl survey (GSBTS): A review. Senckenberg. Marit. 2007, 37, 13–82. [Google Scholar] [CrossRef]
- Barco, A.; Raupach, M.J.; Laakmann, S.; Neumann, H.; Knebelsberger, T. Identification of North Sea molluscs with DNA barcoding. Mol. Ecol. Resour. 2016, 16, 288–297. [Google Scholar] [CrossRef]
- Bakken, T.; Hårsaker, K.; Daverdin, M. Marine Invertebrate Collection NTNU University Museum; Version 1.574; NTNU University Museum: Trondheim, Norway, 2023. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 1994, 3, 294–299. [Google Scholar]
- Palumbi, S.R.; Martin, A.P.; Romano, S.; McMillan, W.O.; Stice, L.; Grabowski, G. The Simple Fool’s Guide to PCR; University of Hawaii: Honolulu, HI, USA, 2002. [Google Scholar]
- Puslednik, L.; Serb, J.M. Molecular phylogenetics of the Pectinidae (Mollusca: Bivalvia) and effect of increased taxon sampling and outgroup selection on tree topology. Mol. Phylogenet. Evol. 2008, 48, 1178–1188. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl. Acid Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis Version 11. Mol. Biol. Evol. 2012, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2011, 21, 1864–1877. [Google Scholar] [CrossRef]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef]
- Pons, J.; Barraclough, T.; Gomez-Zurita, J.; Cardoso, A.; Duran, D.; Hazell, S.; Kamoun, S.; Sumlin, W.D.; Vogler, A.P. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 2006, 55, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Kapli, P.; Lutteropp, S.; Zhang, J.; Kobert, K.; Pavlidis, P.; Stamatakis, A.; Flouri, T. Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics 2017, 33, 1630–1638. [Google Scholar] [CrossRef] [PubMed]
- Martynov, A.V.; Korshunova, T.A. Multilevel organismal diversity in an ontogenetic framework as a solution for the species concept. In Cryptic Species: Morphological Stasis, Circumscription, and Hidden Diversity; Monro, A.K., Mayo, S.J., Eds.; Cambridge University Press: Cambridge, UK, 2022; pp. 78–129. [Google Scholar]
- ICZN. International Code of Zoological Nomenclature; The International Trust for Zoological Nomenclature: London, UK, 1999. [Google Scholar]
- Martynov, A.V.; Korshunova, T.A.; Savinkin, O.V. Shallow-water opisthobranch molluscs of the Murman coast of the Barents Sea, with new distributional data and remarks on biology. Ruthenica 2006, 16, 59–72. [Google Scholar]
- Martynov, A.V.; Korshunova, T.A. Opisthobranch Molluscs of the Seas of Russia. A Colour Guide to Their Taxonomy and Biology; Fiton: Moscow, Russia, 2011. [Google Scholar]
- Kluge, A. Total evidence or taxonomic congruence: Cladistics or consensus classification. Cladistics 1998, 14, 151–158. [Google Scholar] [CrossRef]
- Gómez Daglio, L.; Dawson, M.N. Integrative taxonomy: Ghosts of past, present and future. J. Mar. Biol. Assoc. UK 2019, 99, 1237–1246. [Google Scholar] [CrossRef]
- Dayrat, B. Towards integrative taxonomy. Biol. J. Linn. Soc. 2005, 85, 407–415. [Google Scholar] [CrossRef]
- Padial, J.M.; Miralles, A.; De La Riva, I.; Vences, M. The integrative future of taxonomy. Front. Zool. 2010, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Schlick-Steiner, B.C.; Steiner, F.M.; Seifert, B.; Stauffer, C.; Christian, E.; Crozier, R.J. Integrative taxonomy: A multisource approach to exploring biodiversity. Ann. Rev. Entomol. 2010, 55, 421–438. [Google Scholar] [CrossRef]
- Heethoff, M.; Laumann, M.; Weigmann, G.; Raspotnig, G. Integrative taxonomy: Combining morphological, molecular and chemical data for species delineation in the parthenogenetic Trhypochthonius tectorum complex (Acari, Oribatida, Trhypochthoniidae). Front. Zool. 2011, 8, 2. [Google Scholar] [CrossRef]
- Fritts-Penniman, A.L.; Gosliner, T.M.; Ngurah Mahardika, G.; Barber, P.H. Cryptic ecological and geographic diversification in coral associated nudibranchs. Mol. Phyl. Evol. 2020, 144, 106698. [Google Scholar] [CrossRef]
- Mason, N.A.; Taylor, S.A. Differentially expressed genes match bill morphology and plumage despite largely undifferentiated genomes in a holarctic songbird. Mol. Ecol. 2015, 24, 3009–3025. [Google Scholar] [CrossRef]
- Perez, M.; Lehner, B. Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol. 2019, 21, 143–151. [Google Scholar] [CrossRef]
- Danchin, E.; Pocheville, A.; Rey, O.; Pujol, B.; Blanchet, S. Epigenetically facilitated mutational assimilation: Epigenetics as a hub within the inclusive evolutionary synthesis. Biol. Rev. 2019, 94, 259–282. [Google Scholar] [CrossRef]
- Cerca, J.; Meyer, C.; Purschke, G.; Struck, T.H. Delimitation of cryptic species drastically reduces the geographical ranges of marine interstitial ghost-worms (Stygocapitella, Annelida, Sedentaria). Mol. Phylogen.Evol. 2020, 143, 106663. [Google Scholar] [CrossRef]
- Halasan, L.C.; Lin, H.C. Integrated morphometrics reveals conservatism in the cryptic yellowstripe scad (Perciformes: Carangidae) lineages from the Tropical Western Pacific. Zool. Anz. 2022, 300, 82–91. [Google Scholar] [CrossRef]
- Slager, D.L.; Epperly, K.L.; Ha, R.R.; Rohwer, S.; Wood, C.; Van Hemert, C.; Klicka, J. Cryptic and extensive hybridization between ancient lineages of American crows. Mol. Ecol. 2022, 29, 956–969. [Google Scholar] [CrossRef]
- Karanovic, T.; Djurakic, M.; Eberhard, S.M. Cryptic species or inadequate taxonomy? Implementation of 2D geometric morphometrics based on integumental organs as landmarks for delimitation and description of copepod taxa. Syst. Biol. 2016, 65, 304–327. [Google Scholar] [CrossRef] [PubMed]
- Karanovic, T. Using landmark-based geometric morphometrics for holotype selection in cryptic species: A case study of Western Australian Halicyclops (Copepoda, Cyclopoida). Crustaceana 2022, 95, 631–666. [Google Scholar] [CrossRef]
- Horsáková, V.; Nekola, J.C.; Horsák, M. When is a ‘cryptic’ species not a cryptic species: A consideration from the Holarctic micro-land snail genus Euconulus (Gastropoda: Stylommatophora). Mol. Phylogen. Evol. 2019, 132, 307–320. [Google Scholar] [CrossRef]
- Chou, M.H.; Chu, I.H.; Lau, D.; Huang, J.P. Integrative species delimitation reveals fine-scale allopatric speciation in a good-flying insect: A case study on Cylindera pseudocylindriformis complex (Coleoptera, Cicindelidae). Invert. Syst. 2022, 36, 910–925. [Google Scholar] [CrossRef]
- Gibert, A.; Louty, F.; Buscail, R.; Baguette, M.; Schatz, B.; Bertrand, J.A.M. Extracting quantitative information from images taken in the wild: A case study of two vicariants of the Ophrys aveyronensis species complex. Diversity 2022, 14, 400. [Google Scholar] [CrossRef]
- Pyron, R.A.; Beamer, D.A. Nomenclatural solutions for diagnosing ‘cryptic’ species using molecular and morphological data facilitate a taxonomic revision of the black-bellied salamanders (Urodela, Desmognathus ‘quadramaculatus’) from the southern Appalachian Mountains. Bionomina 2022, 27, 1–43. [Google Scholar] [CrossRef]
- Jirapatrasilp, P.; Backeljau, T.; Prasankok, P.; Ratmanee, C.; Panhab, S. Untangling a mess of worms: Species delimitations reveal morphological crypsis and variability in Southeast Asian semi-aquatic earthworms. Mol. Phyl. Evol. 2019, 139, 106531. [Google Scholar] [CrossRef]
- Hupalo, K.; Copilaș-Ciocianu, D.; Leese, F.; Weiss, M. COI is not always right: Integrative taxonomy reveals striking overestimation of species diversity in a Mediterranean freshwater amphipod. Res. Square 2022, preprint. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-based registry for all animal species: The Barcode Index Number (BIN) system. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef] [PubMed]
- Kijewski, T.K.; Zbawicka, M.; Väinölä, R.; Wenne, R. Introgression and mitochondrial DNA heteroplasmy in the Baltic populations of mussels Mytilus trossulus and M. edulis. Mar. Biol. 2006, 149, 1371–1385. [Google Scholar] [CrossRef]
- Malinsky, M.; Svardal, H.; Tyers, A.; Miska, E.A.; Genner, M.J.; Turner, G.F.; Durbin, R. Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nat. Ecol. Evol. 2018, 2, 1940–1955. [Google Scholar] [CrossRef]
- Flury, J.M.; Haas, A.; Brown, R.M.; Das, I.; Pui, Y.M.; Boon-Hee, K.; Scheidt, U.; Iskandar, D.T.; Jankowski, A.; Hertwig, S.T. Unexpectedly high levels of lineage diversity in Sundaland puddle frogs (Dicroglossidae: Occidozyga Kuhl and van Hasselt, 1822). Mol. Phylogenet. Evol. 2021, 163, 107210. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.O.; Hutter, C.R.; Perry, L.; Wood, J.R.; Su, Y.C.; Brown, R.M. Gene flow increases phylogenetic structure and inflates cryptic species estimations: A case study on widespread Philippine puddle frogs (Occidozyga laevis). Syst. Biol. 2022, 71, 40–57. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.C.; Rossie, J.B. Congruence of molecules and morphology using a narrow allometric approach. Proc. Natl. Acad. Sci. USA 2007, 104, 11910–11914. [Google Scholar] [CrossRef]
- Phillips, J.D.; Gillis, D.J.; Hanner, R.H. Incomplete estimates of genetic diversity within species: Implications for DNA barcoding. Ecol. Evol. 2019, 9, 2996–3010. [Google Scholar] [CrossRef]
- Luo, A.; Ling, C.; Ho, S.Y.W.; Zhu, C. Comparison of methods for molecular species delimitation across a range of speciation scenarios. Syst. Biol. 2018, 67, 830–846. [Google Scholar] [CrossRef]
- Hofmann, E.P.; Nicholson, K.E.; Luque-Montes, I.R.; Köhler, G.; Cerrato-Mendoza, C.A.; Medina-Flores, M.; Wilson, L.D.; Townsend, J.H. Cryptic diversity, but to what extent? Discordance between single-locus species delimitation methods within mainland anoles (Squamata: Dactyloidae) of northern Central America. Front. Genet. 2019, 10, 11. [Google Scholar] [CrossRef]
- Talavera, G.; Dinca, V.; Vila, R. Factors affecting species delimitations with the GMYC model: Insights from a butterfly survey. Methods Ecol. Evol. 2013, 4, 1101–1110. [Google Scholar] [CrossRef]
- Knowlton, N. Sibling species in the sea. Ann. Rev. Ecol. Syst. 1993, 24, 189–216. [Google Scholar] [CrossRef]
- Lindsay, T.; Valdés, Á. The model organism Hermissenda crassicornis (Gastropoda: Heterobranchia) is a species complex. PLoS ONE 2016, 11, e0154265. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, S.B.; Gosliner, T.M. Glossing over cryptic species: Descriptions of four new species of Glossodoris and three new species of Doriprismatica (Nudibranchia: Chromodorididae). Zootaxa 2018, 4444, 501–529. [Google Scholar] [CrossRef]
- Costa-Araújo, R.; Silva-Jr., J.S.; Boubli, J.P.; Rossi, R.V.; Canale, G.R.; Melo, F.R.; Bertuol, F.; Silva, F.E.; Silva, D.A.; Nash, S.D.; et al. An integrative analysis uncovers a new, pseudo-cryptic species of Amazonian marmoset (Primates: Callitrichidae: Mico) from the arc of deforestation. Sci. Rep. 2021, 11, 15665. [Google Scholar] [CrossRef]
- Magpali, L.; Machado, D.R.; Araújo, T.Q.; Garraffoni, A.R. Long distance dispersal and pseudo-cryptic species in Gastrotricha: First description of a new species (Chaetonotida, Chaetonotidae, Polymerurus) from an oceanic island with volcanic rocks. Eur. J. Taxon. 2021, 746, 62–93. [Google Scholar] [CrossRef]
- Cabezas, M.P.; Lasso-Alcalá, O.M.; Quintero, T.E.; Xavier, R.; Giarrizzo, T.; Nunes, J.L.S.; Machado, F.S.; Gómez, J.; Pedroza, W.S.; Jowers, M.J. Clarifying the taxonomy of some cryptic blennies (Blenniidae) in their native and introduced range. Sci. Rep. 2022, 12, 9514. [Google Scholar] [CrossRef] [PubMed]
- Hofman, S.; Cameron, R.A.D.; Proćków, M.; Sîrbu, I.; Osikowski, A.; Jaszczyńska, A.; Sokól, M.; Falniowski, A. Two new pseudocryptic species in the medium-sized common European land snails, Fruticicola Held, 1838; as a result of phylogeographic analysis of Fruticicola fruticum (O. F. Müller, 1774) (Gastropoda: Helicoidea: Camaenidae). Mol. Phyl. Evol. 2022, 168, 107402. [Google Scholar] [CrossRef]
- Paz-Sedano, S.; Álvarez, J.F.M.; Gosliner, T.M.; Pola, M. Reassessing North Eastern Atlantic-Mediterranean species of Trapania (Mollusca, Nudibranchia). Zool. Scr. 2022, 51, 447–459. [Google Scholar] [CrossRef]
- Smith, S.M.; Urvois, T.; Roques, A.; Cognato, A.I. Recognition of the pseudocryptic species Xylosandrus declivigranulatus (Schedl) as distinct from Xylosandrus crassiusculus (Motschulsky) (Coleoptera: Curculionidae: Scolytinae: Xyleborini). Coleopt. Bull. 2022, 76, 367–374. [Google Scholar] [CrossRef]
- Heethoff, M. Cryptic species: Conceptual or terminological chaos? Trends Ecol. Evol. 2018, 33, 310. [Google Scholar] [CrossRef]
- Monro, A.K.; Mayo, S.J. (Eds.) Cryptic Species: Morphological Stasis, Circumscription, and Hidden Diversity; Cambridge University Press: Cambridge, UK, 2022. [Google Scholar]
- Fraga, M.F.; Ballestar, E.; Paz, M.F.; Ropero, S.; Setien, F.; Ballestar, M.L.; Heine-Suñer, D.; Cigudosa, J.C.; Urioste, M.; Benitez, J.; et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. USA 2005, 26, 10604–10609. [Google Scholar] [CrossRef] [PubMed]
- Bierbach, D.; Laskowski, K.; Wolf, M. Behavioural individuality in clonal fish arises despite near-identical rearing conditions. Nat. Comm. 2017, 8, 15361. [Google Scholar] [CrossRef] [PubMed]
- Sáez, A.G.; Probert, I.; Geisen, M.; Quinn, P.; Young, J.R.; Medlin, L.K. Pseudo-cryptic speciation in coccolithophores. Proc. Natl. Acad. Sci. USA 2003, 100, 7163–7168. [Google Scholar] [CrossRef] [PubMed]

| Species Name | Voucher | Locality | COI | 16S |
|---|---|---|---|---|
| Dendronotus arcticus arcticus | ZMMU: Op-561 | Russia: Laptev Sea | KX788140 | KX788129 |
| D. arcticus arcticus | ZMMU: Op-562 | Russia: Laptev Sea | KX788141 | KX788130 |
| D. arcticus arcticus | ZMMU: Op-563 | Russia: Laptev Sea | KX788142 | KX788131 |
| D. arcticus arcticus | ZMMU: Op-564 | Russia: Laptev Sea | OQ588822 | OQ594740 |
| D. arcticus arcticus | ZMMU: Op-356 (the correct voucher number) | Russia: Kara Sea | KP984995 | KP984989 |
| D. arcticus gartensis n. subsp. | ZMMU: Op-804 | Norway | OQ588823 | OQ594741 |
| D. arcticus gartensis n. subsp. | ZMMU: Op-802 | Norway | OQ588824 | OQ594742 |
| D. frondosus | ZMMU: Op-380 | Norway: Gulen | KM396976 | KM397056 |
| D. frondosus | ZMMU: Op-589 | Norway: Gulen | KY391833 | KY391853 |
| D. frondosus | ZMMU: Op-591 | Norway: Gulen | KY391835 | KY391855 |
| D. frondosus | ZMMU: Op-592 | Norway: Gulen | KY391836 | KY391856 |
| D. frondosus | CCMMO069-21 | UK: N. Ireland | OQ588829 | - |
| D. frondosus | CCMMO071-21 | UK: N. Ireland | OQ588830 | - |
| D. kalikal | ZMMU: Op-284.1 | Russia: Kamchatka | KC660026 | KC611285 |
| D. kalikal | ZMMU: Op-284.2 | Russia: Kamchatka | KC660025 | KC611286 |
| D. kalikal | ZMMU: Op-284.3 | Russia: Kamchatka | KM396988 | KM397070 |
| D. kalikal | ZMMU: Op-283 | Russia: Kamchatka | KC660024 | KC611284 |
| D. kalikal | ZMMU: Op-285 | Russia: Bering Strait | KC660027 | KC611287 |
| D. kalikal | ZMMU: Op-349.1 | Russia: Kamchatka | KM396986 | KM397068 |
| D. kalikal | ZMMU: Op-349.2 | Russia: Kamchatka | KM396987 | KM397069 |
| D. kalikal | ZMMU: Op-657 | Russia: Kuril Islands | MK302458 | MK302453 |
| D. kalikal | WS9131 | Russia: Kuril Islands | MN138314 | MN138079 |
| D. kalikal | WS9132 | Russia: Kuril Islands | MN138315 | MN138080 |
| D. kalikal | ZMMU: Op-808 | Russia: Kuril Islands | OQ588825 | OQ594743 |
| D. yrjargul | ZMMU: Op-718 | Norway: Norwegian Sea | MT654641 | MT655313 |
| D. yrjargul | NTNU-VM-76302 | Norway: Norwegian Sea | MT654642 | MT655314 |
| D. yrjargul | NTNU-VM-76306 | Norway: Norwegian Sea | MT654643 | MT655315 |
| D. yrjargul | NTNU-VM-76308 | Norway: Norwegian Sea | MT654644 | MT655316 |
| D. yrjargul | NTNU-VM-76305 | Norway: Norwegian Sea | MT654645 | MT655317 |
| D. yrjargul | WS9116 | Russia: Kara Sea | MN138317 | MN138082 |
| D. yrjargul | WS9113 | Russia: Kara Sea | MN138316 | MN138081 |
| D. yrjargul | WS9117 | Russia: Kara Sea | MN138318 | MN138083 |
| D. yrjargul | WS9103 | Russia: White Sea | MN138311 | MN138076 |
| D. yrjargul | WS9126 | Russia: White Sea | MN138312 | MN138077 |
| D. yrjargul | WS9127 | Russia: White Sea | MN138313 | MN138078 |
| D. keatleyae n. sp. | NTNU-VM-82154 | Norway | OQ588826 | OQ594744 |
| D. keatleyae n. sp. | NTNU-VM-82155 | Norway | OQ588827 | OQ594745 |
| D. keatleyae n. sp. | CCMMO067-21 | UK: N. Ireland | OQ588828 | - |
| D. keatleyae n. sp. | MT09656 | North Sea | KR084744 | - |
| D. keatleyae n. sp. | MT09657 | North Sea | KR084934 | - |
| Duvaucelia plebeia | ZMMU: Op-572 | Norway | KX788134 | KX788122 |
| Duvaucelia odhneri | CASIZ: 176219 | South Africa | HM162716 | HM162641 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korshunova, T.; Grøtan, V.V.; Johnson, K.B.; Bakken, T.; Picton, B.E.; Martynov, A. Similar Ones Are Not Related and Vice Versa—New Dendronotus Taxa (Nudibranchia: Dendronotidae) from the North Atlantic Ocean Provide a Platform for Discussion of Global Marine Biodiversity Patterns. Diversity 2023, 15, 504. https://doi.org/10.3390/d15040504
Korshunova T, Grøtan VV, Johnson KB, Bakken T, Picton BE, Martynov A. Similar Ones Are Not Related and Vice Versa—New Dendronotus Taxa (Nudibranchia: Dendronotidae) from the North Atlantic Ocean Provide a Platform for Discussion of Global Marine Biodiversity Patterns. Diversity. 2023; 15(4):504. https://doi.org/10.3390/d15040504
Chicago/Turabian StyleKorshunova, Tatiana, Viktor V. Grøtan, Kjetil B. Johnson, Torkild Bakken, Bernard E. Picton, and Alexander Martynov. 2023. "Similar Ones Are Not Related and Vice Versa—New Dendronotus Taxa (Nudibranchia: Dendronotidae) from the North Atlantic Ocean Provide a Platform for Discussion of Global Marine Biodiversity Patterns" Diversity 15, no. 4: 504. https://doi.org/10.3390/d15040504
APA StyleKorshunova, T., Grøtan, V. V., Johnson, K. B., Bakken, T., Picton, B. E., & Martynov, A. (2023). Similar Ones Are Not Related and Vice Versa—New Dendronotus Taxa (Nudibranchia: Dendronotidae) from the North Atlantic Ocean Provide a Platform for Discussion of Global Marine Biodiversity Patterns. Diversity, 15(4), 504. https://doi.org/10.3390/d15040504









