Abstract
The planktonic diatom genus Pseudo-nitzschia contains several genetically closely related species that can produce domoic acid, a potent neurotoxin known to cause amnesic shellfish poisoning (ASP). An early identification and an adequate monitoring of the potential toxic Pseudo-nitzschia spp. are necessary. However, effective monitoring programs are time consuming due, in some cases, to the cell morphology similarities among species, determined with light microscopy, that can result in insufficient data to give a definitive species and toxins attribution. In this paper, Whole-Cell Fluorescent In Situ Hybridization (WC-FISH) has been evaluated as a powerful tool to detect and enumerate harmful cryptic and/or pseudo-cryptic Pseudo-nitzschia spp. collected in the Gulf of Naples. Fluorescently labelled probes directed against the ribosomal RNA (rRNA) of the 28S large subunit (LSU) were used. In particular, five probes detecting four cryptic species of Pseudo-nitzschia delicatissima complex and one specific for Pseudo-nitzschia multistriata gave good results for the molecular identification of potentially toxic target species in natural samples. Finally, we can state that the WC-FISH method, to identify Pseudo-nitzschia species, is faster and more cost-effective if compared with other rDNA-based methods.
1. Introduction
The monitoring of the Pseudo-nitzschia spp. is of fundamental importance because of the worldwide distribution of these marine planktonic diatoms producing harmful compounds. Among toxins, domoic acid (DA) is produced by the Pseudo-nitzschia spp. under certain conditions [1] and it can bioaccumulate along the food chain causing damage to mammals and human health with symptoms of different nature, such as ataxia, head weaving, muscle tremor, titanic convulsions, rubbing, and lethargy [2,3,4,5,6].
Multiple toxigenic Pseudo-nitzschia species frequently coexist in the same environment, even during bloom events that appear to be dominated by a single species [7,8,9].
Since the genus Pseudo-nitzschia includes many species, their accurate taxonomic identification is important since they can be associated with domoic acid production [10].
Unfortunately, all Pseudo-nitzschia species have a very similar gross morphology, which makes them difficult and often impossible to identify at the species level with light microscopy. Although species determination, in some cases, can be carried out with electron microscopy, which allows for the observation of the main diagnostic characters, such as the presence/absence of the central larger interspace or the number and structure of fibulae, striae, and poroids, molecular approaches over time have become an increasingly important supplement, constituting as an essential tool for the identification of the several cryptic/pseudocryptic species now belonging to this genus [11,12].
Indeed, new cryptic and pseudo-cryptic species have recently been described within the P. pseudodelicatissima [10,13,14] and P. delicatissima complex [15,16,17].
More recent molecular approaches, such as qPCR, ARISA, microarray, and dot blot hybridization systems have been used for specific and sensitive Pseudo-nitzschia species identification and/or quantification from clonal cultures [18,19,20,21]. Molecular methods are essentially based on the evaluation of the sequence variation of oligonucleotide primers and/or probes in target nucleotide regions and allow the accurate identification of various phytoplanktonic toxic species [22,23,24].
In this paper, Fluorescence In Situ Hybridisation (FISH) has been evaluated as a powerful tool to detect and enumerate harmful microorganisms in the marine environment. Different FISH methods are available and, especially in combination with automated counting techniques and the development and updating of oligonucleotide probes able to discriminate toxic species, can be attainable for routine monitoring of harmful marine microalgae [25,26,27,28,29,30,31,32]. However, FISH-based methods are not yet regularly included in monitoring programs tracking the presence of harmful marine microalgae. A limitation factor of the FISH technique is the currently available number of suited fluorochromes attached to the FISH probes to detect various harmful species in one environmental sample at the same time. However, coupled automated techniques, like biosensors, microarrays, and quantitative polymerase chain reaction (qPCR) can facilitate the analysis of numerous field samples and help to overcome this drawback [33]. A great benefit of FISH compared to other molecular detection methods of harmful algal blooms is the direct visualisation of the hybridised target cells (whole cell FISH), which are not allowed in cell free formats, as DNA-dependent analysis methods [34].
In this study, we applied the Whole-Cell Fluorescent In Situ Hybridization (WC-FISH) on cryptic and/or pseudo-cryptic species of the genus Pseudo-nitzschia collected in the Gulf of Naples. We used fluorescently labelled probes directed against the ribosomal RNA (rRNA) of the 28S large subunit (LSU). Primers designed on DNA ribosomal large subunit (LSU), targeted on its D1 hyper-variable region, identified the most Pseudo-nitzschia species [35]. Oligoprobes, designed on LSU hypervariable region of 28S, of rDNA developed to build up the microarray for toxic Pseudo-nitzschia spp. detection [21] have been used in this study to reveal Pseudo-nitzschia spp. in natural samples through WC-FISH. The work was carried out to obtain a semi-quantitative estimation of the presence of potentially toxic Pseudo-nitzschia cryptic/pseudo-cryptic species in environmental samples by applying the WC-FISH.
2. Materials and Methods
2.1. Pseudo-nitzschia Cultivation
Experiments were carried out with selected strains of Pseudo-nitzschia species established from single chains of cells, isolated at the Long Term Ecological Research Station MareChiara (LTER-MC, 40°48.5′ N, 14°15′ E) in the Gulf of Naples. Nine different species of Pseudo-nitzschia were used: four belonging to the P. delicatissima complex (P. allochrona, P. arenysensis, P. delicatissima, P. dolorosa) and two belonging to the P. pseudodelicatissima complex (P. calliantha, P. pseudodelicatissima) together with non-cryptic P. fraudulenta, P. galaxiae, and P. multistriata. The reason for choosing the strains selected is to develop a method for discriminating between toxic and non-toxic species for a person with no training in the genus Pseudo-nitzschia. In addition, the reason was to also identify among the species belonging to the same complex the so-called cryptic or pseudo-cryptic ones, which often include both toxic and non-toxic species (as in the case of P. delicatissima complex, in which P. delicatissima is toxic while the cryptic P. arenysensis and P. allochrona are not toxic).
All Pseudo-nitzschia species were grown in 50 mL flasks containing 35 mL Guillard’s f/2 medium [36] at 22 °C on a 16:8 h light:dark cycle at an irradiance of 100 μmol photons m−2 s−1. Growth of each species was monitored every 24 h by sedimentation of 1 mL of culture in Sedgewick Rafter Counting Chamber [37] and counted using a Leica DMLD inverted microscope. Pseudo-nitzschia cells at the mid/end of the exponential phase were used for further in situ hybridization experiments.
2.2. Whole-Cell Fluorescent In Situ Hybridization (WC-FISH) on Pseudo-nitzschia Species Monocultures—Probes Testing and Cross-Reactivity Assays
The WC-FISH protocol used in this work was optimised starting from the methods of Miller and Scholin (1998, 2000) [25,38] and Groben and Medlin (2005) [39] with some minor modifications. An aliquot (25 mL) of each culture at mid-exponential phase was filtered on 0.8 μm isopore polycarbonate membrane filters (MilliporeTM, Darmstadt, Germany) using a vacuum filtration apparatus. The filters were incubated at 4 °C for 2 h in a modified saline–ethanol fixative freshly prepared by mixing 22 mL 95% ethanol, 5 mL deionized H2O, and 3 mL 25× SET buffer (3.75 M NaCl, 25 mM EDTA, 0.5 M Tris HCl, at pH 7.8) [38]. Then, the fixative solution was gently vacuum removed (~100 mmHg), and filters were immediately processed for WC-FISH; alternatively, they were stored at −20 °C [38]. For the pre-hybridization step, the filters were incubated in 5× SET buffer for 5 min at RT and treated with dimetil-formamide (DMF, 50%) for 1 h at RT to remove the chlorophyll autofluorescence and washed with 5× SET buffer for 5 min at RT. Then, each filter was cut into 12 sections, and placed on a 24-well plate minibasket (Thermo ScientificTM, Rockford, IL, USA) in hybridization buffer (5× SET buffer, 0.1% v/v IGEPAL-CA630-octylphenoxypolyethoxyethanol, 30 μg mL−1 poly(A), 40% formamide) containing 50 ng μL−1 of the specific probes; then, the filters were incubated at 45 °C for 2 h. During initial testing, cultures were hybridized in separate reactions by using their probes labelled with fluorescein 5-isothiocyanate (FITC) at the 5′ end (Thermo ScientificTM, Rockford, IL, USA). The probes tested and the targeted species are reported in Table 1.

Table 1.
Oligonucleotide probes used for Pseudo-nitzschia WC-FISH. The table shows the complex species, name probe, sequence, target species, target genes, and melting temperature (Tm).
Nine probes tested in dot blot assays for their specificity species were designed to target the large subunit (LSU, 28S) ribosomal DNA of Pseudo-nitzschia species [41]. The ARB (from 2005), now SILVA, database alignment was screened for signature positions for the nine species using the “probe design” function of the program package ARB [42]. The specificity of the potential probes was then tested in silico in ARB and by BLAST searches. The probes were examined for hairpin loops and primer dimer formation using the software Oligo 5 (http://www.oligo.net (accessed on 15 May 2012)). The position of probes in the RNA’s secondary structure was checked within ARB [41]. The samples were also probed with the positive control probe Uni-C (directed toward universally conserved eukaryotic sequences of 18S rRNA), the negative control probe Uni-R (directed toward universally conserved prokaryotic sequences of 16S rRNA) [38], and hybridization without probe. After the hybridization, the unbound probe was removed with post-hybridization washes with 5× SET buffer; concentration of washing buffer, temperature, and washing time were optimised to obtain the most intense and specific fluorescence signal of the probe compared with positive control (Uni-C). Images of cells post-hybridization were captured using a Leica DMRB fluorescence microscopy and Leica camera system at 100× after mounting the filter pieces on slides with Citifluor Antifade Mountant (Thermo Fisher Rockford, IL, USA)/DAPI (1 µg mL−1) 2:1 (v/v). In order to identify probes’ cross-reactivity, each probe was also hybridised against all non-target species in the same condition established in the preliminary probe tests. After the hybridization, the stringency of the post-hybridization washing (i.e., concentration of 5× SET washing buffer and washing temperature) were strictly optimised in order to remove the nonspecific bounds and to avoid the cross-reaction problems.
2.3. WC-FISH on Artificial Samples
WC-FISH test was applied to artificial samples consisting of both unialgal cultures and mixed cultures (simulated field samples). For the unialgal culture samples, 25 mL of monoculture at mid-exponential phase of the cryptic species belonging to the P. delicatissima complex (P. allochrona, P. arenysensis, P. delicatissima) and P. multistriata were respectively filtered and hybridised using the above established protocol. The simulated field samples were prepared by adding 25 mL of the cryptic monoculture P. arenysensis to 25 mL of P. multistriata and also by adding 25 mL of P. allochrona at the previous strain mix. All the monocultures were utilised at a mid-exponential phase. Then, the two different simulated field samples were filtered and hybridised as the above established protocol. The relative cell number of both single monocultures and the simulated field samples was estimated with the Utermöhl method in light microscopy. Counting was carried out at 400× magnification in random visual fields to count a minimum of 500 cells, and the cell density was expressed as mean ± standard error (cells mL−1) (n = 3). After the WC-FISH, the labelled fluorescent cells were counted with a Leica DMRB fluorescence microscopy viewing the entire surface of the filter, and the value (cells mL−1) was expressed as mean ± standard error (n = 3). For the different samples, the detection efficiency (%) of the WC-FISH test was evaluated as the ratio between the cell density detected by the epifluorescence microscope after WC-FISH vs. Utermöhl counting method by light microscope before WC-FISH.
2.4. WC-FISH on Environmental Samples
The screened probes were then tested using field samples from the site Long-Term Ecological Research station MareChiara (40°48.5′ N, 14°15′ E) in the Gulf of Naples (Tyrrhenian Sea) that were routinely collected as part of a long-term ongoing study of phytoplankton monitoring. The samples used were those collected from May 2011 to April 2012. Cell counting was done by using light microscopy on the samples fixed in 0.6% formaldehyde and then stored in the dark at 4 °C. A volume ranging from 1 to 50 mL was left to settle in an Utermöhl chamber for counting and enumeration at 400× magnification along transects (minimum of 200 cells); cell density was expressed as mean ± standard error, cells mL−1 (n = 3). For WC-FISH, volume samples of 50 or 75 mL were filtered on 0.8 μm isopore polycarbonate membrane filters (MilliporeTM, Darmstadt, Germany) fixed in a modified saline–ethanol at 4 °C for 2 h as reported above and hybridised as established by protocol. The probes against the cryptic species P. allochrona, P. arenysensis, P. delicatissima, P. dolorosa labelled with fluorescein isothiocyanate -FITC (Thermo Scientific™, Rockford, IL, USA) were hybridised at the same time but on different filters sections; simultaneously, the probe against P. multistriata was labelled with Cyanine-Cy3 (Thermo Scientific™, Rockford, IL, USA) and used.
3. Results
3.1. Probes Testing and Cross-Reactivity Trials
Species-specific candidate probes labelled with FITC fluorophore were tested on Pseudo-nitzschia target species monocultures (Table 1). Cells of each pure culture were hybridised with their species-specific probes, and the best WC-FISH hybridization conditions for each probe were established (Table 2). Washing temperature and washing buffer concentration were optimised to obtain a good epifluorescent signal as the visual intensity of the probe fluorescence of each target species in comparison with the positive control (Uni-C) (Figure 1). Essentially, in order to use multiplex probes for the subsequent field sample analysis and to ensure the specificity of the probe, even if at a slightly different melting temperature, we decided to maintain a fixed hybridization temperature and FA concentration. For the same reason, we tested different stringency conditions by regulating both the washing and temperature buffer concentration (Table 2). As a result of protocol optimization, the cells showed a uniform distribution of bright fluorescence throughout the cell except for the nuclear region, where fluorescence was confined to nucleoli (Figure 1). Treatments with a negative control probe (Uni-R) showed samples consistently dark, demonstrating no nonspecific binding or autofluorescence (Figure 1).

Table 2.
Probes testing and WC-FISH optimisation. The table shows hybridization temperature, formamide concentration (FA), temperature, and concentration of post-hybridization buffer and epifluorescent signal. Cells with signal intensity similar to the positive control were scored as ‘++’.
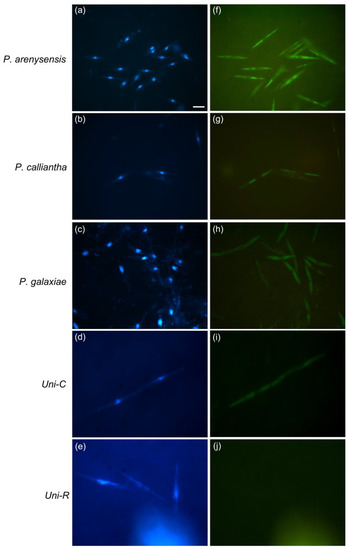
Figure 1.
Images of WC-FISH assays of pure culture of some cryptic Pseudo-nitzschia species hybridised to their species-specific probes. Each row of micrographs displays cells of pure cultures under epifluorescence microscopy in DAPI (a–e) and FITC (f–j) filters. The last two rows of micrographs display the hybridization with Uni-C (positive control) (d,i) and Uni-R (negative control) (e,j) probes. Scale = 10 µm.
To identify any cross-reactivity at this calibrated condition, each probe was hybridised against the different Pseudo-nitzschia species. Determination of cross-hybridizations was based on visual intensity of fluorescence in comparison with non-target cells; the positive (Uni-C) and negative (Uni-R) control treatments defined a range of labelling intensities providing a visual reference to assess the reactivity of specific probes (Table 3). The results showed that probes Pdel4D03_25, Pdel2D01_25, and PmulaD03_25 exclusively detected their target species P. allochrona, P. delicatissima, and P. multistriata, respectively (Table 3); instead, the probes Pdel3D01_25 and Pdel1D01_25 also weakly detected P. fraudulenta; PpdelD02_25 also detected P. galaxiae and P. calliantha; PgalaD02_25 detected both P. pseudodelicatissima and P. calliantha; Pman2D03_25 detected both P. pseudodelicatissima and P. galaxiae; and PfrauD04_25 hybridised with both P. arenysensis and P. dolorosa species (Table 3).

Table 3.
Cross-hybridization trials. Specificity of probes against different Pseudo-nitzschia species. Probes that successfully hybridised were scored as positive “++” (highly visible) and “+” (weakly visible) under the epifluorescent microscope; probes that failed to detect a culture were scored as negative “–”.
After several tests, the recalibration of the WC-FISH condition by increasing post-hybridization temperature and/or buffer washing stringency allowed it to successfully overcome some cross hybridizations (Table 4 and Table 5). In particular, we eliminated the cross-hybridization for all the probes of the P. delicatissima complex (Pdel1D01_25, Pdel2D01_25, Pdel4D02_25, and Pdel4D03_25) which continues to retain a strong signal (++) against their target cells (Table 5); on the contrary, it was very difficult to completely remove the cross-hybridization of P. pseudodelicatissima complex probes (PpdelD02_25, PgalaD02_25, and Pman_25) despite the new hybridization conditions (Table 5). Therefore, for these probes, no other attempts were made to optimise the whole-cell hybridization conditions, and they were excluded from the subsequent trials.

Table 4.
WC-FISH optimization. Hybridization temperature, formamide concentration (FA), temperature, and concentration of post-hybridization buffer to avoid cross-hybridization of the selected probes.

Table 5.
Cross-hybridization trials. Cross-reactivity of the screened probes after WC-FISH optimization conditions. Probes that successfully hybridised were scored as positive “++” (highly visible) and “+” (weakly visible) under the epifluorescent microscope; probes that failed to detect a culture were scored as negative “–”. The species/probes still retaining cross-hybridization signals are in bold.
Based on these results, it may be speculated that by using the new calibrated conditions for WC-FISH experiments, the screened probes could be useful for the molecular identification of the target species and also in natural samples containing many different microalgae. To this aim, we proceeded to their validation as reported below.
3.2. WC-FISH on Artificial Samples
In order to verify if the screened probes would be useful for qualitative and/or quantitative evaluation, the probes Pdel4D02_25, Pdel3D01_25, Pdel2D01_25, and PmulaD03_25, targeted, respectively, on P. allochrona, P. arenysensis, P. delicatissima, and P. multistriata, were hybridised on artificial samples composed of a single and/or mixed Pseudo-nitzschia monoculture species. Specifically, when the Pdel4D02_25, Pdel3D01_25, Pdel2D01_25, and PmulaD03_25 probes were hybridised on samples containing only a single species, a positive hybridization with a well-defined signal was detectable (Figure 2).
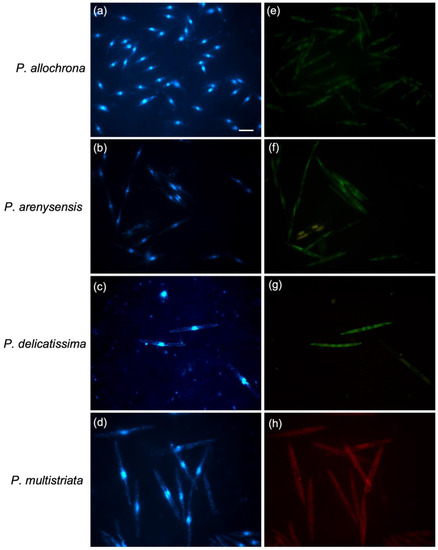
Figure 2.
WC-FISH assays the pure culture of some Pseudo-nitzschia cryptic species (a–c,e–g) and P. multistriata hybridised to their species-specific probes. Each row of micrographs displays cells of pure cultures under fluorescence microscopy in DAPI (a–d) and FITC (e–h) filters. Scale = 10 µm.
For quantitative analyses, the cell density of these samples was evaluated by the Utermöhl method in light microscopy before the WC-FISH test, and the value was compared to that obtained by epifluorescence microscopy after WC-FISH [43,44]. The results, expressed as the detection efficiency (%) of the WC-FISH vs. Utermöhl counting method, showed a good agreement between the two methods in the sample of P. arenysensis and P. multistriata (99% and 89% respectively), slightly an underestimation for P. allochrona (78%), and a very low detection efficiency for P. delicatissima (23%) (Figure 3). This evidence suggests that only the probes Pdel3D01_25 and PmulaD03_25 can also quantitatively reveal their target species P. arenysensis and P. multistriata while the signal from probe Pdel4D03_25 for P. allochrona is weaker and Pdel2D01_25 for P. delicatissima not properly detectable (Figure 3). For this reason, excluding the probe Pdel2D01_25, we evaluated the efficiency of the probes Pdel3D01_25, PmulaD03_25, and Pdel4D03_25 in the artificial mixed samples (simulated field samples).
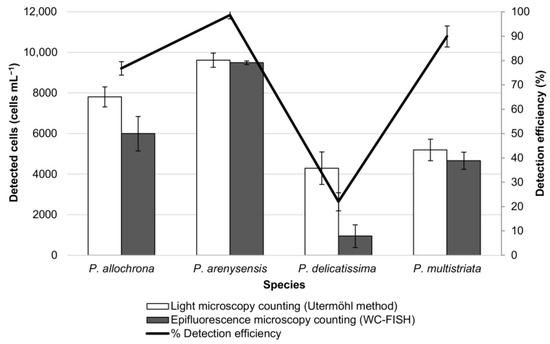
Figure 3.
Artificial samples composed of a single monoculture Pseudo-nitzschia species. Cell density (cells mL−1) was estimated before and after WC-FISH. Detection efficiency was calculated as FISH detectable cells (%) by the ratio between cells detected with rRNA FISH targeted probes vs. cultured cells before hybridisation.
The results showed that the detection efficiency (%) decreased significantly when mixing only two species (i.e., P. arenysensis and P. multistriata (from 99% to 59.1% for P. arenysensis; from 89% to 59.5% for P. multistriata)) (Figure 3 and Figure 4) and dropped when mixing all the three species (i.e., P. arenysensis, P. multistriata, P. allochrona) (Figure 4 and Figure 5). These results confirmed that the detection efficiency of WC-FISH decreases with the increase in the number of species in the sample, probably due to the probe competition at the target sites, even if the probes have been used at saturating concentrations.
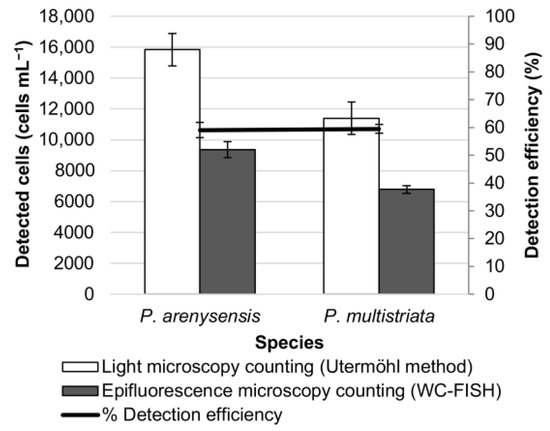
Figure 4.
Simulated natural sample composed of two Pseudo-nitzschia species. Cell density (cells mL−1) was estimated before and after WC-FISH. Detection efficiency was calculated as FISH detectable cells (%) by the ratio between cells detected with rRNA FISH targeted probes vs. cells in culture before hybridization.
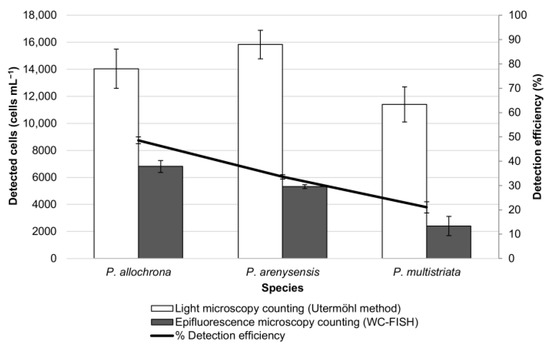
Figure 5.
Simulated natural sample composed of three Pseudo-nitzschia species. Cell density (cells mL−1) was estimated before and after WC-FISH. The detection efficiency (%) of the WC-FISH is calculated by the ratio between epifluorescence counting vs. light microscopy counting.
3.3. WC-FISH on Environmental Samples
To determine if WC-FISH would be able to detect the targeted cells in natural seawater qualitatively and/or quantitatively, we applied the screened probes for the P. delicatissima complex to natural samples (field-test) collected monthly at the LTER station in the Gulf of Naples. After the WC-FISH, it was possible to identify the cryptic species belonging to the P. delicatissima complex; otherwise, it would be impossible to identify the species with light microscopy (LM) (Figure 6). Further, the probe against P. multistriata (non-cryptic species) labelled with a different fluorochrome (Cy3) was used to verify if it would be possible to simultaneously detect almost two target species in the same sample.
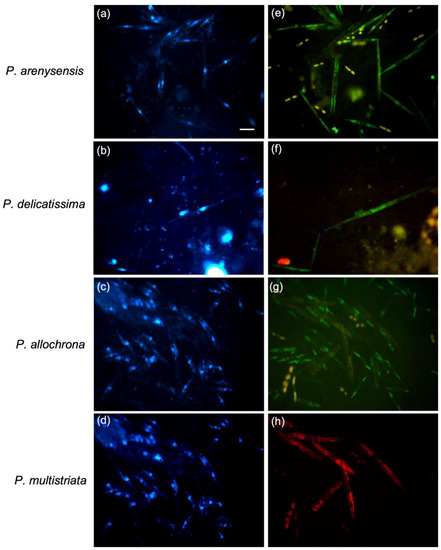
Figure 6.
WC-FISH assays on natural samples targeted to species-specific cryptic Pseudo-nitzschia probes. Each row of micrographs displays cells of natural samples under fluorescence microscopy in DAPI (a–d) and FITC (e–h) filters. The last two rows of micrographs display the same sample processed as multiplex WC-FISH by using the two different probes labelled with FITC and Cy3 respectively. Scale = 10 µm.
The cell density of the different samples was estimated before and after WC-FISH, and the detection efficiency of the WC-FISH was summarised in the Table 6. The highest density of the P. delicatissima complex was detected in April (686,861 cells mL−1) when WC-FISH detected only the cryptic species P. arenysensis and P. delicatissima (Table 6); a conspicuous density has been found in September (288,038 cells mL−1) when the screened probes for the P. delicatissima targeted to all four of the species P. arenysensis, P. delicatissima, P. dolorosa, and P. allochrona (Table 6). As already shown by the test on the artificial samples, while the number of cryptic species increased in the sample, the detection efficiency decreased. Indeed, in the samples where only one cryptic species was detected (June and July), the detection efficiency rose up to the 89%, whereas it decreased up to 14.3% in the samples where all four of the cryptic species were detected (Table 6). In the February sample, the total absence of the P. delicatissima complex corresponded to the total absence of a signal after WC-FISH, confirming the specificity of the method (Table 6).

Table 6.
Cell density (cell mL−1) in natural samples before WC-FISH was estimated as a P. delicatissima complex and cell density of some Pseudo-nitzschia delicatissima cryptic species after WC-FISH. The detection efficiency (%) of the WC-FISH was calculated by the ratio between epifluorescence counting vs. light microscopy counting.
4. Discussion
In this study, for in situ hybridization, we applied the protocols from Miller and Scholin (1998, 2000) [25,38] and Groben and Medlin (2005) [39] with some minor modifications, using specific fluorescently labelled probes to Pseudo-nitzschia species as tested on a microarray rRNA based phylochip [21]. Our results demonstrate that 28S rRNA targeted oligonucleotides are promising tools that can make the identification of Pseudo-nitzschia cryptic and pseudo-cryptic quickly. By using positive and negative control treatments, we defined a range of possible labelling intensities for any sample. Of the nine species-specific probes designed and tested, only six showed a species-specific response with an intensity comparable to that of the positive controls. Unfortunately, the cross-test and the optimization of the WC-FISH hybridization did not eliminate the cross-reaction among the target species of the P. pseudodelicatissima complex, probably due to highly similar target regions on the rRNA of these species because of phylogenetic proximity [2,11]. The presence of a-specific signals is attributed to the relatively conserved 28S region rRNA of ca. 700 bp where the probes were designed to discriminate among and within the groups of closely related species of Pseudo-nitzschia genus; even if in the 28S region of the genus Pseudo-nitzschia, species often differ in single base-pair changes, as reported in the materials and methods [41].
Optimal hybridization conditions of the probes could represent another problem. In particular, the hybridization temperature of probes tested against the P. pseudodelicatissima complex was 45 °C. The high stringency of the hybridization condition cannot be suitable and the best choice for all the probes tested.
Moreover, to verify the accuracy and specificity of the screened probes for P. delicatissima complex and our method developed above, WC-FISH tests were performed on artificial samples composed of both monoculture samples and the mix of two and/or three different monoculture samples (simulated field samples). As far as the monoculture samples, the algal densities determined by LM were in agreement with WC-FISH analysis (Figure 2 and Figure 3). The results were rather similar except for probe Pdel2D01_25, for which the counting by the epifluorescence microscopy after WC-FISH detected only 22% of the counted cells in LM (Figure 3). This discrepancy was probably due to the physiological state of the culture. In fact, the labelling intensity could be caused by an rRNA amount, culture age, and metabolic state of cells that influence the abundance of ribosomes and the target nucleic acid; it has been reported that the fluorescence intensity in cells declined sharply in the late stationary stage of batch culture [45,46]. Indeed, although P. delicatissima cells were analysed at the mid-exponential phase, as in the other samples, the cellular physiologic state likely influences the label intensity, which in turn could significantly wane, not allowing the probes to be visualized. In the natural population, it has been well established that rRNA varies systematically with growth rate; fast growing cells have more RNA per cell than the cells growing at slow rates [45]. Slow growth, associated with phosphorus and nitrogen limitation, resulted in up to a 400% decrease of rRNA intensity of labelled probes compared to nutrient-replete levels with the rRNA probe [46,47].
Even if one of the advantages of using single, fluorochrome labelled oligonucleotides is that labelling and detection is simple and rapid, our results suggest that WC-FISH is not reliable for quantitative analyses. As shown in simulated natural samples and/or in the field samples collected in the Gulf of Naples, the Utermöhl counting and the epifluorescence signal after whole-cell hybridization did not agree well. A quantitative comparison between the species estimates by WC-FISH and microscopic methods revealed that the whole-cell hybridization underestimated the relative abundance of Pseudo-nitzschia species compared to the microscopic results, as the number of species in the simulated natural samples was increasing (Figure 3, Figure 4 and Figure 5). In fact, the WC-FISH detected up to 98.7% of the cells when the simulated samples contained only one species, whereas the detection efficiency decreased to 21.1% when three different cryptic species were mixed (Figure 4 and Figure 5). In the natural field samples, we observed that the WC-FISH method detected between 10% and 85% of the cells, depending on the species richness of the field sample (Table 6) even if the cells with green fluorescence intensity could be clearly and rapidly detected under the epifluorescence microscope (Figure 6). The results on the field-test showed that the efficiency of the WC-FISH depends on the homogeneity of the sample. Detection efficiency decreased when the number of the cryptic species in the sample increased (e.g., in summer samples), but WC-FISH provided a good quantitative estimate when the P. delicatissima complex is likely to be homogeneous in species composition; otherwise, in autumn and early spring, when the P. delicatissima complex increased in biodiversity, the WC-FISH showed a lower percentage of species detection. In order to explain these data, we hypothesise that a competition occurred among different probes simultaneously used at the target sites due to the high percentage of similarity in the nucleotide sequences of the different probes; the weak cross-hybridizations were removed during post-hybridization washing, causing sequestration of the probe to their specific target sites and a partial loss of signal. This process is higher when more target species are in the sample. However, different factors can affect hybridization of cells in environmental natural populations. The high affinity of the probes for closely related species that can cause competition between short and similar sequence probes could result in a-specific hybridization [48,49]. Another parameter that can influence the specificity and strength of signals is the probe length; finding a compromise between probe length and specificity is still a challenging task [50,51]. The strength with which probes label cells is due to the accessibility of the target sequence in the context of the three-dimensional structure of the ribosome [52]. However, analysing low or undetectable fluorescence in natural samples might occur when cells are unhealthy, resulting in fewer ribosomes and, therefore, in a reduced fluorescence or due to the presence of dead Pseudo-nitzschia cells, empty frustules that were free in the sample or hidden within fecal pellets or sediment matrices.
Despite these weaknesses, the WC-FISH method is able to qualitatively detect the different cryptic species present in the samples, confirming that it is a good method for qualitative analyses of a Pseudo-nitzschia delicatissima species complex. In particular, in our study, the probes Pdel1D01_25, Pdel2D01_25, Pdel3D01_25, and Pdel4D03_25 may be useful for molecular identification of the target species in natural samples containing many different potentially toxic microalgae, identifying the specific genus Pseudo-nitzschia and the toxin producing species at their bloom. The limit of WC-FISH for quantitative analysis has not yet been completely overcome due to the effort to design and use probes with similar melting temperatures (Tms) to aid in multiplexing; on the other hand, it could be designed and used in groups of riboprobes with different Tms to be employed in parallel experiments (multiprobing) but with different denaturation and washing temperatures. However, this would make the method longer and perhaps not advantageous compared to other molecular methods (i.e., PCR). Nevertheless, WC-FISH could be a useful tool for a very early qualitative screening method to be combined with classical microscopic analysis or to combine it with other very stable environmental processors. Further, this technique will be most useful for the early detection of single species blooms, especially for HAB, when microscopic analyses are not practical (i.e., cryptic species). That issue becomes particularly prominent when considering that HAB species pose human health and ecological threats often at very low cell densities (~102 cells L−1).
Finally, it should be noted that the WC-FISH is faster and more cost-effective compared with other rDNA-based methods for the identification of Pseudo-nitzschia species. A concrete example of a specific probe hybridization technique used in the environmental field is represented by the company Microbia Environment that patented CARLA technology for Cellular Activity RNA-based eLisA. The assay is designed to detect cyanobacteria and microalgae in environmental water in less than 3 h using sequence-specific hybridization biosensors for a target of microalgae ribosomal RNA (rRNA) (https://www.microbia-environnement.com (accessed on 30 September 2022)).
In this study we have demonstrated that it is possible to discriminate a variety of Pseudo-nitzschia species collected from natural populations in situ and in near real-time. Altogether, the fluorescent oligonucleotide probes tested in our study show great promise as tools that can facilitate the monitoring of Pseudo-nitzschia species in natural samples.
Author Contributions
Conceptualization, M.F., L.R., I.P. and R.C.; methodology, M.F., L.R., I.P. and L.B.; validation, M.F., L.R., I.P. and L.B.; investigation, M.F. and L.R.; writing—original draft preparation, M.F. and L.R.; writing—review and editing, M.F., L.B., A.M., C.G., S.G. and R.C.; supervision, R.C.; project administration, R.C.; funding acquisition, R.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by University of Calabria (ex 60% University Research Funds 2010–2012).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Moreno, A.R.; Anderson, C.; Kudela, R.M.; Sutula, M.; Edwards, C.; Bianchi, D. Development, Calibration, and Evaluation of a Model of Pseudo-nitzschia and Domoic Acid Production for Regional Ocean Modeling Studies. Harmful Algae 2022, 118, 102296. [Google Scholar] [CrossRef] [PubMed]
- Scholin, C.A.; Gulland, F.; Doucette, G.J.; Benson, S.; Busman, M.; Chavez, F.P.; Cordaro, J.; DeLong, R.; De Vogelaere, A.; Harvey, J.; et al. Mortality of Sea Lions along the Central California Coast Linked to a Toxic Diatom Bloom. Nature 2000, 403, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Guillou, L.; Nézan, E.; Cueff, V.; Erard-Le Denn, E.; Cambon-Bonavita, M.-A.; Gentien, P.; Barbier, G. Genetic Diversity and Molecular Detection of Three Toxic Dinoflagellate Genera (Alexandrium, Dinophysis, and Karenia) from French Coasts. Protist 2002, 153, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Zabaglo, K.; Chrapusta, E.; Bober, B.; Kaminski, A.; Adamski, M.; Bialczyk, J. Environmental Roles and Biological Activity of Domoic Acid: A Review. Algal Res. 2016, 13, 94–101. [Google Scholar] [CrossRef]
- Bates, S.S.; Hubbard, K.A.; Lundholm, N.; Montresor, M.; Leaw, C.P. Pseudo-nitzschia, Nitzschia, and Domoic Acid: New Research since 2011. Harmful Algae 2018, 79, 3–43. [Google Scholar] [CrossRef]
- Smodlaka Tanković, M.; Baričević, A.; Gerić, M.; Domijan, A.-M.; Pfannkuchen, D.M.; Kužat, N.; Ujević, I.; Kuralić, M.; Rožman, M.; Matković, K.; et al. Characterisation and Toxicological Activity of Three Different Pseudo-nitzschia Species from the Northern Adriatic Sea (Croatia). Environ. Res. 2022, 214, 114108. [Google Scholar] [CrossRef]
- Trainer, V.L.; Hickey, B.M.; Horner, R.A. Biological and Physical Dynamics of Domoic Acid Production off the Washington Coast. Limnol. Oceanogr. 2002, 47, 1438–1446. [Google Scholar] [CrossRef]
- Orsini, L.; Procaccini, G.; Sarno, D.; Montresor, M. Multiple RDNA ITS-Types within the Diatom Pseudo-nitzschia Delicatissima (Bacillariophyceae) and Their Relative Abundances across a Spring Bloom in the Gulf of Naples. Mar. Ecol. Prog. Ser. 2004, 271, 87–98. [Google Scholar] [CrossRef]
- Sahraoui, I.; Bates, S.S.; Bouchouicha, D.; Mabrouk, H.H.; Hlaili, A.S. Toxicity of Pseudo-nitzschia Populations from Bizerte Lagoon, Tunisia, Southwest Mediterranean, and First Report of Domoic Acid Production by P. Brasiliana. Diatom Res. 2011, 26, 293–303. [Google Scholar] [CrossRef]
- Lundholm, N.; Bates, S.S.; Baugh, K.A.; Bill, B.D.; Connell, L.B.; Léger, C.; Trainer, V.L. Cryptic and pseudo-cryptic diversity in diatoms—with descriptions of Pseudo-nitzschia hasleana sp. nov. and P. fryxelliana sp. nov. J. Phycol. 2012, 48, 436–454. [Google Scholar] [CrossRef]
- Lundholm, N.; Moestrup, Ø.; Kotaki, Y.; Hoef-Emden, K.; Scholin, C.; Miller, P. Inter-and intraspecific variation of the Pseudo-nitzschia delicatissima complex (Bacillariophyceae) illustrated by rRNA probes, morphological data and phylogenetic analyses. J. Phycol. 2006, 42, 464–481. [Google Scholar] [CrossRef]
- Ruggiero, M.V.; Sarno, D.; Barra, L.; Kooistra, W.H.C.F.; Montresor, M.; Zingone, A. Diversity and Temporal Pattern of Pseudo-nitzschia Species (Bacillariophyceae) through the Molecular Lens. Harmful Algae 2015, 42, 15–24. [Google Scholar] [CrossRef]
- Lim, H.C.; Teng, S.T.; Leaw, C.P.; Lim, P.T. Three Novel Species in the Pseudo-nitzschia Pseudodelicatissima Complex: P. Batesiana Sp. Nov., P. Lundholmiae Sp. Nov., and P. Fukuyoi Sp. Nov. (Bacillariophyceae) from the Strait of Malacca, Malaysia. J. Phycol. 2013, 49, 902–916. [Google Scholar] [CrossRef]
- Percopo, I.; Ruggiero, M.V.; Balzano, S.; Gourvil, P.; Lundholm, N.; Siano, R.; Tammilehto, A.; Vaulot, D.; Sarno, D. Pseudo-nitzschia Arctica Sp. Nov., a New Cold-Water Cryptic Pseudo-nitzschia Species within the P. pseudodelicatissima Complex. J. Phycol. 2016, 52, 184–199. [Google Scholar] [CrossRef]
- Quijano-Scheggia, S.I.; Garcés, E.; Lundholm, N.; Moestrup, Ø.; Andree, K.; Camp, J. Morphology, Physiology, Molecular Phylogeny and Sexual Compatibility of the Cryptic Pseudo-nitzschia Delicatissima Complex (Bacillariophyta), Including the Description of P. Arenysensis Sp. Nov. Phycologia 2009, 48, 492–509. [Google Scholar] [CrossRef]
- Ruggiero, M.V.; Kooistra, W.H.C.F.; Piredda, R.; Sarno, D.; Zampicinini, G.; Zingone, A.; Montresor, M. Temporal Changes of Genetic Structure and Diversity in a Marine Diatom Genus Discovered via Metabarcoding. Environ. DNA 2022, 4, 763–775. [Google Scholar] [CrossRef]
- Percopo, I.; Ruggiero, M.V.; Sarno, D.; Longobardi, L.; Rossi, R.; Piredda, R.; Zingone, A. Phenological Segregation Suggests Speciation by Time in the Planktonic Diatom Pseudo-nitzschia Allochrona Sp. Nov. Ecol. Evol. 2022, 12, e9155. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, K.A.; Rocap, G.; Armbrust, E.V. Inter-and intraspecific community structure within the diatom genus Pseudo-nitzschia (Bacillariophyceae). J. Phycol. 2008, 44, 637–649. [Google Scholar] [CrossRef]
- Galluzzi, L.; Cegna, A.; Casabianca, S.; Penna, A.; Saunders, N.; Magnani, M. Development of an Oligonucleotide Microarray for the Detection and Monitoring of Marine Dinoflagellates. J. Microbiol. Methods 2011, 84, 234–242. [Google Scholar] [CrossRef]
- Andree, K.B.; Fernández-Tejedor, M.; Elandaloussi, L.M.; Quijano-Scheggia, S.; Sampedro, N.; Garcés, E.; Camp, J.; Diogène, J. Quantitative PCR Coupled with Melt Curve Analysis for Detection of Selected Pseudo-nitzschia Spp. (Bacillariophyceae) from the Northwestern Mediterranean Sea. Appl. Environ. Microbiol. 2011, 77, 1651–1659. [Google Scholar] [CrossRef]
- Barra, L.; Ruggiero, M.V.; Sarno, D.; Montresor, M.; Kooistra, W.H.C.F. Strengths and Weaknesses of Microarray Approaches to Detect Pseudo-nitzschia Species in the Field. Environ. Sci. Pollut. Res. 2013, 20, 6705–6718. [Google Scholar] [CrossRef] [PubMed]
- Edvardsen, B.; Dittami, S.M.; Groben, R.; Brubak, S.; Escalera, L.; Rodríguez, F.; Reguera, B.; Chen, J.; Medlin, L.K. Molecular Probes and Microarrays for the Detection of Toxic Algae in the Genera Dinophysis and Phalacroma (Dinophyta). Environ. Sci. Pollut. Res. 2013, 20, 6733–6750. [Google Scholar] [CrossRef]
- Kegel, J.U.; Del Amo, Y.; Medlin, L.K. Introduction to Project MIDTAL: Its Methods and Samples from Arcachon Bay, France. Environ. Sci. Pollut. Res. 2013, 20, 6690–6704. [Google Scholar] [CrossRef]
- McCoy, G.R.; Touzet, N.; Fleming, G.T.; Raine, R. An Evaluation of the Applicability of Microarrays for Monitoring Toxic Algae in Irish Coastal Waters. Environ. Sci. Pollut. Res. 2013, 20, 6751–6764. [Google Scholar] [CrossRef] [PubMed]
- Miller, P.E.; Scholin, C.A. Identification and enumeration of cultured and wild Pseudo-nitzschia (Bacillariophyceae) using species-specific LSU rRNA-targeted fluorescent probes and filter-based whole cell hybridization. J. Phycol. 1998, 34, 371–382. [Google Scholar] [CrossRef]
- Parsons, M.L.; Scholin, C.A.; Miller, P.E.; Doucette, G.J.; Powell, C.L.; Fryxell, G.A.; Dortch, Q.; Soniat, T.M. Pseudo-nitzschia species (Bacillariophyceae) in Louisiana coastal waters: Molecular probe field trials, genetic variability, and domoic acid analyses. J. Phycol. 1999, 35, 1368–1378. [Google Scholar] [CrossRef]
- Sako, Y.; Hosoi-Tanabe, S.; Uchida, A. fluorescence in situ hybridization using rRNA-targeted probes for simple and rapid identification of the toxic dinoflagellates Alexandrium tamarense and Alexandrium catenella. J. Phycol. 2004, 40, 598–605. [Google Scholar] [CrossRef]
- Anderson, D.M.; Kulis, D.M.; Keafer, B.A.; Gribble, K.E.; Marin, R.; Scholin, C.A. Identification and Enumeration of Alexandrium Spp. from the Gulf of Maine Using Molecular Probes. Deep. Sea Res. Part II Top. Stud. Oceanogr. 2005, 52, 2467–2490. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, C.; Zhang, B.; Wang, G.; Lu, D.; Xu, Z.; Yan, P. Development of a PNA Probe for Fluorescence In Situ Hybridization Detection of Prorocentrum Donghaiense. PLoS ONE 2011, 6, e25527. [Google Scholar] [CrossRef]
- Caracciolo, A.B.; Dejana, L.; Fajardo, C.; Grenni, P.; Martin, M.; Mengs, G.; Sánchez-Fortún, S.; Lettieri, T.; Saccà, M.L.; Medlin, L.K. A New Fluorescent Oligonucleotide Probe for In-Situ Identification of Microcystis Aeruginosa in Freshwater. Microchem. J. 2019, 148, 503–513. [Google Scholar] [CrossRef]
- Morais, S.L.; Barros, P.; Santos, M.; Delerue-Matos, C.; Gomes, A.C.; Fátima Barroso, M. Electrochemical Genosensor for the Detection of Alexandrium Minutum Dinoflagellates. Talanta 2021, 222, 121416. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, C.; Wang, Y.; Chen, G. A Review of the Current and Emerging Detection Methods of Marine Harmful Microalgae. Sci. Total Environ. 2022, 815, 152913. [Google Scholar] [CrossRef] [PubMed]
- Medlin, L.K.; Orozco, J. Molecular Techniques for the Detection of Organisms in Aquatic Environments, with Emphasis on Harmful Algal Bloom Species. Sensors 2017, 17, 1184. [Google Scholar] [CrossRef] [PubMed]
- Toebe, K. Whole Cell Hybridisation for Monitoring Harmful Marine Microalgae. Environ. Sci. Pollut. Res. 2013, 20, 6816–6823. [Google Scholar] [CrossRef]
- McDonald, S.M.; Sarno, D.; Scanlan, D.J.; Zingone, A. Genetic Diversity of Eukaryotic Ultraphytoplankton in the Gulf of Naples during an Annual Cycle. Aquat. Microb. Ecol. 2007, 50, 75–89. [Google Scholar] [CrossRef]
- Guillard, R.R.L. Culture of Phytoplankton for Feeding Marine Invertebrates. In Culture of Marine Invertebrate Animals: Proceedings —1st Conference on Culture of Marine Invertebrate Animals Greenport; Smith, W.L., Chanley, M.H., Eds.; Springer: Boston, MA, USA, 1975; pp. 29–60. ISBN 978-1-4615-8714-9. [Google Scholar]
- Zingone, A.; Totti, C.; Sarno, D.; Cabrini, M.; Caroppo, C.; Giacobbe, M.; Lugliè, A.; Nuccio, C.; Socal, G. Fitoplancton: Metodiche Di Analisi Quali-Quantitativa. In Metodologie di Studio del Plancton Marino. Manuali e Linee Guida 56/2010; ISPRA: Roma, Italy, 2010; pp. 213–237. ISBN 978-88-448-0427-5. [Google Scholar]
- Miller, P.E.; Scholin, C.A. On Detection of Pseudo-nitzschia (Bacillariophyceae) Species Using Whole Cell Hybridization: Sample Fixation and Stability. J. Phycol. 2000, 36, 238–250. [Google Scholar] [CrossRef]
- Groben, R.; Medlin, L. In Situ Hybridization of Phytoplankton Using Fluorescently Labeled RRNA Probes. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2005; Volume 395, pp. 299–310. ISBN 0076-6879. [Google Scholar]
- Field, C.B.; Behrenfeld, M.J.; Randerson, J.T.; Falkowski, P. Primary Production of the Biosphere: Integrating Terrestrial and Oceanic Components. Science 1998, 281, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Barra, L.; Ruggiero, M.V.; Chen, J.; Kooistra, W.H.C.F. Specificity of LSU rRNA-Targeted Oligonucleotide Probes for Pseudo-nitzschia Species Tested through Dot-Blot Hybridisation. Environ. Sci. Pollut. Res. 2013, 21, 548–557. [Google Scholar] [CrossRef]
- Ludwig, W.; Strunk, O.; Westram, R.; Richter, L.; Meier, H.; Yadhukumar; Buchner, A.; Lai, T.; Steppi, S.; Jobb, G.; et al. ARB: A Software Environment for Sequence Data. Nucleic Acids Res. 2004, 32, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.Y. The Errors of the Sedgwick-Rafter Counting Chamber in the Enumeration of Phytoplankton. Trans. Am. Microsc. Soc. 1942, 61, 217–226. [Google Scholar] [CrossRef]
- Lund, J.W.G.; Kipling, C.; Le Cren, E.D. The Inverted Microscope Method of Estimating Algal Numbers and the Statistical Basis of Estimations by Counting. Hydrobiologia 1958, 11, 143–170. [Google Scholar] [CrossRef]
- DeLong, E.; Wickham, G.; Pace, N. Phylogenetic Stains: Ribosomal RNA-Based Probes for the Identification of Single Cells. Science 1989, 243, 1360–1363. [Google Scholar] [CrossRef]
- Anderson, D.M.; Kulis, D.M.; Keafer, B.A.; Berdalet, E. Detection of the Toxic Dinoflagellate Alexandrium Fundyense (Dinophyceae) with Oligonucleotide and Antibody Probes: Variability in Labeling Intensity with Physiological Condition. J. Phycol. 1999, 35, 870–883. [Google Scholar] [CrossRef]
- Dittami, S.M.; Edvardsen, B. Culture conditions influence cellular RNA content in ichthyotoxic flagellates of the genus Pseudochattonella (Dictyochophyceae). J. Phycol. 2012, 48, 1050–1055. [Google Scholar] [CrossRef] [PubMed]
- Medlin, L.K.; Kegel, J.U. Validation of the Detection of Pseudo-nitzschia Spp. Using Specific RNA Probes Tested in a Microarray Format: Calibration of Signal Based on Variability of RNA Content with Environmental Conditions. Harmful Algae 2014, 37, 183–193. [Google Scholar] [CrossRef]
- Wright, E.S.; Yilmaz, L.S.; Corcoran, A.M.; Ökten, H.E.; Noguera, D.R. Automated Design of Probes for RRNA-Targeted Fluorescence In Situ Hybridization Reveals the Advantages of Using Dual Probes for Accurate Identification. Appl. Environ. Microbiol. 2014, 80, 5124–5133. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Ono, N.; Furusawa, C.; Kashiwagi, A.; Yomo, T. Experimental Optimization of Probe Length to Increase the Sequence Specificity of High-Density Oligonucleotide Microarrays. BMC Genom. 2007, 8, 373. [Google Scholar] [CrossRef] [PubMed]
- Metfies, K.; Medlin, L.K. Feasibility of Transferring Fluorescent in Situ Hybridization Probes to an 18S RRNA Gene Phylochip and Mapping of Signal Intensities. Appl. Environ. Microbiol. 2008, 74, 2814–2821. [Google Scholar] [CrossRef] [PubMed]
- Behrens, S.; Fuchs, B.M.; Mueller, F.; Amann, R. Is the In Situ Accessibility of the 16S RRNA of Escherichia Coli for Cy3-Labeled Oligonucleotide Probes Predicted by a Three-Dimensional Structure Model of the 30S Ribosomal Subunit? Appl. Environ. Microbiol. 2003, 69, 4935–4941. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).