Abstract
Medicinal plants maintain structures and diversities of bacteria, fungi, and arbuscular mycorrhizal fungi (AMF) that can interact to promote growth and therapeutic properties. Therefore, the purpose of this research was to evaluate the microbiome of Lippia alba and Petiveria alliacea, species known for their high potential for medicinal benefits in Colombia. To achieve this, rhizosphere soils and roots were sampled from five departments in Colombia: Boyacá, Cundinamarca, Tolima, Putumayo, and Valle del Cauca. The results revealed that the dominant bacterial groups in both plants were primarily Proteobacteria, Acidobacteriota, and Actinobacteriota, with the first phylum showing the highest number of differentially abundant genera between the sampling points. In fungi, Ascomycota tended to dominate in most of the sampled locations, while Mortierellomycota was particularly abundant in roots of P. alliacea in Valle. Furthermore, the study of AMF indicated differentiation in the colonization for both plants, with the genera Glomus and Paraglomus being predominant. Differences in the Shannon diversity index were recorded between sampling types within these sampling points, possibly influenced by local and environmental factors. Our findings reveal that the microbiomes of both medicinal plants exhibit distinct community assemblies, which could be a significant factor for their future therapeutic use.
1. Introduction
The soil microbiome serves essential functions, including aiding in soil regulation and fertility. Moreover, their active participation in various biogeochemical cycles enables the microbiome to perform other functions, such as enhancing nutrient absorption and contributing to pest and phytopathogen control. Consequently, the plant-microbiome system stands as the primary interaction involved in plant growth and survival [1,2,3,4].
This interaction is influenced by a diverse array of abiotic factors, including soil type and biogeographical characteristics such as altitude and seasonal climate variations [5,6]. Additionally, biotic factors, like the specific plant species, particularly impact this relationship through root secretions [7,8]. The latter aspect has garnered much interest in exploring the soil microbiome associated with medicinal plants, known to secrete and synthesize a wide range of distinctive secondary metabolites, such as organic, fatty, and phenolic acids, as well as terpenes and alkaloids [9,10,11,12].
In Colombia, medicinal plants have been a source of therapeutic products throughout history for treating various diseases [13]. Among these, Lippia alba and Petiveria alliacea, belonging, respectively, to the Verbenaceae and Petiveriaceae families and native to Latin America, have gained prominence for their numerous medicinal benefits, particularly their anti-carcinogenic properties. Several studies have examined the anti-carcinogenic effects of their essential oils, in the case of L. alba, research has shown its potential in halting the cell cycle and inducing apoptosis in liver and lung cancer cells [14], colon and prostate cancer cells [15], leukemia cells [16], and gastric carcinoma cells [17]. Similarly, P. alliacea has been found to trigger cell apoptosis and reduce the growth capability of breast cancer cell colonies [18], along with diminishing tumor burden and myeloid leukemia metastasis [19]. Despite the significance of these plants and the evidence suggesting that the interaction between medicinal plants and the microbiome results in the production of phytotherapeutic compounds [20,21,22], no studies have been conducted on the microbiome associated with L. alba and P. alliacea.
This research constitutes one of the initial approaches to microbiome evaluations in the medicinal plants L. alba and P. alliacea, representing a first effort to identify those microbial elements that could be involved in plant growth and maintaining the therapeutic potential of these plants. We aimed to determine the structure and diversity of bacteria, fungi, and arbuscular mycorrhizal fungi (AMF) in rhizosphere soil and root associated with L. alba and P. alliacea. Additionally, to investigate taxonomic relationships with soil physicochemical properties in areas where these plants predominate. We hypothesize that the microbiome would exhibit differential structures and diversities of microorganisms between both medicinal plants in several sampling points of Colombia. We hope to find taxa with variations in abundance both in rhizosphere soil and roots of plants, according to the ecological characteristics of the microorganisms. Furthermore, we anticipate that microbial taxonomic associations with physicochemical properties will reveal significant relationships, providing valuable information about the ecological aspects of these medicinal plants.
2. Materials and Methods
2.1. Soil Sampling of Medicinal Plants
The sampling process started with the taxonomic identification of plant species in five departments of Colombia, including primarily wild individuals found within home gardens and natural areas, and a few specimens categorized as cultivated (Figure 1 and Table S1). Plants of L. alba were identified from the locations of Palmira (Valle del Cauca), Sibundoy (Putumayo), and Lérida (Tolima), with 3–4 replicates at each sampling point (Figure 1 and Table S1). For P. alliacea, sampling was conducted in Palmira (Valle del Cauca), Pacho (Cundinamarca), Ambalema (Tolima), Armero (Tolima), and Villa de Leyva (Boyacá), with 2–6 replicates at each of these localities (Figure 1 and Table S1). For both plant species, rhizosphere soil and root samples were collected to study bacteria and fungi, while only root samples were collected for AMF analysis. Secondary roots and rhizosphere soil samples were collected from each individual after the subterranean portion had been extracted from the ground. Additionally, approximately 1 kg samples of the top 20 cm of soil were collected from these locations for physicochemical analysis. The microbiome study samples were stored at −80 °C until processing.
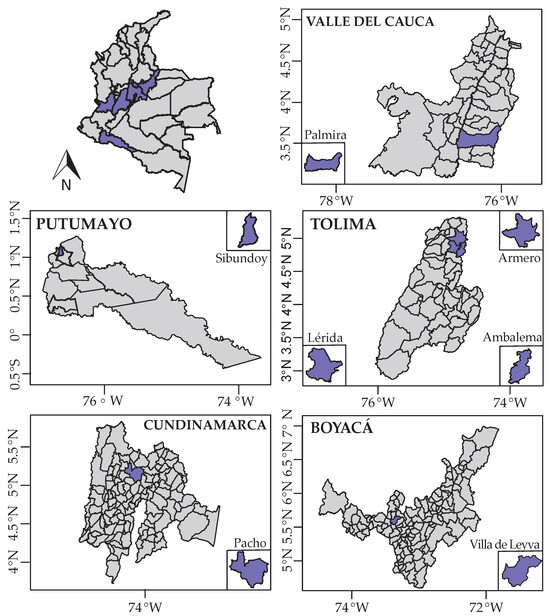
Figure 1.
Colombian departments and municipalities sampled for the medicinal plants L. alba and P. allicaea. Purple surfaces represent the departments and municipalities where the medicinal plants were found.
2.2. Physicochemical Measurements of Soils
The soil samples for physicochemical analysis were sent to the AGRI-LAB laboratory, where measurements of macro and microelements, such as pH, effective cation exchange capacity (ECEC), cation exchange capacity (CEC), electrical conductivity (EC), soil organic matter (SOM), soil organic carbon (SOC), total nitrogen (N), phosphorus (P), sulfur (S), exchangeable calcium (Ca), exchangeable magnesium (Mg), exchangeable potassium (K), exchangeable sodium (Na), boron (B), iron (Fe), copper (Cu), manganese (Mn), and zinc (Zn), were conducted. Additionally, physical variables including bulk density (BD), texture, and average moisture saturation (AMS) were assessed. The data were analyzed using a non-parametric Kruskal-Wallis test with the Jamovi 2.2.5 software [23].
2.3. DNA Extraction and Metabarcoding from Bacteria, Fungi, and AMF
The DNA of soil microorganisms was extracted in the Pontificia Universidad Javeriana according to Doyle and Doyle [24]. The quality of the extractions was assessed using spectrophotometry and 0.8% agarose gels. Subsequently, library preparation and sequencing were conducted at the Argonne National Laboratory of the U.S. Department of Energy. For bacteria, the V4-V5 region of the 16S ARN ribosomal gene was amplified following the methodology of Caporaso et al. [25]. Primers ITS1F/ITS2 [26,27] were employed for fungi and NS31/aML2 [28] for AMF. Sequencing was performed using Illumina HiSeq2000 and MiSeq technology. The raw data were deposited in the European Nucleotide Archive (ENA) under project number PRJEB67294 and accession numbers ERS16459293 to ERS16459433.
2.4. Bioinformatic and Diversity Analyses
The bioinformatic began with QIIME2 software version 2023.7 [29], where a quality control step was run with the DEMUX plugin, including noise and chimera sequence removal using DADA2 [30]. Next, abundance tables were normalized to 16,000 reads for bacteria and 10,774 reads for fungi. Taxonomic assignment was carried out using the SILVA v. 13_8 database for bacteria, UNITE v. 8_99 for fungi, and Maarjam 18S for AMF [31]. The analysis was then continued in the R software version 4.2.3, using packages such as qiime2R v. 0.99.6 and phyloseq [32]. Taxonomic composition graphs for bacteria and fungi were generated considering the top 10 most abundant taxa at the phylum and class levels. Additionally, differential abundance analyses at the genus level were conducted using the DESeq2—Bioconductor package [33]. Alpha diversity indices, including Shannon diversity and Berger–Parker dominance, were calculated using the microbiome package v. 1.18.0 in Bioconductor. Beta diversity was visualized through principal coordinate analysis (PcoA) with Bray–Curtis distances. Furthermore, a multiple factor analysis (MFA) was performed using the factoextra package to explore the relationships between the 10 most abundant taxa for bacteria and fungi at each plant and soil physicochemical measurements.
3. Results
3.1. Soil Physicochemical Analysis
The results of the physicochemical parameters revealed significant differences in soil pH among the study locations (Table S2). In both plants, locations with strongly acidic soils (Sibundoy), moderately acidic soils (Ambalema, Armero, and Pacho), and neutral soils (Palmira, Lérida, and Villa de Leyva) were observed (Table S2). These variations led to significant differences in effective cation exchange capacity (ECEC), cation exchange capacity (CEC), and ion concentrations such as Ca, Mg, and Fe (Table S2). Soil organic matter and organic carbon content varied among the sampling points, ranging from 1% to 4%, but no significant differences were found according to the statistical test. Total soil nitrogen did not show significant differences in both plants, but phosphorus levels were notably high, particularly in P. alliacea locations (Table S2). Regarding soil texture, the results indicated clayey soils with good water retention capacity but potential drainage issues. Additionally, the moisture saturation percentages were similar, with values around 20%, and the bulk density recorded values exceeding one, with no significant differences among the sampling points.
3.2. Comparative Analysis of Soil and Root Microbiome
3.2.1. Rhizosphere Soil and Root Bacteria Associated with Medicinal Plants
Initially, a total of 2,301,354 reads were obtained for bacteria, with 789,094 reads from L. alba (averaging 39,454 ± 9433 reads per sample) and 1,512,260 from P. alliacea (averaging 47,258 ± 15,630 reads per sample). After the quality control and normalization processes, 282,977 reads for bacteria were retained for L. alba, and 572,271 reads for P. alliacea. At the taxonomic level, the reads from L. alba generated 8539 ASVs assigned to 30 phyla (100% assignment), 86 classes (98%), 183 orders (92%), 234 families (78%), and 363 genera (48%) (Figure S1). On the other hand, P. alliacea yielded 12,318 ASVs with assignments to 37 phyla (99%), 98 classes (98%), 201 orders (91%), 270 families (75%), and 399 genera (47%) (Figure S2).
At the phylum level, the taxa depicted in Figure 2 were the most abundant overall, accounting for approximately 92% of the total reads. Specifically, Proteobacteria was the most abundant in the locations of both medicinal plants. For L. alba, it was found to be more abundant in the rhizosphere soils of Palmira (50% soil vs. 42% root), Sibundoy (60% vs. 48%), and Lérida (41% vs. 16%) (Figure 2a). Additionally, for P. allicaea, Proteobacteria showed variation between sample types, with a greater number of reads assigned to the roots of Ambalema (17% vs. 35%) and the rhizosphere soil of Villa de Leyva (52% vs. 31%) (Figure 2c). On the other hand, Acidobacteriota tended to have more reads assigned in root samples than in rhizosphere soil in locations like Palmira (5% rhizosphere soil vs. 17% root) and Lérida (9% vs. 28%) for L. alba, and Palmira (8.0% vs. 17%) and Villa de Leyva (5% vs. 17%) for P. alliacea (Figure 2). However, in Ambalema for the latter plant, this trend was reversed, with a 25% abundance in rhizosphere soil versus 13% in root. In other phyla, such as Bacteroidota, there was greater abundance in the roots of Lérida and Sibundoy in L. alba and Palmira and Pacho in P. alliacea but was more abundant in the rhizosphere soils of Ambalema and Villa de Leyva for the latter plant (Figure 2). Actinobacteriota was more prevalent in the rhizosphere soil of Palmira and Lérida for L. alba and in the soil of Armero and Villa de Leyva for P. alliacea, while Verrucomicrobiota was more abundant in the root of Lérida for L. alba and Armero and Villa de Leyva for P. alliacea (Figure 2).
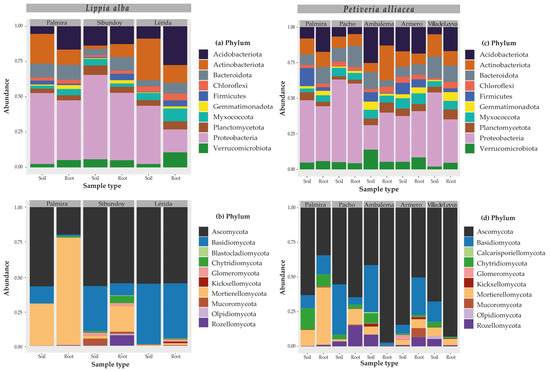
Figure 2.
Taxonomic composition at the phylum level for bacteria (a,c) and fungi (b,d) associated with the rhizosphere soil and roots of the medicinal plants L. alba (a,b) and P. alliacea (c,d).
In terms of class, Gammaproteobacteria and Alphaproteobacteria were the most abundant classifications overall. The former showed higher reads in the rhizosphere soils of Sibundoy (54% soil vs. 31% root) for L. alba, while differences were reported in soils from Palmira (36% vs. 28%) and Villa de Leyva (34% vs. 24%), and in the roots of Ambalema (12% vs. 19%) for P. allicaea (Figure S3). The latter remained more abundant at 54% in rhizosphere soil samples from Sibundoy for L. alba and in both root and soil samples from Ambalema and Villa de Leyva, respectively, for P. alliacea (Figure S3). Furthermore, Bacteroidia from the phylum Bacteroidetes maintained its abundance in the roots of Sibundoy and Lérida for L. alba and in the roots of Palmira and Pacho and soil of Ambalema for P. alliacea. On the other hand, Actinobacteria had higher allocations in the rhizosphere soils of Palmira and Lérida for L. alba and both the soil and roots of Villa de Leyva and Ambalema for P. alliacea (Figure S3).
The differential abundance analyses at the genus level revealed that in the L. alba location of Palmira, the majority were predominantly Proteobacteria such as Stenotrophomonas, Pseudomonas, and Lysobacter, and Actinobacteriota like Streptomyces (Figure S4a,b). In Lérida, the genera Bryobacter, Rickettsiela from Acidobacteriota, Bacillus from Firmicutes, and Candidatus Udeobacter from Verrucomicrobiota were more prevalent (See Figure S4b,c). Meanwhile, in Sibundoy, Proteobacteria genera like Pseudoxanthomonas, Brevundimonas, and Ellin6067 distinguished themselves from other localities (Figure S4a,c). In P. alliacea of Palmira, Proteobacteria remained with distinct abundances in genera such as Sphingomonas, and Firmicutes like Bacillus and Clostridium sensu stricto 8 (Figure S5a–d). Additionally, Pacho recorded the highest number of differentially abundant genera compared to Villa de Leyva (Figure S5g), while Ambalema showed differences from Armero (Figure S5h).
3.2.2. Rhizosphere Soil and Root Fungi Associated with Medicinal Plants
For fungi, 2.341,906 reads were obtained between the two plants, with 972.170 and 1.369.736 reads, respectively, for L. alba and P. alliacea. After filtering and normalization processes, 144,219 reads remained for the first plant and 249,248 reads for the second. At the taxonomic level, 3417 ASVs from L. alba were assigned to 11 phyla (100%), 37 classes (90%), 88 orders (86%), 181 families (70%), and 315 genera (53%) (Figure S1). For P. alliacea, 4150 ASVs were assigned to 11 phyla (100%), 35 classes (75%), 86 orders (73%), 171 families (54%), and 286 genera (49%) (Figure S2).
At the phylum classification level, Ascomycota accounted for more than 50% of the total reads in both plants. In L. alba, the ascomycetes only showed differences in the relative abundances of Palmira (57% rhizosphere soil vs. 20% root). In contrast, for P. alliacea, dominant abundances were recorded in the roots of Pacho (55% vs. 65%), Ambalema (41% vs. 97%), and Villa de Leyva (68% vs. 93%), and the rhizosphere soils of Palmira (63% vs. 34%), and Armero (84% vs. 50%) (Figure 2). Additionally, Basidiomycota tended to be more abundant in rhizosphere soil samples at most sampling points, except in the P. alliacea samples from Palmira (9% soil vs. 14% root) and Armero (6% vs. 27%), where the highest abundance to roots was recorded (Figure 2). In contrast, Mortierellomycota showed the highest read assignation in the roots of L. alba from Palmira (30% soil vs. 78% root) and Sibundoy (2% vs. 19%) and in P. alliacea from Palmira (12% vs. 41%) and Pacho (2% vs. 11%). Rozellomycota, on the other hand, reached greater abundances in the root samples of L. alba from Sibundoy (0.3% vs. 8%), in P. alliacea roots from Pacho (4% vs. 15%) and Armero (1% vs. 7%), and in the rhizosphere soils of Ambalema (8% vs. 1%) and Villa de Leyva (5% vs. 1%) (Figure 2). Lastly, Chytridiomycota was predominantly abundant in the roots of Sibundoy (1% soil vs. 5% root) for L. alba and in the soils of Palmira (15% vs. 9%), Ambalema (7% vs. 0.1%), and Villa de Leyva (4% vs. 1%) for P. alliacea (Figure 2).
In terms of class, Sardariomycetes, Dothideomycetes, Leotiomycetes, and Eurotiomycetes were the Ascomycota categories with the highest number of assigned reads. Sardariomycetes had higher abundances in the rhizosphere soils of Palmira (36% soil vs. 7% root) and Lérida (23% vs. 19%) for L. alba, and in Palmira (32% vs. 20%), Armero (64% vs. 45%), and Villa de Leyva (56% vs. 15%) for P. allicaea (Figure S3). However, in Pacho (35% vs. 50%) and Ambalema (31% vs. 52%) of the same species, most reads were assigned to roots (Figure S3). Dothideomycetes in L. alba recorded higher reads in the rhizosphere soils of Palmira (19% soil vs. 9% root) and in the roots of Sibundoy (2% vs. 29%) and Lérida (19% vs. 35%), while P. alliacea had higher abundances in the soils of Palmira (30% vs. 13%) and roots of Ambalema (6% vs. 17%). Leotiomycetes, on the other hand, had most of its assignments in Sibundoy for L. alba with 31% in roots and 8% in soil. Meanwhile, the class Eurotiomycetes was highly dominant in Villa de Leyva for P. allicaea, with 77% abundance in roots compared to 8% in rhizosphere soil. On the other hand, Agaricomycetes from Basidiomycota consistently showed higher abundances in the soils of L. alba localities (32% in soil vs. 13% roots) and in P. allicaea from Pacho (38% vs. 7%), Ambalema (40% vs. 22%), and Villa de Leyva (13% vs. 0.3%), but higher in the roots of Armero (12% vs. 31%) (Figure S3). Conversely, Mortierellomycetes from Mortierellomycota tended to have a higher number of reads in the roots of Palmira and Sibundoy for L. alba and in Palmira (12% vs. 45%) and Pacho (2% vs. 14%) for P. alliacea (Figure S3).
The fungal genera that showed differential abundance in the L. alba plants from Palmira were Mortierella from Mortierellomycota and Periconia, Curvularia, and Tetracladium from Ascomycota (Figure S4d,e). In Sibundoy, differences were noted in Gorgomyces and Neptunomyces compared to Palmira (Figure S4d), while Lérida had no record of differentially abundant genera (Figure S4e,f). For the P. alliacea plants, the only comparisons that showed genera with differential abundances were Palmira versus Ambalema, Villa de Leyva versus Ambalema, and Villa de Leyva versus Pacho, with genera such as Trichoderma, Penicillium, Eleutherascus, and Mortierella (Figure S6).
3.2.3. Taxonomic Assignation of AMF to Medicinal Plants
In L. alba, Glomeromycetes was the most abundant class in Lérida and Sibundoy, with 78% and 76%, respectively (Figure S7). In Palmira, this class and Paraglomeromycetes maintained similar abundances, 53% and 47%, respectively (Figure S7). In P. allicaea, both Pacho and Villa de Leyva were dominated by Glomeromycetes, each with an abundance close to 100%. In contrast, Armero showed a higher proportion of Glomeromycetes at 78% compared to Paraglomeromycetes at 22% (Figure S7).
At the family level for L. alba, Glomeraceae and Paraglomeraceae accounted for 98.8% of the reads in Lérida, with an abundance of 76% for the former and 23% for the latter (Figure S7). In Sibundoy, these two families displayed a more even abundance, with 47% for Paraglomeraceae and 40% for Glomeraceae (Figure S7). Palmira was the only location where Diversisporaceae was highly represented, accounting for 30% of the abundance. In P. alliacea, Paraglomeraceae dominated Palmira with an abundance of 67% compared to 33% of Glomeraceae (Figure S7). In contrast, Armero had a higher abundance of Glomeraceae at 74% compared to Paraglomeraceae at 26%. Both Pacho and Villa de Leyva showed readings predominantly assigned to Glomeraceae, with values close to 100% (Figure S7).
For the genera associated with L. alba, Glomus, and Paraglomus dominated in Lérida with 76% and 23%, respectively (Figure S7). In contrast, the roots from Palmira showed a majority abundance towards Paraglomus at 47% and Glomus at 40% (Figure S7). Additionally, in Sibundoy there was an even distribution in the abundances of the genera: Glomus with 28%, Paraglomus with 24%, and Diversispora with 29% (Figure S7). For P. alliacea, Palmira was the only location where Paraglomus displayed greater abundance at 67% compared to Glomus with 33%. As such, Armero, Pacho, and Villa de Leyva all exhibited dominance of Glomus (Figure S7).
3.3. Diversity Analysis by Sample Type and Medicinal Plants
The alpha diversity results for L. alba’s Shannon index revealed that for most of the rhizosphere soil and root samples from the three locations, bacteria displayed a diversity range between 5 and 6. The sole exception was the soil from Lérida, which had lower values due to the dominance of the phyla Proteobacteria and Actinobacteriota (Figure 3a). For fungi, most of the samples had Shannon values hovering around 2 and 3, except for root samples from Sibundoy and soil from Lérida, which exceeded 3 (Figure 3c). Additionally, the Berger–Parker dominance index for L. alba bacteria highlighted that the soil samples from Sibundoy exhibited the highest dominance, whereas, for fungi, it was the rhizosphere soil and root samples from Palmira.

Figure 3.
Comparative representation of Shannon Diversity and Berger–Parker Dominance indices for bacteria (panels (a,b,e,f)) and fungi (panels (c,d,g,h)) in the rhizosphere soil and root samples of L. alba and P. alliacea.
For P. alliacea, the Shannon results for bacteria revealed diversity differences for most study locations based on the sample type (Figure 3b). The roots from Pacho, Armero, and Villa de Leyva showed higher diversities compared to their corresponding rhizosphere soil samples. Only the soil from Ambalema registered higher diversity than its root (Figure 3b). In the case of fungi, diversity values were recorded around three for most locations, except for the soil from Ambalema and the root from Villa de Leyva, which had values lower or close to two (Figure 3d). Moreover, the dominance index for bacteria was mostly close to zero, reflecting the low dominance of species (Figure 3f). Meanwhile, fungi maintained high dominance in those samples where low diversity was recorded (Figure 3h).
In terms of beta diversity, the PCoA for bacteria in L. alba shows that samples from the three sampling points tended to cluster together, with greater differences noted between the rhizosphere soil and root samples from Sibundoy and Lérida (Figure 4a). Regarding fungi, L. alba displayed sample clustering tendencies by location, where Palmira was mainly positioned on the positive axis 1, while Lérida and Sibundoy were on the negative side (Figure 4b). In Palmira, however, a separation on axis 2 was observed for three soil samples. For P. allicacea’s bacteria, Palmira was the only location where rhizosphere soil and root appeared close in the ordination, as the other locations showed mixed groupings of locations and sample types (Figure 4c). Similarly, for fungi in P. alliacea, it was evident that four soil root samples from Palmira managed to maintain differentiation from samples that mainly grouped on the positive side of axis 1 (Figure 4d).
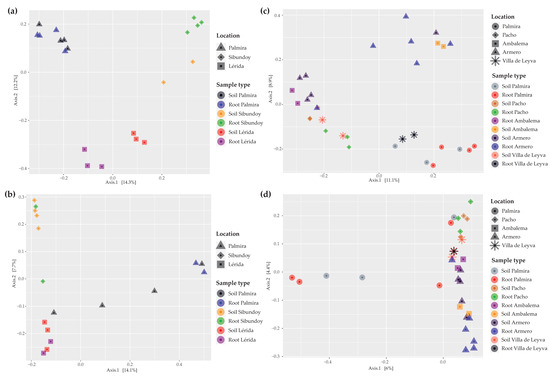
Figure 4.
PCoA by Bray–Curtis distances of bacteria (a,c) and fungi (b,d) associated with medicinal plants L. alba (a,b) and P. alliacea (c,d).
3.4. Multiple Factor Analysis for Bacteria and Fungi
The results of the physicochemical soil parameters along with the most abundant genera of bacteria and fungi were analyzed in multiple component graphs (Figure 5). In the MFA for L. alba bacteria, dimension 1 accounted for 30.1% of the variance, separating most of the samples from Valle del Cauca from Tolima and Putumayo, with high contributions from the genera Acinetobacter (9.69) and Pseudomonas (9.69) and measurements of Fe (7.22), ECEC (7.16), and texture (7.06) (Figure 5a and Table S3). The second dimension, accounting for 20.9% of the variance, had the highest contributions in bacteria RB41 (27.69) and Candidatus Udaeobacter (23.93), and the measurement of Ca (1.11) (Figure 5a and Table S3). In the case of P. alliacea, Steroidobacter (10.48), Lysobacter (8.35), pH (6.78), and P (6.34) reported the highest contributions for dimension 1, which separated the Valle del Cauca and Boyacá samples from those of Cundinamarca and some from Tolima (Figure 5c and Table S3). Haliangium (16.25), Nitrospira (15.26), and S (3.99) from dimension 2 marked the highest contributions (Figure 5c and Table S3).
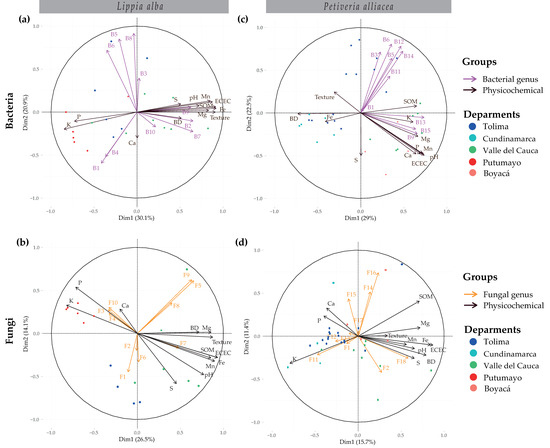
Figure 5.
Multiple factor analysis (MFA) for bacteria (a,c) and fungi (b,d), together with physicochemical parameters for the plants L. alba (a,b) and P. alliacea (c,d). Bacterial genus: B1: Acidibacter, B2: Acinetobacter, B3: Bacillus, B4: Bradyrhizobium, B5: Candidatus Udaeobacter, B6: Haliangium, B7: Pseudomonas, B8: RB41, B9: Sphingomonas, B10: Streptomyces, B11: MND1, B12: Nitrospira, B13: Steroidobacter, B14: Candidatus Latescibacter, B15: Lysobacter. Fungal genus: F1: Mortierella, F2: Pyrenochaetopsis, F3: Lectera, F4: Leucocoprinus, F5: Coprinellus, F6: Gorgomyces, F7: Ilyonectria, F8: Aspergillus, F9: Pseudopithomyces, F10: Plectosphaerella, F11: Cercophora, F12: Paecilomyces, F13: Apiotrichum, F14: Parasola, F15: Clitopilus, F16: Fusarium, F17: Trichoderma, F18: Simplicillium.
In terms of fungi, for L. alba, dimension 1 separated the Valle del Cauca samples from those of Tolima and Putumayo, with the largest contributions from the genera Coprinellus (12.32) and Pseudopithomyces (11.4), and the parameters ECEC (6.54) and Mn (6.05) (Figure 5b and Table S3). In the second dimension, Coprinellus (20.23) and Mortierella (10.78), along with pH (3.25) and S (4.81), marked the highest contributions (Figure 5b and Table S3). For P. alliacea, dimension 1 recorded the greatest contributions from the fungal genera Simplicillium (17.1) and Cercophora (11.37), and the measurements of ECEC (8.28) and Fe (7.76) (Figure 5b and Table S3). In dimension 2, Fusarium (37.94), Parasola (18.48), and Pyrenochaetopsis (12.37), along with SOM (2.51), presented the highest contributions (Figure 5b and Table S3).
4. Discussion
In this study, soil microorganisms from both rhizosphere soil and roots were analyzed in medicinal plants of interest to Colombia. Our primary findings suggest and demonstrate differences in the structure and diversity of bacteria, fungi, and AMF in the L. alba and P. alliacea plants. These plants were found in municipalities throughout Colombia where variations were recorded in the physicochemical measurements of the soil, reflecting the diversity of environmental microorganisms (Table S1). Notably, the pH showed average values per location as either acidic or basic, which allows us to infer plant adaptations, with implications for nutrient availability, microorganism adsorption, and cellular growth [34]. Thus, the microbiome in Sibundoy, Ambalema, Armero, and Pacho might indicate differences compared to other locations due to their acidic conditions.
At the bacterial level, the results found here showed the same phylogenetic taxon groups in both rhizosphere soils and roots, but with marked differences in abundance between the sample types. Proteobacteria was the phylum with the most readings in both plants and had the highest number of differentially abundant genera reported among the sampling locations, which is consistent with previous records in the medicinal plant Fritillaria thunbergii [35]. This phylum has been found enriched in the rhizosphere soil of the medicinal plant Astragalus mongholicus [6] and the oak [36], which is consistent with our abundance results for most of the locations sampled in Colombia. The differences in sample type might be attributed to the abundance of bacteria that prefer copiotroph environments, with the highest abundances of taxa being found in environments with high availability of energy resources and labile carbon [37,38]. Additionally, Acidobacteriota was more abundantly present in the roots of both medicinal plants and may be associated with the decomposition processes of carbon elements, facilitating optimal nutrient recycling rates [39]. The previous research suggests that plant exudates alter the pH in the rhizosphere soil and the roots, influencing the ecological dynamics of Acidobacteriota in these environments [40,41].
In fungi, dominances were reported in certain rhizosphere soil and root locations of the phylum Ascomycota. This phylum is considered one of the key soil groups due to its role as a decomposer of organic matter in soils and for the assimilation of exudates in the rhizospheres, which is why it can be highly abundant in both types of samples as was found in this study [42]. The differences in fungal abundance across sample types are primarily due to the organisms’ preferences for metabolizing rhizosphere exudates or decomposing plant material. While most Ascomycota taxa can be involved in the decomposition of plant litter, some fungi, such as those from the Sordariomycetes class, can also derive energy from consuming root exudates [43,44]. Therefore, it’s possible that in the roots of P. alliacea in Ambalema and Villa de Leyva, the dominance of ascomycetes emerged due to the generation of specific functions associated with the roots.
On the other hand, the Basidiomycota phylum was found to be more abundant in rhizosphere soils compared to the roots, which is consistent with previous findings that suggest this group of fungi is favored in environments with high concentrations of degrading plant material since some taxa achieve complete degradation of lignin [45,46]. Additionally, Mortierella from the Mortierellomycota phylum may be highly abundant in the roots from Palmira because this genus is associated with easily accessible carbon sources such as glucose proportionated as an energy source for the plants [47].
Also, both medicinal plants were reported to have AMF groups colonizing their roots, but with clear differences in taxonomic abundance. The AMF report for L. alba and P. alliacea is consistent with the associations these fungi maintain with at least 17 species of medicinal plants, with Glomus being one of the genera most frequently found in multiple roots [48]. This taxon is recognized for its broad versatility, which gives it the ability to colonize various plant roots without needing a specific affinity. Additionally, the higher presence of Paraglomus in the soils of Palmira for both plants could be linked to factors of the plant growth environment since the genus has been associated with soils rich in plant diversity and forested areas [49,50]. Moreover, the higher abundance of Diversispora in Sibundoy for L. alba, compared to other sampling points, could be associated with reduced precipitation levels, which have previously been described as a factor for the genus’s high abundance and potentially diminishing the dominance of Glomus [51].
At the diversity level, we observed that the Shannon index indicated certain trends at various sampling points, showing higher values in rhizosphere soils or roots, or presenting similar numbers for both types of samples (Figure 3). Our findings may be influenced by certain factors previously reported in the literature. In an analysis of community complexity, studies have demonstrated that microbial assemblages in rhizosphere soils exhibit greater complexity compared to those in root environments, a phenomenon attributable to the selective pressures exerted by plant root systems on resident microorganisms [52,53]. However, several factors can influence the diversity trends between these two types of samples [6,42]. Some predominant factors include soil temperature, moisture, pH, and chemicals secreted by roots [54,55]. These results provide a deeper understanding of the ecological dynamics of the rhizosphere and roots, emphasizing the importance of considering a broad range of environmental and microbial factors in future research.
5. Conclusions
This research evaluated the differences in composition and diversity of the rhizosphere soil and roots microbiome of the medicinal plants L. alba and P. alliacea. The findings suggest that the microbial structure in both medicinal plants showed groups of bacteria, fungi, and AMF that predominated the sample types, but with variations in their abundances at different sampling points and plant species in Colombia. Some bacterial groups, such as Proteobacteria and Acidobacteriota, and fungal groups like Ascomycota, Basidiomycota, and Mortierellomycota, as well as AMF genera like Glomus and Paraglomus, showed the greatest differences in abundance. Consequently, variations in compositional makeup exhibited differences in alpha and beta diversity metrics for both plant types, potentially elucidating the distinct microbial community structures driven by plant species and geographic locations. Future efforts should consider the relationships between microorganisms and the medicinal metabolic characteristics of the plants.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15121167/s1. Figure S1: Venn diagrams for bacteria (a–f) and fungi (g–l) for the medicinal plant L. alba; Figure S2: Venn diagrams for bacteria (a–f) and fungi (g–l) for the medicinal plant P. alliacea; Figure S3: Taxonomic composition at the class level for bacteria (a,b) and fungi (c,d) associated with the soil and roots of the medicinal plants L. alba (a–c) and P. alliacea (b–d); Figure S4: Differential abundance plots at the genus level for bacteria (a–c) and fungi (d–f) from the L. alba plant; Figure S5: Differential abundance plots at the genus level for bacteria from the P. alliacea plant; Figure S6: Differential abundance plots at the genus level for fungi from the P. alliacea plant; Figure S7: Taxonomic composition at the class, family and genus level for AMF associated with the roots of the medicinal plants L. alba (a–c) and P. alliacea (b–f); Table S1: Sample locations and results of physicochemical parameter measurements for soils in four departments of Colombia; Table S2: Kruskal-Wallis test of the soil’s physicochemical parameters. Values are presented as averages per location with their respective standard deviation. Table S3: Contributions of the dimensional variables in the multiple factor analyses for bacteria and fungi associated with L. alba and P. alliacea.
Author Contributions
All authors participated in the conception of the idea; S.A.D.G., L.A.D.A., J.E.M.F. and D.L.-Á. designed the methodology; S.A.D.G. and L.A.D.A. compiled the data; G.A.V.-M., J.D.D.-Z., W.L.R.-A. and D.L.-Á. analyzed the data; G.A.V.-M., J.D.D.-Z. and W.L.R.-A. led the writing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was conducted with the financial support of Pontificia Universidad Javeriana, Ministerio de Ciencia, Tecnología e Innovación, Ministerio de Educación Nacional, Ministerio de Industria, Comercio y Turismo e ICETEX, 2a Convocatoria Ecosistema científico—Colombia Científica 792-2017, Programa “Generación de alternativas terapéuticas en cáncer a partir de plantas a través de procesos de investigación y desarrollo traslacional, articulados en sistemas de valor sostenibles ambiental y económicamente” (Contrat no. FP44842-221-2018).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Acknowledgments
The authors thank the Ministerio de Ambiente y Desarrollo Sostenible for allowing the use of genetic resources and derivative products (CARG no. 212/2018; Resolution 210/2020), Agencia Nacional de Licencias Ambientales (ANLA) for the technical concept N° 5445, within the file NTC0204-00-2019.
Conflicts of Interest
The authors declare that they have no known competing commercial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Lee, S.-J.; Kong, M.; St-Arnaud, M.; Hijri, M. Arbuscular Mycorrhizal Fungal Communities of Native Plant Species under High Petroleum Hydrocarbon Contamination Highlights Rhizophagus as a Key Tolerant Genus. Microorganisms 2020, 8, 872. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Peng, T.; Liu, X.; Wang, H.; Huang, T.; Gu, J.-D.; Hu, Z. Ecological Role of Bacteria Involved in the Biogeochemical Cycles of Mangroves Based on Functional Genes Detected through GeoChip 5.0. mSphere 2022, 7, e00936-21. [Google Scholar] [CrossRef] [PubMed]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Ismail, S.; Nasrulhaq Boyce, A. Role of Plant Growth Promoting Rhizobacteria in Agricultural Sustainability—A Review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef]
- Banerjee, S.; van der Heijden, M.G.A. Soil Microbiomes and One Health. Nat. Rev. Microbiol. 2023, 21, 6–20. [Google Scholar] [CrossRef]
- Busby, R.R.; Barbato, R.A.; Jung, C.M.; Bednar, A.J.; Douglas, T.A.; Ringelberg, D.B.; Indest, K.J. Alaskan Plants and Their Assembled Rhizosphere Communities Vary in Their Responses to Soil Antimony. Appl. Soil Ecol. 2021, 167, 104031. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.; Wu, T.; Zhang, H.; Wei, G.; Li, Z. Rhizosphere Bacterial and Fungal Spatial Distribution and Network Pattern of Astragalus Mongholicus in Representative Planting Sites Differ the Bulk Soil. Appl. Soil Ecol. 2021, 168, 104114. [Google Scholar] [CrossRef]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion. Front. Plant Sci. 2020, 10, 1741. [Google Scholar] [CrossRef]
- Zhang, W.; Mason, G.A. Modulating the Rhizosphere Microbiome by Altering the Cocktail of Root Secretions. Plant Physiol. 2022, 188, 12–13. [Google Scholar] [CrossRef]
- Wu, L.; Wang, J.; Huang, W.; Wu, H.; Chen, J.; Yang, Y.; Zhang, Z.; Lin, W. Plant-Microbe Rhizosphere Interactions Mediated by Rehmannia Glutinosa Root Exudates under Consecutive Monoculture. Sci. Rep. 2015, 5, 15871. [Google Scholar] [CrossRef]
- Amani Machiani, M.; Javanmard, A.; Habibi Machiani, R.; Sadeghpour, A. Arbuscular Mycorrhizal Fungi and Changes in Primary and Secondary Metabolites. Plants 2022, 11, 2183. [Google Scholar] [CrossRef]
- Goodwin, P.H. The Rhizosphere Microbiome of Ginseng. Microorganisms 2022, 10, 1152. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Yang, J.; Zhao, S.; Yu, Q.; Weng, L.; Xiao, C. Root Exudates Influence Rhizosphere Fungi and Thereby Synergistically Regulate Panax Ginseng Yield and Quality. Front. Microbiol. 2023, 14, 1194224. [Google Scholar] [CrossRef] [PubMed]
- Castañeda, R.; Cáceres, A.; Velásquez, D.; Rodríguez, C.; Morales, D.; Castillo, A. Medicinal Plants Used in Traditional Mayan Medicine for the Treatment of Central Nervous System Disorders: An Overview. J. Ethnopharmacol. 2022, 283, 114746. [Google Scholar] [CrossRef] [PubMed]
- Montero-Villegas, S.; Crespo, R.; Rodenak-Kladniew, B.; Castro, M.A.; Galle, M.; Cicció, J.F.; García de Bravo, M.; Polo, M. Cytotoxic Effects of Essential Oils from Four Lippia Alba Chemotypes in Human Liver and Lung Cancer Cell Lines. J. Essent. Oil Res. 2018, 30, 167–181. [Google Scholar] [CrossRef]
- Morais, S.M.D.; Sobrinho, A.C.N.; Liberato, H.R.; Pereira, R.D.C.A.; Pessoa, C.; Alves, D.R.; Fontenelle, R.O.D.S. Biotechnological potential of essential oils from different chemotypes of Lippia alba(Mill.) N.E.Br. ex Britton & P. Wilson. Boletín Latinoam. Y Caribe Plantas Med. Y Aromáticas 2022, 21, 725–736. [Google Scholar] [CrossRef]
- García, L.T.; Leal, A.F.; Moreno, É.M.; Stashenko, E.E.; Arteaga, H.J. Differential Anti-Proliferative Effect on K562 Leukemia Cells of Lippia Alba (Verbenaceae) Essential Oils Produced under Diverse Growing, Collection and Extraction Conditions. Ind. Crops Prod. 2017, 96, 140–148. [Google Scholar] [CrossRef]
- Ortiz, N.; Jiménez, M.F.; Chaverri, C.; Cicció, J.F.; Díaz, C. Effect on Cell Growth, Viability and Migration of Geraniol and Geraniol-Containing Essential Oil from Lippia Alba (Verbenaceae) on Gastric Carcinoma Cells. J. Essent. Oil Res. 2022, 34, 65–76. [Google Scholar] [CrossRef]
- Hernández, J.F.; Urueña, C.P.; Cifuentes, M.C.; Sandoval, T.A.; Pombo, L.M.; Castañeda, D.; Asea, A.; Fiorentino, S. A Petiveria Alliacea Standardized Fraction Induces Breast Adenocarcinoma Cell Death by Modulating Glycolytic Metabolism. J. Ethnopharmacol. 2014, 153, 641–649. [Google Scholar] [CrossRef]
- Murillo, N.; Lasso, P.; Urueña, C.; Pardo-Rodriguez, D.; Ballesteros-Ramírez, R.; Betancourt, G.; Rojas, L.; Cala, M.P.; Fiorentino, S. Petiveria Alliacea Reduces Tumor Burden and Metastasis and Regulates the Peripheral Immune Response in a Murine Myeloid Leukemia Model. Int. J. Mol. Sci. 2023, 24, 12972. [Google Scholar] [CrossRef]
- Vaghela, N.; Gohel, S. Medicinal Plant-Associated Rhizobacteria Enhance the Production of Pharmaceutically Important Bioactive Compounds under Abiotic Stress Conditions. J. Basic Microbiol. 2023, 63, 308–325. [Google Scholar] [CrossRef]
- Rizvi, A.; Ahmed, B.; Khan, M.S.; El-Beltagi, H.S.; Umar, S.; Lee, J. Bioprospecting Plant Growth Promoting Rhizobacteria for Enhancing the Biological Properties and Phytochemical Composition of Medicinally Important Crops. Molecules 2022, 27, 1407. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-S.; Liu, J.-M.; Sun, J.; Huang, Y.-T.; Jin, N.; Li, M.-M.; Liang, Y.-T.; Fan, B.; Wang, F.-Z. Analysis of Endophytic Bacterial Diversity From Different Dendrobium Stems and Discovery of an Endophyte Produced Dendrobine-Type Sesquiterpenoid Alkaloids. Front. Microbiol. 2022, 12, 775665. [Google Scholar] [CrossRef] [PubMed]
- The Jamovi Project Jamovi. Available online: https://www.jamovi.org/ (accessed on 2 October 2023).
- Doyle, J.J.; Doyle, J. Isolation of Plant DNA from Fresh Tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S rRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS Primers with Enhanced Specificity for Basidiomycetes—Application to the Identification of Mycorrhizae and Rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J.; Innis, M.; Gelfand, D.; Sninsky, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In Pcr Protocols: A Guide to Methods and Applications; Academic Press, Inc.: New York, NY, USA, 1990; Volume 31, pp. 315–322. [Google Scholar]
- Morgan, B.S.T.; Egerton-Warburton, L.M. Barcoded NS31/AML2 Primers for Sequencing of Arbuscular Mycorrhizal Communities in Environmental Samples. Appl. Plant Sci. 2017, 5, 1700017. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Öpik, M.; Vanatoa, A.; Vanatoa, E.; Moora, M.; Davison, J.; Kalwij, J.M.; Reier, Ü.; Zobel, M. The Online Database MaarjAM Reveals Global and Ecosystemic Distribution Patterns in Arbuscular Mycorrhizal Fungi (Glomeromycota). New Phytol. 2010, 188, 223–241. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Jackson, R.B. The Diversity and Biogeography of Soil Bacterial Communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.-Y.; Yuan, X.-F.; Lin, H.-R.; Yang, Y.-Q.; Li, Z.-Y. Differences in Soil Properties and Bacterial Communities between the Rhizosphere and Bulk Soil and among Different Production Areas of the Medicinal Plant Fritillaria Thunbergii. Int. J. Mol. Sci. 2011, 12, 3770–3785. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Buée, M.; Murat, C.; Frey-Klett, P.; Martin, F. Pyrosequencing Reveals a Contrasted Bacterial Diversity between Oak Rhizosphere and Surrounding Soil. Environ. Microbiol. Rep. 2010, 2, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Ni, T.; Xun, W.; Huang, X.; Huang, Q.; Ran, W.; Shen, B.; Zhang, R.; Shen, Q. Influence of Straw Incorporation with and without Straw Decomposer on Soil Bacterial Community Structure and Function in a Rice-Wheat Cropping System. Appl. Microbiol. Biotechnol. 2017, 101, 4761–4773. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an Ecological Classification of Soil Bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Ward, N.L.; Challacombe, J.F.; Janssen, P.H.; Henrissat, B.; Coutinho, P.M.; Wu, M.; Xie, G.; Haft, D.H.; Sait, M.; Badger, J.; et al. Three Genomes from the Phylum Acidobacteria Provide Insight into the Lifestyles of These Microorganisms in Soils. Appl. Environ. Microbiol. 2009, 75, 2046–2056. [Google Scholar] [CrossRef]
- Sait, M.; Davis, K.E.R.; Janssen, P.H. Effect of pH on Isolation and Distribution of Members of Subdivision 1 of the Phylum Acidobacteria Occurring in Soil. Appl. Environ. Microbiol. 2006, 72, 1852–1857. [Google Scholar] [CrossRef]
- Navarrete, A.A.; Kuramae, E.E.; de Hollander, M.; Pijl, A.S.; van Veen, J.A.; Tsai, S.M. Acidobacterial Community Responses to Agricultural Management of Soybean in Amazon Forest Soils. FEMS Microbiol. Ecol. 2013, 83, 607–621. [Google Scholar] [CrossRef]
- Hugoni, M.; Luis, P.; Guyonnet, J.; Haichar, F. el Z. Plant Host Habitat and Root Exudates Shape Fungal Diversity. Mycorrhiza 2018, 28, 451–463. [Google Scholar] [CrossRef]
- Zhang, N.; Castlebury, L.; Miller, A.; Huhndorf, S.M.; Schoch, C.; Seifert, K.; Rossman, A.; Rogers, J.; Kohlmeyer, J.; Volkmann-Kohlmeyer, B.; et al. An Overview of the Systematics of the Sordariomycetes Based on a Four-Gene Phylogeny. Mycologia 2006, 98, 1076–1087. [Google Scholar] [CrossRef] [PubMed]
- Buée, M.; De Boer, W.; Martin, F.; van Overbeek, L.; Jurkevitch, E. The Rhizosphere Zoo: An Overview of Plant-Associated Communities of Microorganisms, Including Phages, Bacteria, Archaea, and Fungi, and of Some of Their Structuring Factors. Plant Soil 2009, 321, 189–212. [Google Scholar] [CrossRef]
- Moll, J.; Hoppe, B.; König, S.; Wubet, T.; Buscot, F.; Krüger, D. Spatial Distribution of Fungal Communities in an Arable Soil. PLoS ONE 2016, 11, e0148130. [Google Scholar] [CrossRef] [PubMed]
- Baldrian, P.; Voříšková, J.; Dobiášová, P.; Merhautová, V.; Lisá, L.; Valášková, V. Production of Extracellular Enzymes and Degradation of Biopolymers by Saprotrophic Microfungi from the Upper Layers of Forest Soil. Plant Soil 2011, 338, 111–125. [Google Scholar] [CrossRef]
- Van der Wal, A.; Geydan, T.D.; Kuyper, T.W.; de Boer, W. A Thready Affair: Linking Fungal Diversity and Community Dynamics to Terrestrial Decomposition Processes. FEMS Microbiol. Rev. 2013, 37, 477–494. [Google Scholar] [CrossRef]
- Bhargav, V.; Kumar, A.; Dhiman, H.; Aggarwal, A. Biodiversity of Vesicular-Arbuscular Mycorrhiza in the Rhizosphere of Some Medicinal Plants of Kurukshetra University. Res. J. Biotechnol. 2023, 18, 24–30. [Google Scholar] [CrossRef]
- Peña-Venegas, C.P.; Sterling, A.; Andrade-Ramírez, T.K. Arbuscular Mycorrhization in Colombian and Introduced Rubber (Hevea brasiliensis) Genotypes Cultivated on Degraded Soils of the Amazon Region. Agriculture 2021, 11, 361. [Google Scholar] [CrossRef]
- Marinho, F.; Ramalho da Silva, I.; Oehl, F.; Maia, L. Checklist of Arbuscular Mycorrhizal Fungi in Tropical Forests. Sydowia 2018, 70, 107–127. [Google Scholar] [CrossRef]
- Zheng, Z.; Ma, X.; Zhang, Y.; Liu, Y.; Zhang, S. Soil Properties and Plant Community-Level Traits Mediate Arbuscular Mycorrhizal Fungal Response to Nitrogen Enrichment and Altered Precipitation. Appl. Soil Ecol. 2022, 169, 104245. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The Rhizosphere Microbiome: Significance of Plant Beneficial, Plant Pathogenic, and Human Pathogenic Microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Richardson, A.E.; Kawasaki, A.; Condron, L.M.; Ryan, P.R.; Gupta, V.V.S.R. Root Microbiome Structure and Microbial Succession in the Rhizosphere. In Rhizosphere Biology: Interactions between Microbes and Plants; Gupta, V.V.S.R., Sharma, A.K., Eds.; Rhizosphere Biology; Springer: Singapore, 2021; pp. 109–128. ISBN 9789811561252. [Google Scholar]
- Cui, Y.; Bing, H.; Fang, L.; Wu, Y.; Yu, J.; Shen, G.; Jiang, M.; Wang, X.; Zhang, X. Diversity Patterns of the Rhizosphere and Bulk Soil Microbial Communities along an Altitudinal Gradient in an Alpine Ecosystem of the Eastern Tibetan Plateau. Geoderma 2019, 338, 118–127. [Google Scholar] [CrossRef]
- Misra, P.; Maji, D.; Awasthi, A.; Pandey, S.S.; Yadav, A.; Pandey, A.; Saikia, D.; Babu, C.S.V.; Kalra, A. Vulnerability of Soil Microbiome to Monocropping of Medicinal and Aromatic Plants and Its Restoration through Intercropping and Organic Amendments. Front. Microbiol. 2019, 10, 2604. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).