Characterization of the Root Nodule Microbiome of the Exotic Tree Falcataria falcata (Fabaceae) in Guangdong, Southern China
Abstract
1. Introduction
2. Materials and Methods
2.1. Nodule Collection
2.2. Nodule Surface Sterilisation
2.3. 16S rRNA Gene Amplification and PacBio Sequencing
2.4. Sequence Data Analyses
3. Results
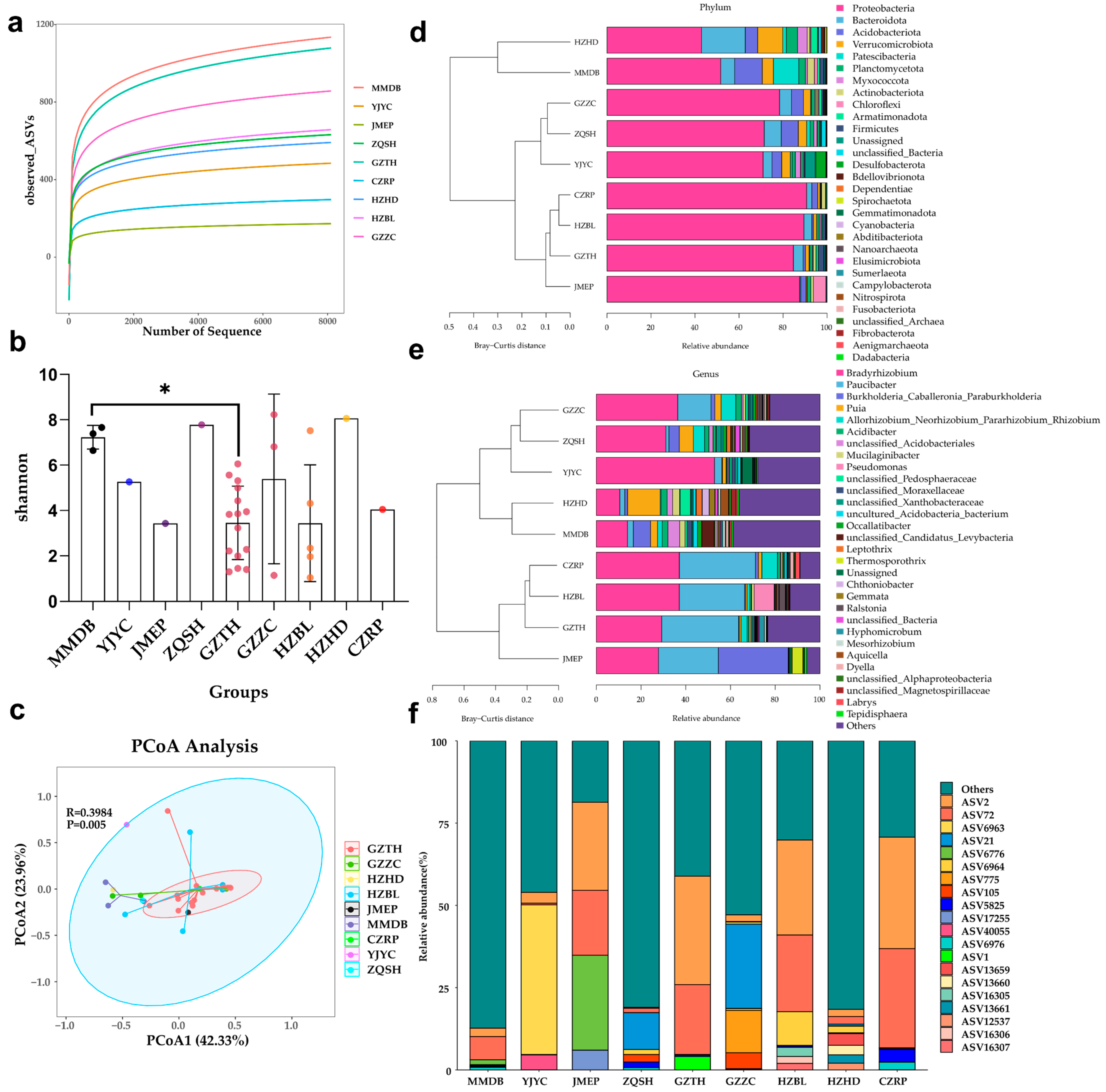
3.1. Microbial Diversity and Microbiome Composition of the Root Nodules
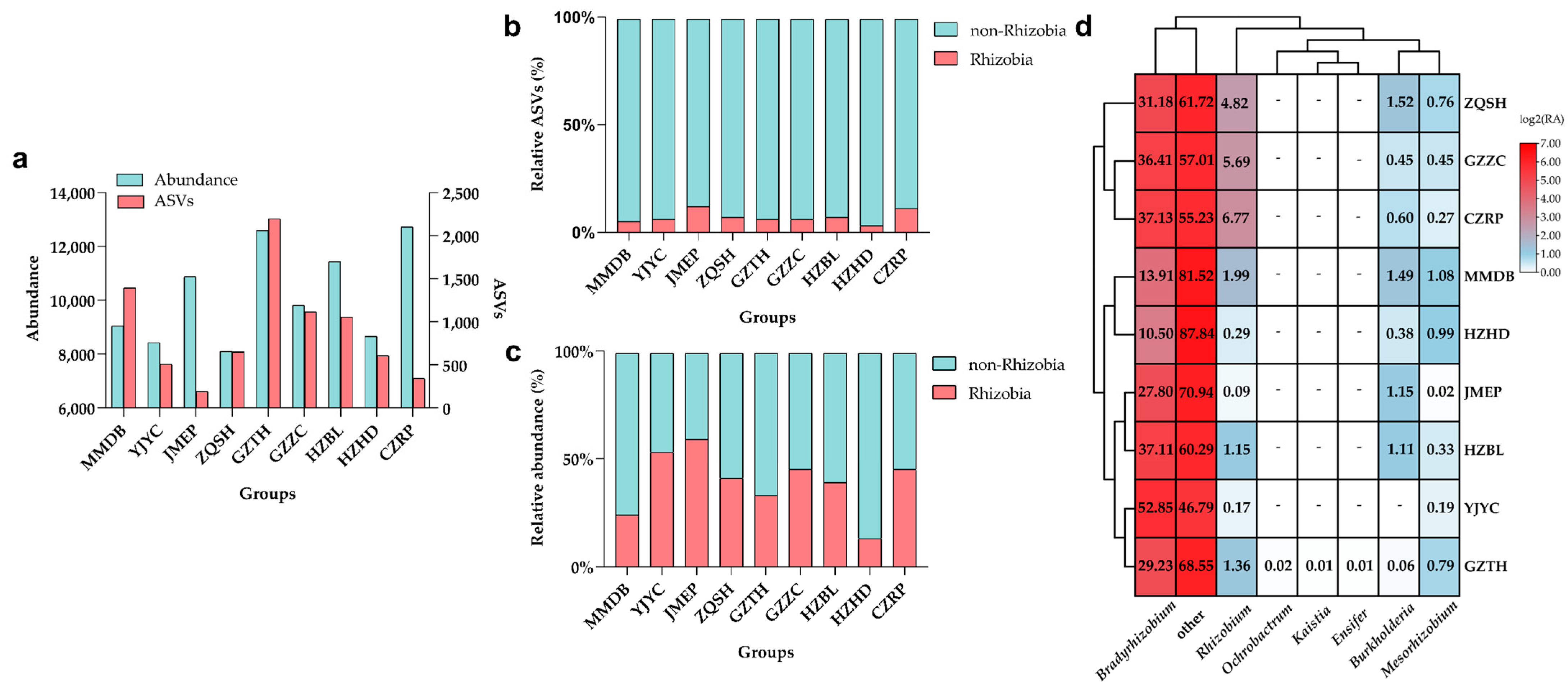
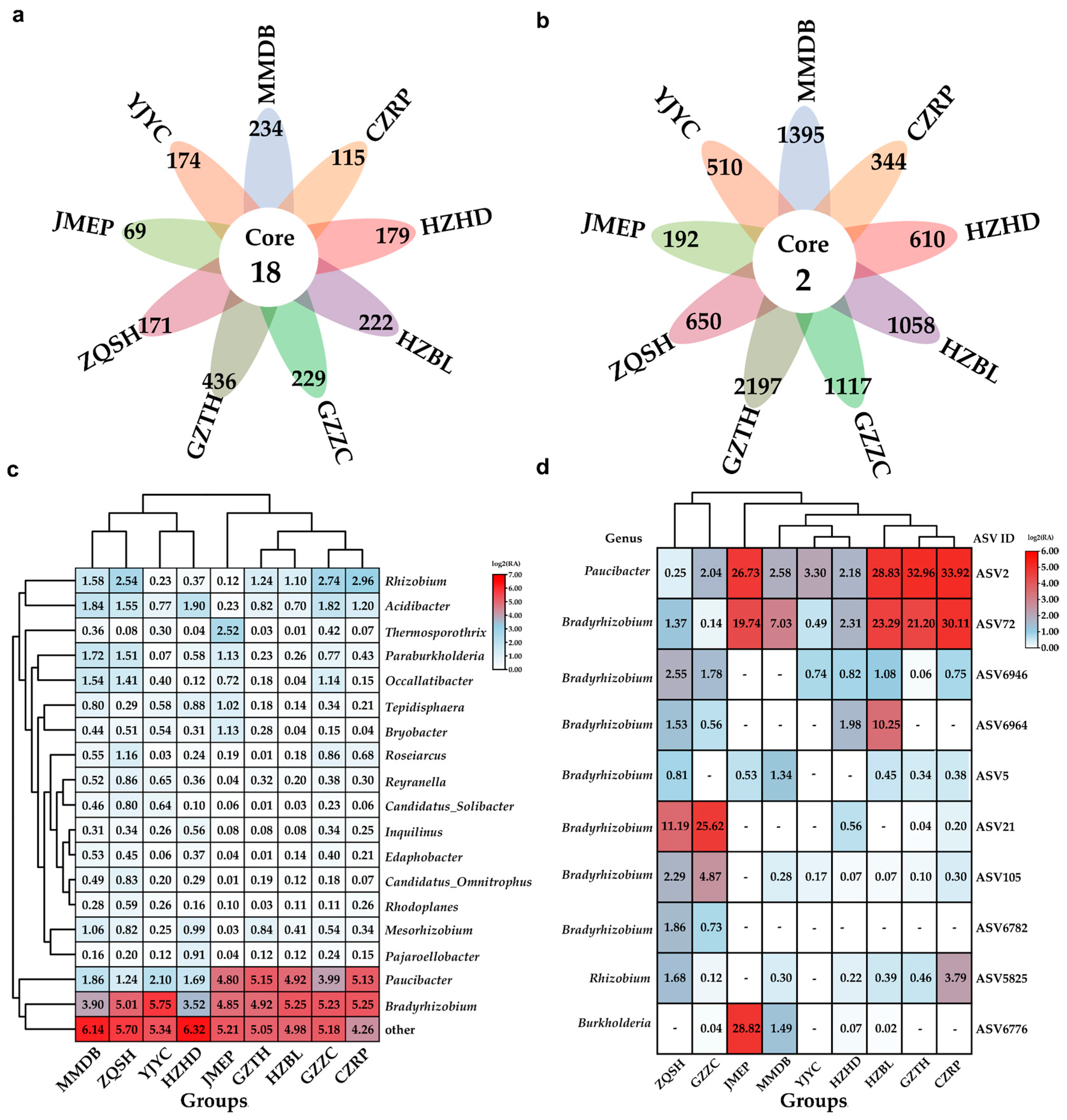
3.2. The Dominant Endophytic Rhizobia for F. falcata Root Nodules
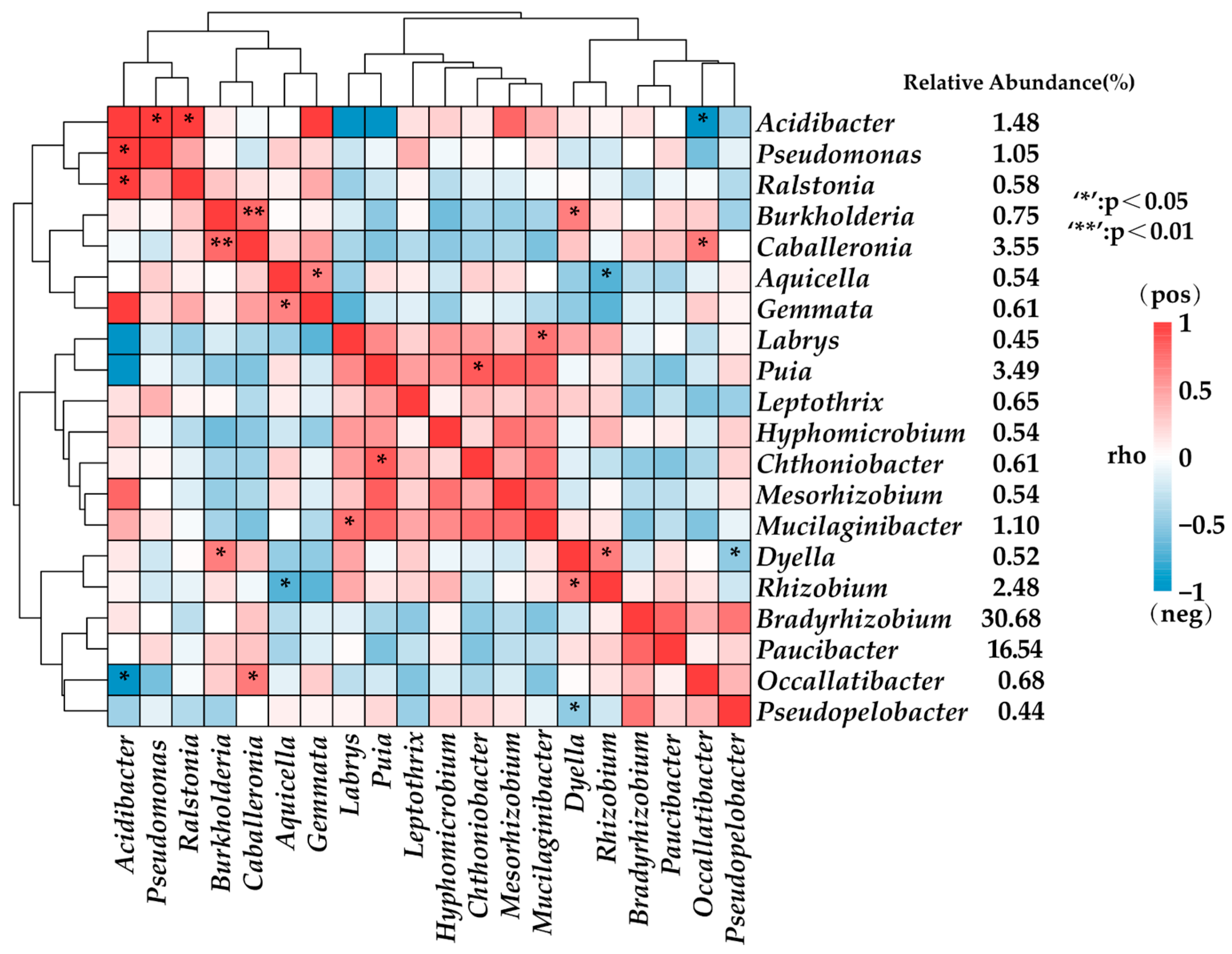
3.3. Limited Symbiotic Correlations between Rhizobia and Other Endophytic Bacteria in Root Nodules
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Kondorosi, E.; Mergaert, P.; Kereszt, A. A Paradigm for Endosymbiotic Life: Cell Differentiation of Rhizobium Bacteria Provoked by Host Plant Factors. Annu. Rev. Microbiol. 2013, 67, 611–628. [Google Scholar] [CrossRef] [PubMed]
- Vitousek, P.M.; Menge, D.N.L.; Reed, S.C.; Cleveland, C.C. Biological nitrogen fixation: Rates, patterns and ecological controls in terrestrial ecosystems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20130119. [Google Scholar] [CrossRef] [PubMed]
- Gage, D.J. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 2004, 68, 280–300. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Sprent, J.I.; Ardley, J.; James, E.K. Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytol. 2017, 215, 40–56. [Google Scholar] [CrossRef]
- Andrews, M.; Andrews, M.E. Specificity in Legume-Rhizobia Symbioses. Int. J. Mol. Sci. 2017, 18, 705. [Google Scholar] [CrossRef]
- Ngom, A.; Nakagawa, Y.; Sawada, H.; Tsukahara, J.; Wakabayashi, S.; Uchiumi, T.; Nuntagij, A.; Kotepong, S.; Suzuki, A.; Higashi, S.; et al. A novel symbiotic nitrogen-fixing member of the Ochrobactrum clade isolated from root nodules of Acacia mangium. J. Gen. Appl. Microbiol. 2004, 50, 17–27. [Google Scholar] [CrossRef]
- Adhikari, D.; Kaneto, M.; Itoh, K.; Suyama, K.; Pokharel, B.B.; Gaihre, Y.K. Genetic diversity of soybean-nodulating rhizobia in Nepal in relation to climate and soil properties. Plant Soil 2012, 357, 131–145. [Google Scholar] [CrossRef]
- Han, L.L.; Wang, E.T.; Han, T.X.; Liu, J.; Sui, X.H.; Chen, W.F.; Chen, W.X. Unique community structure and biogeography of soybean rhizobia in the saline-alkaline soils of Xinjiang, China. Plant Soil 2009, 324, 291–305. [Google Scholar] [CrossRef]
- Sharaf, H.; Rodrigues, R.R.; Moon, J.; Zhang, B.; Mills, K.; Williams, M.A. Unprecedented bacterial community richness in soybean nodules vary with cultivar and water status. Microbiome 2019, 7, 63. [Google Scholar] [CrossRef] [PubMed]
- Shiro, S.; Matsuura, S.; Saiki, R.; Sigua, G.C.; Yamamoto, A.; Umehara, Y.; Hayashi, M.; Saeki, Y. Genetic Diversity and Geographical Distribution of Indigenous Soybean-Nodulating Bradyrhizobia in the United States. Appl. Environ. Microbiol. 2013, 79, 3610–3618. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- O’Brien, S.L.; Gibbons, S.M.; Owens, S.M.; Hampton-Marcell, J.; Johnston, E.R.; Jastrow, J.D.; Gilbert, J.A.; Meyer, F.; Antonopoulos, D.A. Spatial scale drives patterns in soil bacterial diversity. Environ. Microbiol. 2016, 18, 2039–2051. [Google Scholar] [CrossRef]
- Siregar, U.; Rachmi, A.; Massijaya, M.Y.; Ishibashi, N.; Ando, K. Economic analysis of sengon (Paraserianthes falcataria) community forest plantation, a fast growing species in East Java, Indonesia. For. Policy Econ. 2007, 9, 822–829. [Google Scholar] [CrossRef]
- Garcia-Montiel, D.C.; Binkley, D. Effect of Eucalyptus saligna and Albizia falcataria on soil processes and nitrogen supply in Hawaii. Oecologia 1998, 113, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Clapp, J.P.; Mansur, I.; Dodd, J.C.; Jeffries, P. Ribotyping of rhizobia nodulating Acacia mangium and Paraserianthes falcataria from different geographical areas in Indonesia using PCR-RFLP-SSCP (PRS) and sequencing. Environ. Microbiol. 2001, 3, 273–280. [Google Scholar] [CrossRef]
- Aagot, N.; Nybroe, O.; Nielsen, P.; Johnsen, K. An altered Pseudomonas diversity is recovered from soil by using nutrient-poor Pseudomonas-selective soil extract media. Appl. Environ. Microbiol. 2001, 67, 5233–5239. [Google Scholar] [CrossRef]
- Khan, M.M.T.; Pyle, B.H.; Camper, A.K. Specific and rapid enumeration of viable but nonculturable and viable-culturable gram-negative bacteria by using flow cytometry. Appl. Environ. Microbiol. 2010, 76, 5088–5096. [Google Scholar] [CrossRef]
- Bai, Y.; D’Aoust, F.; Smith, D.L.; Driscoll, B.T. Isolation of plant-growth-promoting Bacillus strains from soybean root nodules. Can. J. Microbiol. 2002, 48, 230–238. [Google Scholar] [CrossRef]
- Zhao, L.; Xu, Y.; Lai, X. Antagonistic endophytic bacteria associated with nodules of soybean (Glycine max L.) and plant growth-promoting properties. Braz. J. Microbiol. 2018, 49, 269–278. [Google Scholar] [CrossRef] [PubMed]
- de Almeida Lopes, K.B.; Carpentieri-Pipolo, V.; Oro, T.H.; Stefani Pagliosa, E.; Degrassi, G. Culturable endophytic bacterial communities associated with field-grown soybean. J. Appl. Microbiol. 2016, 120, 740–755. [Google Scholar] [CrossRef] [PubMed]
- Mwenda, G.M.; Hill, Y.J.; O’Hara, G.W.; Reeve, W.G.; Howieson, J.G.; Terpolilli, J.J. Competition in the Phaseolus vulgaris-Rhizobium symbiosis and the role of resident soil rhizobia in determining the outcomes of inoculation. Plant Soil 2023, 487, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, B.; Dong, Y.; Lin, H. Endophyte colonization enhanced cadmium phytoremediation by improving endosphere and rhizosphere microecology characteristics. J. Hazard. Mater. 2022, 434, 128829. [Google Scholar] [CrossRef] [PubMed]
- Newcombe, G.; Shipunov, A.; Eigenbrode, S.D.; Raghavendra, A.K.H.; Ding, H.; Anderson, C.L.; Menjivar, R.; Crawford, M.; Schwarzländer, M. Endophytes influence protection and growth of an invasive plant. Commun. Integr. Biol. 2009, 2, 29–31. [Google Scholar] [CrossRef]
- Klemetsen, T.; Willassen, N.P.; Karlsen, C.R. Full-length 16S rRNA gene classification of Atlantic salmon bacteria and effects of using different 16S variable regions on community structure analysis. Microbiologyopen 2019, 8, e898. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Xiang, S.-Y.; Wang, Y.-T.; Chen, C.-X.; Liao, C.-M.; Li, T.; Pan, X.-X.; Zhu, S.-S.; Yang, M.-Z. Dominated “inheritance” of endophytes in grapevines from stock plants via in vitro-cultured plantlets: The dawn of plant endophytic modifications. Horticulturae 2023, 9, 180. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Alm, E.J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef] [PubMed]
- Hartman, K.; van der Heijden, M.G.; Roussely-Provent, V.; Walser, J.-C.; Schlaeppi, K. Deciphering composition and function of the root microbiome of a legume plant. Microbiome 2017, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Peng, S.; Li, S.; Song, J.; Brunel, B.; Wang, E.; James, E.K.; Chen, W.; Andrews, M. Arachis hypogaea L. from Acid Soils of Nanyang (China) Is Frequently Associated with Bradyrhizobium guangdongense and Occasionally with Bradyrhizobium ottawaense or Three Bradyrhizobium Genospecies. Microb. Ecol. 2022, 84, 556–564. [Google Scholar] [CrossRef]
- Parker, M.A.; Kennedy, D.A. Diversity and relationships of bradyrhizobia from legumes native to eastern North America. Can. J. Microbiol. 2006, 52, 1148–1157. [Google Scholar] [CrossRef]
- Stępkowski, T.; Watkin, E.; McInnes, A.; Gurda, D.; Gracz, J.; Steenkamp, E.T. Distinct Bradyrhizobium communities nodulate legumes native to temperate and tropical monsoon Australia. Mol. Phylogenet. Evol. 2012, 63, 265–277. [Google Scholar] [CrossRef]
- Fonseca, M.B.; Peix, A.; de Faria, S.M.; Mateos, P.F.; Rivera, L.P.; Simões-Araujo, J.L.; França, M.G.C.; Isaias, R.M.d.S.; Cruz, C.; Velázquez, E.; et al. Nodulation in Dimorphandra wilsonii Rizz. (Caesalpinioideae), a threatened species native to the Brazilian Cerrado. PLoS ONE 2012, 7, e49520. [Google Scholar] [CrossRef]
- Perrineau, M.-M.; Galiana, A.; de Faria, S.; Béna, G.; Duponnois, R.; Reddell, P.; Prin, Y. Monoxenic nodulation process of Acacia mangium (Mimosoideae, Phyllodineae) by Bradyrhizobium sp. Symbiosis 2012, 56, 87–95. [Google Scholar] [CrossRef][Green Version]
- Lafay, B.; Bullier, E.; Burdon, J.J. Bradyrhizobia isolated from root nodules of Parasponia (Ulmaceae) do not constitute a separate coherent lineage. Int. J. Syst. Evol. Microbiol. 2006, 56, 1013–1018. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]

| Group | Sample | Location | Latitude (° E) | Longitude (° N) | Number of Sampled Trees |
|---|---|---|---|---|---|
| MMDB | MM01, MM02, MM03 | Dianbai, Maoming, Guangdong | 111.2487 | 21.62869 | 3 |
| YJYC | YC01 | Yangjiang, Yangchun, Guangdong | 111.8722 | 22.29952 | 1 |
| JMEP | JM01 | Enping, Jiangmen, Guangdong | 112.2234 | 22.33324 | 1 |
| ZQSH | ZQ01 | Sihui, Zhaoqing, Guangdong | 112.7064 | 23.33962 | 1 |
| GZTH | GZ01, GZ02, GZ03, GZ04, GZ05, GZ06, M1, M2, M3, M4, M5, M6, T1, T2, T3 | Tianhe, Guangzhou, Guangdong | 113.876 | 23.30645 | 15 |
| GZZC | GZ07, GZ08, GZ09 | Zengcheng, Guangzhou, Guangdong | 113.876 | 23.30645 | 3 |
| HZBL | HZ01, HZ02, HZ03, HZ04, HZ05 | Boluo, Huizhou, Guangdong | 114.2419 | 23.20189 | 5 |
| HZHD | HD01 | Huidong, Huizhou, Guangdong | 114.7853 | 23.18096 | 1 |
| CZRP | RP01 | Raoping, Chaozhou, Guangdong | 116.9247 | 23.75595 | 1 |
| Sample | Relative Abundance (%) | |||||
|---|---|---|---|---|---|---|
| Bradyrhizobium | Paucibacter | Pseudomonas | Rhizobium | Puia | Caballeronia | |
| MMDB | 13.75 ** | 2.63 | 0.00 | 1.99 | 3.13 | 2.39 |
| YJYC | 52.40 *** | 3.30 | 0.00 | 0.17 | 1.66 | 0.00 |
| JMEP | 27.72 ** | 26.79 ** | 0.00 | 0.09 | 0.00 | 28.92 ** |
| ZQSH | 29.69 ** | 1.36 | 0.07 | 4.82 | 6.25 * | 0.37 |
| GZTH | 28.91 ** | 34.44 *** | 0.17 | 1.36 | 0.78 | 0.02 |
| GZZC | 36.33 *** | 14.89 ** | 0.03 | 5.69 * | 2.81 | 0.06 |
| HZBL | 36.95 *** | 29.18 ** | 9.01 * | 1.15 | 0.66 | 0.02 |
| HZHD | 10.50 ** | 2.23 | 0.18 | 0.29 | 14.59 ** | 0.10 |
| CZRP | 37.08 *** | 34.07 *** | 0.00 | 6.77 * | 1.54 | 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, S.; Yan, S.; Lin, Q.; Huang, R.; Wang, R.; Wei, R.; Wu, G.; Zheng, H. Characterization of the Root Nodule Microbiome of the Exotic Tree Falcataria falcata (Fabaceae) in Guangdong, Southern China. Diversity 2023, 15, 1092. https://doi.org/10.3390/d15101092
Xiang S, Yan S, Lin Q, Huang R, Wang R, Wei R, Wu G, Zheng H. Characterization of the Root Nodule Microbiome of the Exotic Tree Falcataria falcata (Fabaceae) in Guangdong, Southern China. Diversity. 2023; 15(10):1092. https://doi.org/10.3390/d15101092
Chicago/Turabian StyleXiang, Siyu, Shu Yan, Qianxi Lin, Rong Huang, Runhui Wang, Ruping Wei, Guandi Wu, and Huiquan Zheng. 2023. "Characterization of the Root Nodule Microbiome of the Exotic Tree Falcataria falcata (Fabaceae) in Guangdong, Southern China" Diversity 15, no. 10: 1092. https://doi.org/10.3390/d15101092
APA StyleXiang, S., Yan, S., Lin, Q., Huang, R., Wang, R., Wei, R., Wu, G., & Zheng, H. (2023). Characterization of the Root Nodule Microbiome of the Exotic Tree Falcataria falcata (Fabaceae) in Guangdong, Southern China. Diversity, 15(10), 1092. https://doi.org/10.3390/d15101092







