Abstract
Traditional species distribution modelling relies on the links between species and their environments, but often such information is unavailable or unreliable. The objective of our research is to take a machine learning (ML) approach to estimate ant species richness in data-poor countries based on published data on the broader distribution of described ant species. ML is a novel black box method that does not consider functional links between species and their environment. Its prediction accuracy is limited only by the quality and quantity of species records data. ML modelling is applied to calculate the global distribution of ant species richness and achieves 71.78% (decision tree), 70.62% (random forest), 71.09% (logistic regression), and 75.18% (neural network) testing accuracy. The results show that in some West African countries, the species predicted by ML are 1.99 times as many as the species currently recorded. These West African countries have many ant species but lack observational data, and policymakers may be overlooking areas that require protection.
1. Introduction
The distribution of species richness is an essential research field in biodiversity conservation. Based on a species’ response to environmental factors, scientists generally employ species distribution modelling (SDM) [1,2]. SDM is a numerical tool that links species observation with environmental estimations [3,4]. Typically, SDM consists of five steps: (1) environmental data and species data, including climate and presence/absence; (2) model structure, meaning model algorithm selection, and specification; (3) model calibration and model fitting; (4) model validation, including verification of the model’s performance; and (5) model projection, suggesting predictions under different scenarios [3,4,5]. Normally, SDM uses three approaches: correlative models (statistical links between spatial data and distribution of species richness), process-based models (mechanistic links between the functional traits of organisms and their environments), and hybrid models [6]. However, these methods have limitations: for example, process-based mechanistic models are built on existing links that lead to bias and they cannot guarantee an accurate outcome [6,7,8].
To decrease the effect of links and bias, scientists consider using novel “black box” machine learning (ML) methods to make predictions about the distribution of species richness based on field data [9]. ML specialises in processing massive continual and categorical data to acquire accurate prediction results [10,11,12,13]. It has been applied with complex modelling skills to correctly solve undiscovered nonlinear relationships by working with massive and intractable variables in animal science [14,15]. ML has also been confirmed to be an effective tool for predicting animal distribution, including avian [16], mosquito [17], fish [18], and great grey shrike [19]. Since ML uses high-quality and extensive data, its accuracy would overcome the disadvantages associated with SDM and discover more unknown links between the functional traits of organisms and their environments (Table 1). Therefore, ML is an intelligent, rapid, and accurate method that has great potential for use in predicting the distribution of species richness.

Table 1.
Comparison of SDM and ML modelling.
To examine the application of ML methods in the distribution of species richness, we select ants as the object of this paper for the following reasons. First, this is a global-scale study, and ants are widely distributed in most countries [27,28]. Second, ants are small and difficult to capture, causing databases to vary widely from country to country [29]. Third, more than 14,000 classified species of ants exist, and scientists can assess comprehensive databases of these insects [30]. Fourth, ants are an ecologically dominant faunal group globally [31,32]. Finally, ants greatly impact humans, such as by suppressing pest populations in the soil or providing biological control for citrus cultivation [33,34]. Kass et al. applied ML with high resolution (20 km) to predict ant distribution in comparison with the distribution of vertebrate groups globally. They suggested that landscape conservation would benefit both vertebrate and invertebrate groups because the existence of these animals relies heavily on certain environmental conditions [35]. Kass et al.’s study inspired us to further explore ant distribution, specifically in West African countries since these countries have been recognised to have a scarcity of ant species, and in these countries, it is difficult to obtain observational information. Moreover, we introduced some new variables (e.g., temperature, precipitation, forest area, GDP per capita, and human development index) to ascertain which factors influence ant distribution the most [36,37,38,39,40,41], as well as to utilise the neural network method to make predictions. Numerous studies have addressed the distribution of ants geographically through fieldwork and observation [42,43,44,45]. However, these data are mostly from developed rather than underdeveloped countries [46,47]. Consequently, scientists are often confused about whether or not a given species exists in an underdeveloped country. In such cases, policymakers occasionally equate situations in which data are missing with the absence of a given species when they designate protected areas [48].
The objective of our research is to take an ML approach to estimate species richness in data-poor countries based on published data on the broader distribution of described ant species. The specific aims are (1) to compile the number of described species and total species records for all countries. It is important to note that our study deals with described species only, and does not take into account differences among countries in numbers of undescribed species; (2) to correlate these with environmental and socioeconomic factors (e.g., temperature, precipitation, forest area, GDP per capita, human development index); (3) to use ML to estimate the likely number of described species in each country; and (4) to map the discrepancy between the number of recorded species and the number of likely species [49].
2. Material and Methods
The ML method followed the previous studies with minor modifications [50,51,52]. It captured linear and nonlinear patterns via mathematical expressions. Briefly, ML involved three steps: (1) collecting data, (2) using the data to train the model, and (3) applying the model to predict the species richness (Figure 1).
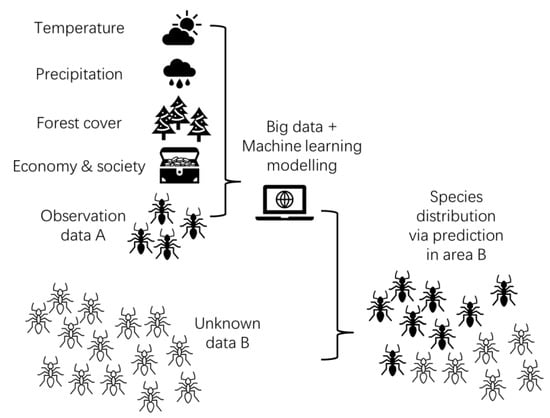
Figure 1.
Framework showing how ML used the experimental data and predicted distribution of species richness in a region that had not yet been investigated. Temperature, precipitation, and forest cover have a significant effect on the distribution of species richness. The socioeconomic factors can affect the intensity of sampling ants.
First, the ant database was constructed using AntWiki (https://www.antwiki.org, accessed on 1 May 2022) and with 33,655 records and labelled with four distribution categories: “Uncertain”, “Introduced”, “Likely”, and “Present” [53]. The environmental data were obtained from governmental websites and the WorldClim database (https://www.worldclim.org/, accessed on 1 May 2022) [54,55]. The temperature data came from the Global Temperature and World Bank databases [56,57]. The precipitation data came from the Global Precipitation Climatology Centre and World Bank databases [57,58]. The forest data came from the World Bank database [57]. “Temperature”, “Precipitation”, and “Forest area” were the data that most directly affected the environment and the distribution of species richness. The GDP per capita data were obtained from the International Monetary Fund [59] and World Bank databases [57]. The human development index was a composite index of life expectancy, education, and income from the human development report of the United Nations Development Programme [60].
Second, the model input the dataset with 33,655 records. Of the data set, 80% was used to train the model and the remaining 20% was used for testing. Then the model was trained based on supervised learning, in which a function was learned based on input–output pairs [26]. Unless otherwise stated, we trained the ML via the following environment: Python 3.7.1 (Python Software Foundation, Wilmington, DE, USA), Scikit-learn 0.20.0, Graphviz 0.8.4, Numpy 1.15.3, Pandas 0.23.4, Matplotlib 3.0.1, and SciPy 1.1.0 [61,62]. The coding library was based on Scikit-learn (Sklearn), Keras, Pandas, and Matplotlib. Four ML methods for calculation were decision tree [63,64], random forest [65,66], logistic regression [64], and neural network [9]. ML methods were based on previous studies with default methods in the Scikit-learn module [21,22,23]. For the neural network, there were 100 hidden layers in total [67], and each layer had 100 nodes [68]. The maximum number of iterations was 50. The activation function was the rectified linear unit (“relu”). The solver for the neural network algorithm was Adam optimisation (“adam”). The Python coding was obtained from GitHub with minor modifications (Tables S2–S6; permissions under MIT license) [21,22,23].
Third, after the model was trained, we examined the training accuracy and testing accuracy. Furthermore, we applied the ML model to make predictions on the data in unknown areas. For the convenience of calculation, the distribution of species richness was quantified and standardised using a distribution index. The distribution index was the ratio of the total likely number of species and all known ant species in a certain country with a value from 0 (the lowest number of the documented species) to 1 (the highest possible number of the observed species). All countries all over the world were finally calculated to a value from 0 to 1. According to the number for each country, the global distribution index of ants was visualised using the software Heatmapper, Canada (http://www.heatmapper.ca/, accessed on 1 May 2022) [69].
3. Results
3.1. Observation Data Analysis
The “present” category had the most common category of data, accounting for approximately 60–70% of all data (Figure 2). The amount of data on the distribution of species richness varied greatly among the different countries (Figure 2). A large amount of the observed data on the distribution of species richness was from Argentina, China, Brazil, Mexico, and the United States. In this case, the number of described species has been comprehensively documented. There were few observed data on species occurrence in Niger, Mauritania, Mali, Chad, and Bhutan. For example, Mauritania had records for only 15 native species, whereas neighbouring Algeria (with a similar landscape and area) had records for 339 native species (https://antmaps.org/).
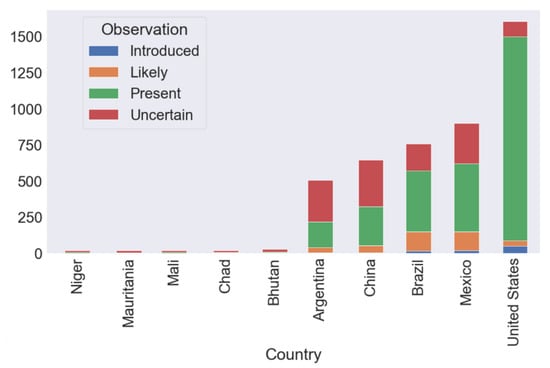
Figure 2.
Total number of species records for bottom 5 countries and top 5 countries. “Introduced” means a certain species is not native but has been migrated or carried by human activity to that country. “Likely” means the ant species probably exist in that country. “Present” means the ant species has been observed and recorded in that country. “Uncertain” means the ant species has not been confirmed in that country. These are four data categories according to the AntWiki database.
3.2. Observation versus Prediction via ML
The neural network achieved the best overall accuracy in the training (75.77%) and testing (75.18%; Table 2). As a result, we applied the neural network for next-step analysis.

Table 2.
ML training results in comparison among different methods.
In the temperature analysis, we found that there were two peaks above the annual average temperature, 7 °C and 25 °C. In the precipitation analysis, most species of ants were observed in the range of approximately 700 mm to 2000 mm of annual precipitation (see Figure 3a,b). The data showed that most species of ants were distributed between warm and humid regions. In the forest area analysis, most ant species were observed in countries with 30% to 60% of the forest area and 0.65 to 0.95 of the human development index (see Figure 3c,d). This indicates that developed countries with high vegetation coverage contribute positively to the distribution of species richness. The prediction frequency (see Figure 3e–h) showed a similar pattern to the observation frequency from the overall data structure.
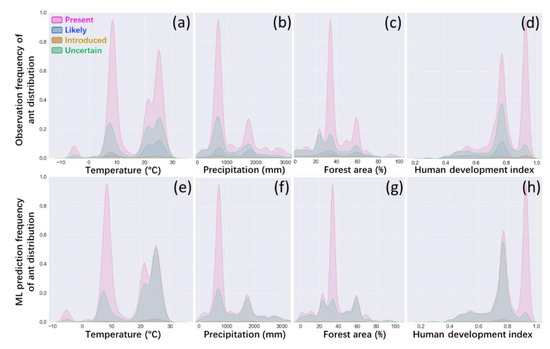
Figure 3.
Observation frequency (a–d) versus prediction frequency (e–h) of ant distribution. The Y-axis shows the frequency from 0 (the amount of data equal to 0) to 1 (the highest possible amount of data). Coloured areas indicate that the observation frequency (upper Y-axis) and ML prediction frequency (lower Y-axis) of ant distribution are currently present (pink) or likely (blue) or introduced (yellow) or unknown (green) under certain conditions of temperature, precipitation, forest area, and human development index (X-axis).
From the correlation heatmap analysis (see Figure 4), the main factors affecting observation frequency were “Forest area” (+0.13), “GDP per capita” (+0.23), and “Human development index” (+0.21). These results suggest that the natural environment (forest area) plays an important role in the distribution of species richness. A higher GDP per capita and human development index may lead to an adequate food supply, leading to higher ant population development. In feature importance analysis, we ranked all features affecting the predictions “GDP per capita” (0.546), “Human development index” (0.309), “Temperature” (0.090), “Precipitation” (0.034), and “Forest area” (0.022). These results suggest that economic factors may play a more important role in the distribution of species richness than environmental factors (Figure S2).
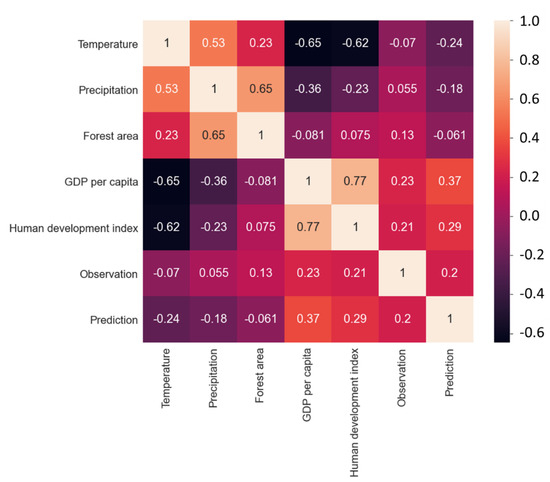
Figure 4.
Heatmap analysis showing the relationship between “Temperature”, “Precipitation”, “Forest area”, “GDP per capita”, “Human development index”, “Observation”, and “Prediction”. The brighter the colour, the stronger the positive correlation between the two data.
We also found that observation and prediction showed a correlation of +0.2. This result showed that there was a weak positive correlation between the predicted results and the observed results. In the distribution of these species, ants may actually exist in some areas but lack observed data.
When we carefully checked the test results of the neural network, the predicted result of 25,301 was consistent with the observed result in a total dataset of 33,655 (see Table 3). Of the 20,659 “presented” species in the observation, 17,896 were accurately predicted (see Table 3). From a global perspective, the predicted result was roughly consistent with the observed result and the accuracy was good. However, the deviation was large when we paid attention to a country or a small area.

Table 3.
The prediction and observation in the testing set.
The distribution of observed species richness is mapped in Figure 5a, and the full dataset is provided in Supplementary Materials Figure S1. These figures show that the United States has a high number of observed species, which indicates the privilege of research support and favourable conditions for research in the United States. On the contrary, few species have been observed in some countries in West Africa, which suggests that these underdeveloped countries have not received enough research support in the past due to their fragile economic status and instability [35]. Both the observed species richness and the ML-predicted results capture a lot of variations among countries. Most countries present little difference between observation and ML prediction, and this is a special case in Europe (Figure 5b). However, a few countries in West Africa show a large difference between observation and ML prediction.
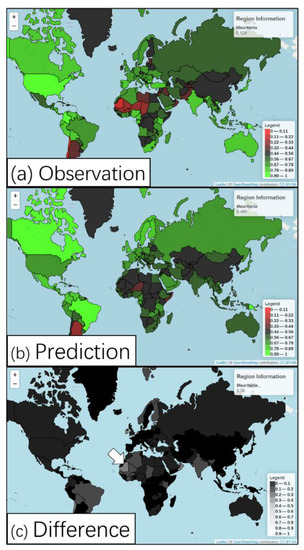
Figure 5.
(a) Distribution index via observation; (b) distribution index via ML prediction; red colour represents areas with a lower distribution of species richness, while green colour represents areas with a higher distribution of species richness; (c) the absolute value of the difference between observation and prediction; dark colour indicates a small difference, while light colour suggests a large difference. The white arrow points to West Africa, an area with a large difference. The data are provided in Supplementary Materials Table S1.
4. Discussion
4.1. Distribution of Species Richness in Data-Poor Countries
One specific aim of this study is to use ML to estimate the likely number of described species in each country by overcoming the limitations of low sampling intensity in underdeveloped countries. Upon examining the global distribution of species richness, we found significant differences between the observed data and the ML prediction results in certain countries.
Specifically, this paper carefully examined several countries with the smallest observed index. Five of these countries were in West Africa, two were in Central Africa, two were in West Asia, one was in South Asia, and one was in South America. In these countries, the predicted index was much higher than the observed index. The observed data for these countries were highly suspicious, as very few species were observed (Table 4). We believe that this suspicion was probably due to insufficient observational fieldwork rather than a lack of species richness [70].

Table 4.
Eleven countries with the smallest observed index. For the observed index and predicted index, 0 means that the number of species is zero, and 1 means the highest possible value of the number of species (all known ant species are found in that country).
Based on the above analysis, we suggest that scientists and policymakers should not underestimate the distribution of species richness in underdeveloped countries due to a lack of observational data. In some data-poor countries in West Africa in particular, scientists have naturally considered the existence of very few species, and policymakers have ignored these areas when formulating conservation plans [71]. When observation data are insufficient, ML prediction has a wide application and can provide effective suggestions for policymakers to determine areas requiring protection [72,73].
ML will always show a little discrepancy between observed and predicted richness in island states such as Australia (and indeed North America), and also in regions of very low ant diversity such as in the boreal zone. We acknowledge that our analysis considers described species only, and that very many undescribed ant species are known to occur. However, the proportion of undescribed species is likely to be higher in underdeveloped countries, which would further accentuate the influence of socioeconomic status on documented patterns of ant diversity.
4.2. Further Improvements to ML Models
One of this paper’s goals was to find ML models that can efficiently and accurately predict the global distribution of species richness. To achieve this goal, this paper utilised four popular ML models (i.e., decision tree, random forest, logistic regression, and neural network) [74]. The decision tree, random forest, and logistic regression models achieve fast predictions with around 70% accuracy. In contrast, the neural network model is more flexible. We can adjust the layers, nodes, and other parameters of these models to optimise their algorithms and produce higher prediction accuracy (~75%). Therefore, this paper recommends the use of the neural network to process ML models.
To further improve the ML model, there are three recommendations for the future. First, the present study’s geographic resolution can be improved. Due to limitations in computational capability, we selected countries’ data for calculation. A higher resolution may lead to new findings if a higher resolution of the data (e.g., 5 km × 5 km) can be provided, enabling policymakers to more precisely locate areas that require protection [75,76]. Second, if additional data and factors (such as population migration, vegetation distribution in various regions, and related species in the food chains) can be added to future ML models, more extensive databases and more complex systems can lead to predictions with higher accuracy [77]. Third, by increasing the power of the computation, attempts at prediction can be made using several novel artificial intelligence (AI) methods [78], including classification [79], nearest neighbour [80,81], linear discriminant analysis [82,83], K-means [84,85], hidden Markov [86], and hierarchical planning [87].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d14090706/s1, Table S1: Predicted index and observed index in different countries (regions); Table S2: Coding for the machine learning; Table S3: Coding for the data analysis; Table Table S4: Coding for the feature importance; Table S5: Randomised search result of random forest hyperparameter tuning; Table S6: Coding for column drawing; Figure S1: Heatmap to check correlation; Figure S2: Feature importance.
Author Contributions
S.C. worked on the manuscript edition, designed the structure of the paper, and supported on the visualisation. Y.D. conceived of the study, gathered the data, performed the statistical analysis, and prepared the manuscript draft. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data has been included in this study.
Acknowledgments
The authors thank the knowledge and computation support from the School of Geography and the Environment, University of Oxford, United Kingdom.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zimmermann, N.E.; Edwards, T.C., Jr.; Graham, C.H.; Pearman, P.B.; Svenning, J.C. New trends in species distribution modelling. Ecography 2010, 33, 985–989. [Google Scholar] [CrossRef]
- Araújo, M.B.; Guisan, A. Five (or so) challenges for species distribution modelling. J. Biogeogr. 2006, 33, 1677–1688. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R. Species distribution models: Ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- Babar, S.; Amarnath, G.; Reddy, C.S.; Jentsch, A.; Sudhakar, S. Species distribution models: Ecological explanation and prediction of an endemic and endangered plant species (Pterocarpus santalinus Lf). Curr. Sci. 2012, 102, 1157–1165. [Google Scholar]
- Macias-Fauria, M.; Johnson, E.A. Warming-induced upslope advance of subalpine forest is severely limited by geomorphic processes. Proc. Natl. Acad. Sci. USA 2013, 110, 8117–8122. [Google Scholar] [CrossRef] [PubMed]
- Kearney, M.; Porter, W. Mechanistic niche modelling: Combining physiological and spatial data to predict species’ ranges. Ecol. Lett. 2009, 12, 334–350. [Google Scholar] [CrossRef]
- Cristóbal-Salas, A.; Santiago-Vicente, B.; Estrada-Contreras, I.; Ibarra-Zavaleta, S. A Parallel Implementation of the Species Distribution Modeling Algorithm. In Proceedings of the 2019 IEEE International Autumn Meeting on Power, Electronics and Computing (ROPEC), Ixtapa, Mexico, 13–15 November 2019; pp. 1–6. [Google Scholar]
- Pagel, J.; Schurr, F.M. Forecasting species ranges by statistical estimation of ecological niches and spatial population dynamics. Glob. Ecol. Biogeogr. 2012, 21, 293–304. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S. A Review of Machine Learning Based Species’ Distribution Modelling. In Proceedings of the 2017 International Conference on Industrial Informatics-Computing Technology, Intelligent Technology, Industrial Information Integration (ICIICII), Wuhan, China, 2–3 December 2017; pp. 199–206. [Google Scholar]
- Zhou, L.; Pan, S.; Wang, J.; Vasilakos, A.V. Machine learning on big data: Opportunities and challenges. Neurocomputing 2017, 237, 350–361. [Google Scholar] [CrossRef]
- Shakhovska, N.; Vovk, O.; Hasko, R.; Kryvenchuk, Y. The method of big data processing for distance educational system. In Proceedings of the Conference on Computer Science and Information Technologies, Yerevan, Armenia, 25–29 September 2017; pp. 461–473. [Google Scholar]
- Shakhovska, N.; Kaminskyy, R.; Zasoba, E.; Tsiutsiura, M. Association Rules Mining in Big Data. Int. J. Comput. 2018, 17, 25–32. [Google Scholar] [CrossRef]
- Fedushko, S.; Ustyianovych, T. Predicting pupil’s successfulness factors using machine learning algorithms and mathematical modelling methods. In Proceedings of the International Conference on Computer Science, Engineering and Education Applications, Toronto, ON, Canada, 19–21 August 2019; pp. 625–636. [Google Scholar]
- Valletta, J.J.; Torney, C.; Kings, M.; Thornton, A.; Madden, J. Applications of machine learning in animal behaviour studies. Anim. Behav. 2017, 124, 203–220. [Google Scholar] [CrossRef]
- Martín, B.; González–Arias, J.; Vicente–Vírseda, J. Machine learning as a successful approach for predicting complex spatio–temporal patterns in animal species abundance. Mach. Learn. 2021, 44, 289–301. [Google Scholar] [CrossRef]
- Wellmann, T.; Lausch, A.; Scheuer, S.; Haase, D. Earth observation based indication for avian species distribution models using the spectral trait concept and machine learning in an urban setting. Ecol. Indic. 2020, 111, 106029. [Google Scholar] [CrossRef]
- Früh, L.; Kampen, H.; Kerkow, A.; Schaub, G.A.; Walther, D.; Wieland, R. Modelling the potential distribution of an invasive mosquito species: Comparative evaluation of four machine learning methods and their combinations. Ecol. Model. 2018, 388, 136–144. [Google Scholar] [CrossRef]
- Ahmad, H. Machine learning applications in oceanography. Aquat. Res. 2019, 2, 161–169. [Google Scholar] [CrossRef]
- Dormann, C.F.; Purschke, O.; Marquez, J.R.G.; Lautenbach, S.; Schroeder, B. Components of uncertainty in species distribution analysis: A case study of the great grey shrike. Ecology 2008, 89, 3371–3386. [Google Scholar] [CrossRef]
- Miller, J. Species distribution modeling. Geogr. Compass 2010, 4, 490–509. [Google Scholar] [CrossRef]
- Hao, J.; Ho, T.K. Machine learning made easy: A review of scikit-learn package in python programming language. J. Educ. Behav. Stat. 2019, 44, 348–361. [Google Scholar] [CrossRef]
- Bisong, E. More supervised machine learning techniques with scikit-learn. In Building Machine Learning and Deep Learning Models on Google Cloud Platform; Springer: Berlin/Heidelberg, Germany, 2019; pp. 287–308. [Google Scholar]
- Nelli, F. Machine Learning with scikit-learn. In Python Data Analytics; Springer: Berlin/Heidelberg, Germany, 2018; pp. 313–347. [Google Scholar]
- Beery, S.; Cole, E.; Parker, J.; Perona, P.; Winner, K. Species distribution modeling for machine learning practitioners: A review. In Proceedings of the ACM SIGCAS Conference on Computing and Sustainable Societies, New York, NY, USA, 28 June–2 July 2021; pp. 329–348. [Google Scholar]
- Guillera-Arroita, G.; Lahoz-Monfort, J.J.; Elith, J.; Gordon, A.; Kujala, H.; Lentini, P.E.; McCarthy, M.A.; Tingley, R.; Wintle, B.A. Is my species distribution model fit for purpose? Matching data and models to applications. Glob. Ecol. Biogeogr. 2015, 24, 276–292. [Google Scholar] [CrossRef]
- Gobeyn, S.; Mouton, A.M.; Cord, A.F.; Kaim, A.; Volk, M.; Goethals, P.L.M. Evolutionary algorithms for species distribution modelling: A review in the context of machine learning. Ecol. Model. 2019, 392, 179–195. [Google Scholar] [CrossRef]
- McGlynn, T.P. The worldwide transfer of ants: Geographical distribution and ecological invasions. J. Biogeogr. 1999, 26, 535–548. [Google Scholar] [CrossRef]
- Borowiec, M.L.; Rabeling, C.; Brady, S.G.; Fisher, B.L.; Schultz, T.R.; Ward, P.S. Compositional heterogeneity and outgroup choice influence the internal phylogeny of the ants. Mol. Phylogenetics Evol. 2019, 134, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Guénard, B.; Weiser, M.D.; Gomez, K.; Narula, N.; Economo, E.P. The Global Ant Biodiversity Informatics (GABI) database: Synthesizing data on the geographic distribution of ant species (Hymenoptera: Formicidae). Myrmecol. News/Osterreichische Ges. Fur. Entomofaunist. 2017, 24, 83–89. [Google Scholar]
- Moreau, C.S.; Bell, C.D.; Vila, R.; Archibald, S.B.; Pierce, N.E. Phylogeny of the ants: Diversification in the age of angiosperms. Science 2006, 312, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Tiede, Y.; Schlautmann, J.; Donoso, D.A.; Wallis, C.I.B.; Bendix, J.; Brandl, R.; Farwig, N. Ants as indicators of environmental change and ecosystem processes. Ecol. Indic. 2017, 83, 527–537. [Google Scholar] [CrossRef]
- Parr, C.L.; Bishop, T.R. The response of ants to climate change. Global Chang. Biol. 2022, 28, 3188–3205. [Google Scholar] [CrossRef]
- Huang, H.T.; Yang, P. The ancient cultured citrus ant. BioScience 1987, 37, 665–671. [Google Scholar] [CrossRef]
- Haddad Junior, V.; Cardoso, J.L.C.; Moraes, R.H.P. Description of an injury in a human caused by a false tocandira (Dinoponera gigantea, Perty, 1833) with a revision on folkloric, pharmacological and clinical aspects of the giant ants of the genera Paraponera and Dinoponera (sub-family Ponerinae). Rev. Inst. Med. Trop. São Paulo 2005, 47, 235–238. [Google Scholar] [CrossRef]
- Kass, J.M.; Guénard, B.; Dudley, K.L.; Jenkins, C.N.; Azuma, F.; Fisher, B.L.; Parr, C.L.; Gibb, H.; Longino, J.T.; Ward, P.S. The global distribution of known and undiscovered ant biodiversity. Sci. Adv. 2022, 8, eabp9908. [Google Scholar] [CrossRef]
- Corro, E.J.; Ahuatzin, D.A.; Jaimes, A.A.; Favila, M.E.; Ribeiro, M.C.; López-Acosta, J.C.; Dáttilo, W. Forest cover and landscape heterogeneity shape ant–plant co-occurrence networks in human-dominated tropical rainforests. Landsc. Ecol. 2019, 34, 93–104. [Google Scholar] [CrossRef]
- Ahuatzin, D.A.; Corro, E.J.; Jaimes, A.A.; Valenzuela González, J.E.; Feitosa, R.M.; Ribeiro, M.C.; Acosta, J.C.L.; Coates, R.; Dáttilo, W. Forest cover drives leaf litter ant diversity in primary rainforest remnants within human-modified tropical landscapes. Biodivers. Conserv. 2019, 28, 1091–1107. [Google Scholar] [CrossRef]
- Purcell, J.; Avilés, L. Gradients of precipitation and ant abundance may contribute to the altitudinal range limit of subsocial spiders: Insights from a transplant experiment. Proc. R. Soc. B Biol. Sci. 2008, 275, 2617–2625. [Google Scholar] [CrossRef] [PubMed]
- Stringer, L.D.; Haywood, J.; Lester, P.J. The influence of temperature and fine-scale resource distribution on resource sharing and domination in an ant community. Ecol. Entomol. 2007, 32, 732–740. [Google Scholar] [CrossRef]
- Wahlqvist, M.L.; Specht, R.L. Food variety and biodiversity: Econutrition. Asia Pac. J. Clin. Nutr. 1998, 7, 314–319. [Google Scholar]
- Tscharntke, T.; Clough, Y.; Wanger, T.C.; Jackson, L.; Motzke, I.; Perfecto, I.; Vandermeer, J.; Whitbread, A. Global food security, biodiversity conservation and the future of agricultural intensification. Biol. Conserv. 2012, 151, 53–59. [Google Scholar] [CrossRef]
- Yamaguchi, T. Influence of urbanization on ant distribution in parks of Tokyo and Chiba City, Japan I. Analysis of ant species richness. Ecol. Res. 2004, 19, 209–216. [Google Scholar] [CrossRef]
- Jackson, D.A. Ant distribution patterns in a Cameroonian cocoa plantation: Investigation of the ant mosaic hypothesis. Oecologia 1984, 62, 318–324. [Google Scholar] [CrossRef]
- Carter, W.G. Ant distribution in North Carolina. J. Elisha Mitchell Sci. Soc. 1962, 78, 150–204. [Google Scholar]
- Torres, J.A. Diversity and distribution of ant communities in Puerto Rico. Biotropica 1984, 16, 296–303. [Google Scholar] [CrossRef]
- Miravete, V.; Roura-Pascual, N.; Dunn, R.R.; Gómez, C. How many and which ant species are being accidentally moved around the world? Biol. Lett. 2014, 10, 20140518. [Google Scholar] [CrossRef]
- Gibb, H.; Dunn, R.R.; Sanders, N.J.; Grossman, B.F.; Photakis, M.; Abril, S.; Agosti, D.; Andersen, A.N.; Angulo, E.; Armbrecht, I. A global database of ant species abundances. Ecology 2017, 98, 883–884. [Google Scholar] [CrossRef]
- Liu, C.; White, M.; Newell, G. Measuring and comparing the accuracy of species distribution models with presence–absence data. Ecography 2011, 34, 232–243. [Google Scholar] [CrossRef]
- Johnson, V.M.; Rogers, L.L. Accuracy of neural network approximators in simulation-optimization. J. Water Resour. Plan. Manag. 2000, 126, 48–56. [Google Scholar] [CrossRef]
- Nokeri, T.C. Solving Economic Problems Applying Artificial Neural Networks. In Econometrics and Data Science; Springer: Berlin/Heidelberg, Germany, 2022; pp. 161–188. [Google Scholar]
- Beinrohr, L.; Kail, E.; Piros, P.; Tóth, E.; Fleiner, R.; Kolev, K. Anatomy of a Data Science Software Toolkit That Uses Machine Learning to Aid ‘Bench-to-Bedside’Medical Research—With Essential Concepts of Data Mining and Analysis Explained. Appl. Sci. 2021, 11, 12135. [Google Scholar] [CrossRef]
- Khansari, S.M.; Arbabi, F.; Moazen Jamshidi, M.H.; Soleimani, M.; Ebrahimi, P. Health Services and Patient Satisfaction in IRAN during the COVID-19 Pandemic: A Methodology Based on Analytic Hierarchy Process and Artificial Neural Network. J. Risk Financ. Manag. 2022, 15, 288. [Google Scholar] [CrossRef]
- Urbani, B.; Andrade, D. AntWiki. The Ants—Online. Available online: https://www.antwiki.org/wiki/Downloadable_Data (accessed on 1 May 2022).
- Cerasoli, F.; D’Alessandro, P.; Biondi, M. Worldclim 2.1 versus Worldclim 1.4: Climatic niche and grid resolution affect between-version mismatches in Habitat Suitability Models predictions across Europe. Ecol. Evol. 2022, 12, e8430. [Google Scholar] [CrossRef]
- WorldClim. WorldClim Database; WorldClim-Global Climate Data: Berkeley, CA, USA, 2022; Available online: https://www.worldclim.org/data/index.html (accessed on 1 May 2022).
- Lindsey, R.; Dahlman, L. Climate Change: Global Temperature. Available online: https://www.climate.gov/ (accessed on 1 May 2022).
- World Bank. World Bank Database; World Bank: Washington, DC, USA, 2022. [Google Scholar]
- Schneider, U.; Fuchs, T.; Meyer-Christoffer, A.; Rudolf, B. Global precipitation analysis products of the GPCC. Glob. Precip. Climatol. Cent. GPCC DWD Internet Publ. 2008, 1, 1–112. [Google Scholar]
- International Monetary Fund. International Monetary Fund Database; International Monetary Fund: Bretton Woods, NH, USA, 2022. [Google Scholar]
- United Nations Development Programme. Human Development Index; United Nations Development Programme: New York, NY, USA, 2022. [Google Scholar]
- Indriani, D.; Nasution, A.H.; Monika, W.; Nasution, S. Towards a Sentiment Analyser for Low-resource Languages. In Proceedings of the International Conference on Smart Computing and Cyber Security, Gangwon-do, Korea, 7–8 July 2020; pp. 109–118. [Google Scholar]
- Yasenko, L.; Klyatchenko, Y.; Tarasenko-Klyatchenko, O. Image noise reduction by denoising autoencoder. In Proceedings of the 2020 IEEE 11th International Conference on Dependable Systems, Services and Technologies (DESSERT), Kyiv, Ukraine, 14–18 May 2020; pp. 351–355. [Google Scholar]
- Kadiyala, A.; Kumar, A. Applications of python to evaluate the performance of decision tree-based boosting algorithms. Environ. Prog. Sustain. Energy 2018, 37, 618–623. [Google Scholar] [CrossRef]
- Noviyarto, H. Comparation Logistic Regression and Decision Tree Method to Distribution Type of Works in Jakarta. Int. J. Multidiscip. Res. Publ. 2020, 2, 26–29. [Google Scholar]
- Polimis, K.; Rokem, A.; Hazelton, B. Confidence intervals for random forests in python. J. Open Source Softw. 2017, 2, 124. [Google Scholar] [CrossRef]
- Adugna, T.; Xu, W.; Fan, J. Comparison of Random Forest and Support Vector Machine Classifiers for Regional Land Cover Mapping Using Coarse Resolution FY-3C Images. Remote Sens. 2022, 14, 574. [Google Scholar] [CrossRef]
- Guo, K.; Han, S.; Yao, S.; Wang, Y.; Xie, Y.; Yang, H. Software-hardware codesign for efficient neural network acceleration. IEEE Micro 2017, 37, 18–25. [Google Scholar] [CrossRef]
- Chellapilla, K.; Puri, S.; Simard, P. High performance convolutional neural networks for document processing. HAL Open Sci. 2006, 1, 1–10. Available online: https://hal.inria.fr/inria-00112631/document (accessed on 1 May 2022).
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Zhao, D.-M.; Jiao, Y.-M.; Wang, J.-L.; Ding, Y.-P.; Liu, Z.-L.; Liu, C.-J.; Qiu, Y.-M.; Zhang, J.; Xu, Q.-E.; Wu, C.-R. Comparative performance assessment of landslide susceptibility models with presence-only, presence-absence, and pseudo-absence data. J. Mt. Sci. 2020, 17, 2961–2981. [Google Scholar] [CrossRef]
- Barbet-Massin, M.; Jiguet, F.; Albert, C.H.; Thuiller, W. Selecting pseudo-absences for species distribution models: How, where and how many? Methods Ecol. Evol. 2012, 3, 327–338. [Google Scholar] [CrossRef]
- Guisan, A.; Tingley, R.; Baumgartner, J.B.; Naujokaitis-Lewis, I.; Sutcliffe, P.R.; Tulloch, A.I.T.; Regan, T.J.; Brotons, L.; McDonald-Madden, E.; Mantyka-Pringle, C. Predicting species distributions for conservation decisions. Ecol. Lett. 2013, 16, 1424–1435. [Google Scholar] [CrossRef]
- Maris, V.; Huneman, P.; Coreau, A.; Kéfi, S.; Pradel, R.; Devictor, V. Prediction in ecology: Promises, obstacles and clarifications. Oikos 2018, 127, 171–183. [Google Scholar] [CrossRef]
- Tso, G.K.F.; Yau, K.K.W. Predicting electricity energy consumption: A comparison of regression analysis, decision tree and neural networks. Energy 2007, 32, 1761–1768. [Google Scholar] [CrossRef]
- Gonthier, D.J.; Ennis, K.K.; Farinas, S.; Hsieh, H.-Y.; Iverson, A.L.; Batáry, P.; Rudolphi, J.; Tscharntke, T.; Cardinale, B.J.; Perfecto, I. Biodiversity conservation in agriculture requires a multi-scale approach. Proc. R. Soc. B Biol. Sci. 2014, 281, 20141358. [Google Scholar] [CrossRef] [PubMed]
- Du Toit, J.T. Considerations of scale in biodiversity conservation. Anim. Conserv. 2010, 13, 229–236. [Google Scholar] [CrossRef]
- Gui, G.; Liu, F.; Sun, J.; Yang, J.; Zhou, Z.; Zhao, D. Flight delay prediction based on aviation big data and machine learning. IEEE Trans. Veh. Technol. 2019, 69, 140–150. [Google Scholar] [CrossRef]
- Abiodun, O.I.; Jantan, A.; Omolara, A.E.; Dada, K.V.; Mohamed, N.A.; Arshad, H. State-of-the-art in artificial neural network applications: A survey. Heliyon 2018, 4, e00938. [Google Scholar] [CrossRef]
- Kotsiantis, S.B.; Zaharakis, I.D.; Pintelas, P.E. Machine learning: A review of classification and combining techniques. Artif. Intell. Rev. 2006, 26, 159–190. [Google Scholar] [CrossRef]
- Samworth, R.J. Optimal weighted nearest neighbour classifiers. Ann. Stat. 2012, 40, 2733–2763. [Google Scholar] [CrossRef]
- Duivesteijn, W.; Feelders, A. Nearest neighbour classification with monotonicity constraints. In Proceedings of the Machine Learning and Knowledge Discovery in Databases, European Conference, ECML/PKDD 2008, Antwerp, Belgium, 15–19 September 2008; pp. 301–316. [Google Scholar]
- Izenman, A.J. Linear discriminant analysis. In Modern Multivariate Statistical Techniques; Springer: Berlin/Heidelberg, Germany, 2013; pp. 237–280. [Google Scholar]
- Xanthopoulos, P.; Pardalos, P.M.; Trafalis, T.B. Linear discriminant analysis. In Robust Data Mining; Springer: Berlin/Heidelberg, Germany, 2013; pp. 27–33. [Google Scholar]
- Likas, A.; Vlassis, N.; Verbeek, J.J. The global k-means clustering algorithm. Pattern Recognit. 2003, 36, 451–461. [Google Scholar] [CrossRef]
- Li, L.; Wang, J.; Li, X. Efficiency analysis of machine learning intelligent investment based on K-means algorithm. IEEE Access 2020, 8, 147463–147470. [Google Scholar] [CrossRef]
- Manogaran, G.; Vijayakumar, V.; Varatharajan, R.; Malarvizhi Kumar, P.; Sundarasekar, R.; Hsu, C.-H. Machine learning based big data processing framework for cancer diagnosis using hidden Markov model and GM clustering. Wirel. Pers. Commun. 2018, 102, 2099–2116. [Google Scholar] [CrossRef]
- Mohr, F.; Wever, M.; Hüllermeier, E. ML-Plan: Automated machine learning via hierarchical planning. Mach. Learn. 2018, 107, 1495–1515. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).