The First Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Pistachio (Pistacia vera)
Abstract
1. Introduction
2. Materials and Methods
2.1. Chloroplast Genome Assembly and Validation
2.2. Gene Annotation and Sequence Analysis
2.3. Simple Sequence Repeats (SSR) Analysis
2.4. Phylogenetic Analysis
3. Results
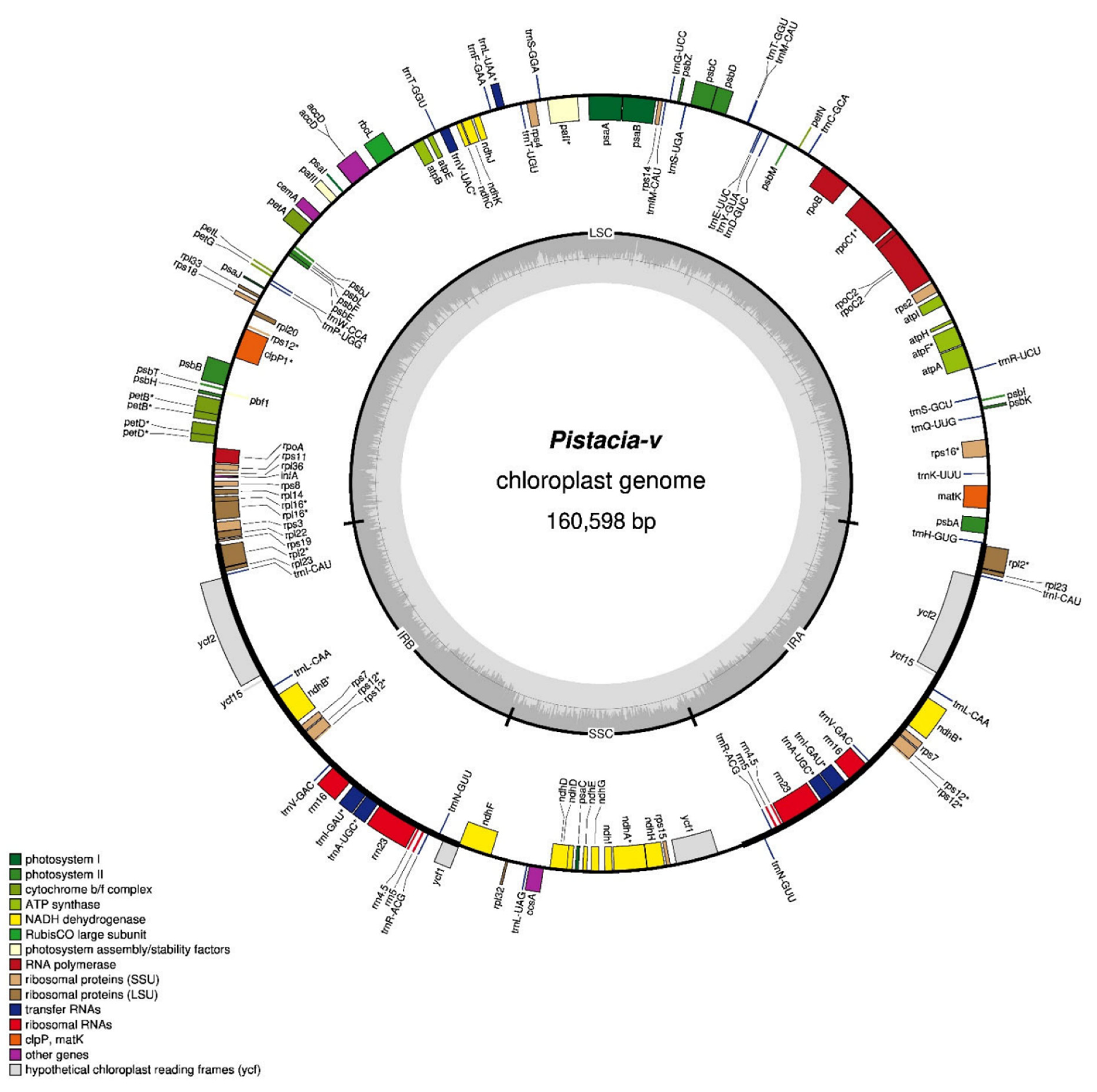
3.1. Structural Features of P. vera Chloroplast Genome
3.2. Comparison of P. vera Chloroplast Genome to Other Members of Anacardiaceae
3.3. Repeat Sequences Analysis
3.4. Phylogenetic Analysis
4. Discussion
4.1. Structural Features of P. vera Chloroplast Genome
4.2. Repeat Sequences
4.3. Phylogenetic Reconstruction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zohary, M. A monographical study of the genus Pistacia. Palest. J. Bot. Jerus. Ser. 1952, 5, 187–228. [Google Scholar]
- Kafkas, S.; Özkan, H.; Ak, B.E.; Acar, L.; Atli, H.S.; Koyuncu, S. Detecting NA polymorphism and genetic diversity in a wide pistachio germplasm: Comparison of AFLP, ISSR and RAPD markers. J. Am. Soc. Hortic. Sci. 2006, 131, 522–529. [Google Scholar] [CrossRef]
- Zarei, A.; Erfani-Moghadam, J. SCoT markers provide insight into the genetic diversity, population structure and phylogenetic relationships among three Pistacia species of Iran. Genet. Resour. Crop. Evol. 2021, 68, 1625–1643. [Google Scholar] [CrossRef]
- Sagheb Talebi, K.; Sajedi, T.; Pourhashemi, M. Forests of Iran: A Treasure from the Past, a Hope for the Future; Springer: Dordrecht, The Netherlands, 2014; p. 152. [Google Scholar] [CrossRef]
- Food and Agriculture Organization. FAO. 2019. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 19 February 2019).
- Hernaddez-Alonso, P.; Bullo, M.; Salas-Salvado, J. Pistachios for health; what do we know about this multifaceted nut? Nutr. Today 2016, 51, 133–138. [Google Scholar] [CrossRef]
- Askan, E. Economic analysis and marketing margin of pistachios in Turkey. Bull. Natl. Res. Cent. 2019, 43, 177. [Google Scholar] [CrossRef][Green Version]
- Dagan, T.; Roettger, M.; Stucken, K.; Landan, G.; Koch, R.; Major, P.; Gould, S.B.; Goremykin, V.V.; Rippka, R.; Tandeau de Marsac, N.; et al. Genomes of Stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol. Evol. 2013, 5, 31–44. [Google Scholar] [CrossRef]
- Neuhaus, H.E.; Ernes, M.J. Nonphotosyntiietic metabolism in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 111–140. [Google Scholar] [CrossRef]
- Yoo, Y.H.; Hong, W.J.; Jung, K.H. A systematic view exploring the role of chloroplasts in plant abiotic stress responses. BioMed Res. Int. 2019, 2019, 6534745. [Google Scholar] [CrossRef]
- Stavridou, E.; Michailidis, M.; Gedeon, S.; Ioakeim, A.; Kostas, S.; Chronopoulou, E.; Labrou, N.E.; Edwards, R.; Day, A.; Nianiou-Obeidat, I.; et al. Tolerance of transplastomic tobacco plants overexpressing a theta class glutathione transferase to abiotic and oxidative stresses. Front. Plant Sci. 2019, 9, 1861. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, N.; Wu, H. Analyzing and characterizing the chloroplast genome of Salix Wilsonii. BioMed Res. Int. 2019, 5190425. [Google Scholar] [CrossRef]
- McCauley, D.E. The use of chloroplast DNA polymorphism in studies of gene flow in plants. Trends Ecol. Evol. 1995, 10, 198–202. [Google Scholar] [CrossRef]
- Bi, Y.; Zhang, M.; Xue, J.; Dong, R.; Du, Y.; Zhang, X. Chloroplast genomic resources for phylogeny and DNA barcoding: A case study on Fritillaria. Sci. Rep. 2018, 8, 1184. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Wu, P.; Cheng, T.; Yu, J.; Zhou, S.; Hong, D.Y. Resolving the systematic positions of enigmatic taxa: Manipulating the chloroplast genome data of Saxifragales. Mol. Phylogenet. Evol. 2018, 126, 321–330. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, X.; Yu, Y.; Yuan, S.; Jiang, D.; Zhang, Y.; Zhang, T.; Zhong, W.; Yuan, Q.; Huang, L. Complete chloroplast genome sequences of Dioscorea: Characterization, genomic resources, and phylogenetic analyses. PeerJ 2018, 6, e6032. [Google Scholar] [CrossRef]
- Welch, J.; Collins, K.; Ratan, A.; Drautz-Moses, D.I.; Schuster, S.C.; Lindqvist, C. The quest to resolve recent radiations: Plastid phylogenomics of extinct and endangered Hawaiian endemicmints (Lamiaceae). Mol. Phylogenet. Evol. 2016, 99, 16–33. [Google Scholar] [CrossRef]
- Xue, J.H.; Dong, W.P.; Cheng, T.; Zhou, S.L. Nelumbonaceae: Systematic position and species diversification revealed by the complete chloroplast genome. J. Syst. Evol. 2012, 50, 477–487. [Google Scholar] [CrossRef]
- Thode, V.A.; Lohmann, L.G. Comparative chloroplast genomics at low taxonomic levels: A case study using Amphilophium (Bignonieae, Bignoniaceae). Front. Plant Sci. 2019, 10, 796. [Google Scholar] [CrossRef]
- Magee, A.M.; Aspinall, S.; Rice, D.W.; Cusack, B.P.; Sémon, M.; Perry, A.S.; Stefanović, S.; Milbourne, D.; Barth, S.; Palmer, J.D.; et al. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 2010, 20, 1700–1710. [Google Scholar] [CrossRef]
- Ochoterena, H. Homology in coding and non-coding DNA sequences: A parsimony perspective. Plant Sys. Evol. 2009, 282, 151–168. [Google Scholar] [CrossRef]
- Khodaeiaminjan, M.; Kafkas, S.; Motalebipour, E.Z.; Coban, N. In silico polymorphic novel SSR marker development and the first SSR-based genetic linkage map in pistachio. Tree Genet. Genomes 2018, 14, 1–14. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 35, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high-confidence variant calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Liu, C.; Shi, L.; Zhu, Y.; Chen, H.; Zhang, J.; Lin, X.; Guan, X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012, 13, 715. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Zheng, W.; Li, K.; Wang, W.; Xu, X. The complete chloroplast genome of the threatened Pistacia weinmannifolia, an economically and horticulturally important evergreen plant. Conserv. Genet. Resour. 2017, 10, 535–538. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, I.; Kim, J.K.; Park, J.Y.; Joh, H.J.; Park, H.S.; Lee, H.O.; Lee, S.C.; Hur, Y.J.; Yang, T.J. The complete chloroplast genome sequence of Rhus chinensis Mill Anacardiaceae). Mitochondrial DNA B Resour. 2016, 1, 696–697. [Google Scholar] [CrossRef]
- Rabah, S.O.; Lee, C.; Hajrah, N.H.; Makki, R.M.; Alharby, H.F.; Alhebshi, A.M.; Sabir, J.S.; Jansen, R.K.; Ruhlman, T.A. Plastome sequencing of ten nonmodel crop species uncovers a large insertion of mitochondrial DNA in cashew. Plant Genome 2017, 10. [Google Scholar] [CrossRef]
- Santose, V.; Almeida, C. The complete chloroplast genome sequences of three Spondias species reveal close relationship among the species. Genet. Mol. Biol. 2019, 42, 132–138. [Google Scholar] [CrossRef]
- Khan, A.L.; Al-Harrasi, A.; Asaf, S.; Park, C.E.; Park, G.S.; Khan, A.R.; Lee, I.J.; Al-Rawahi, A.; Shin, J.H. The first chloroplast genome sequence of Boswellia sacra, a resin-producing plant in Oman. PLoS ONE 2017, 12, e0169794. [Google Scholar] [CrossRef]
- Yang, B.; Li, M.; Ma, J.; Fu, Z.; Xu, X.; Chen, Q. The complete chloroplast genome sequence of Sapindus mukorossi. Mitochondrial DNA A 2016, 27, 1825–1826. [Google Scholar]
- Logacheva, M.D.; Shipunov, A.B. Phylogenomic analysis of Picramnia, Alvaradoa, and Leitneria supports the independent Picramniales: Phylogenomics of Picramniales. J. Sys. Evol. 2017, 55, 171–176. [Google Scholar] [CrossRef]
- Mader, M.; Pakull, B.; Blanc-Jolivet, C.; Paulini-Drewes, M.; Bouda, Z.H.; Degen, B.; Small, I.; Kersten, B. Complete chloroplast genome sequences of four Meliaceae species and comparative analyses. Int. J. Mol. Sci. 2018, 19, 701. [Google Scholar] [CrossRef]
- Bausher, M.G.; Singh, N.D.; Lee, S.B.; Jansen, R.K.; Daniell, H. The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var ‘Ridge Pineapple’: Organization and phylogenetic relationships to other angiosperms. BMC Plant Biol. 2006, 6, 21. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, A. The complete chloroplast genome sequence of an economically important plant, Zanthoxylum bungeanum (Rutaceae). Conserv. Genet. Resour. 2017, 9, 25–27. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, J.; Ling, Q.; Fan, L.; Zhang, M. Complete chloroplast genome sequence of Prunus davidiana (Rosaceae). Mitochondrial DNA B Resour. 2018, 3, 890–891. [Google Scholar] [CrossRef]
- Shinozaki, K.; Ohme, M.; Tanaka, M.; Wakasugi, T.; Hayashida, N.; Matsubayashi, T.; Zaita, N.; Chunwongse, J.; Obokata, J.; Yamaguchi-Shinozaki, K.; et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986, 5, 2043–2049. [Google Scholar] [CrossRef]
- Zuo, L.H.; Shang, A.Q.; Zhang, S.; Yu, X.Y.; Ren, Y.C.; Yang, M.S.; Wang, J.M. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: Genome comparative and taxonomic position analysis. PLoS ONE 2017, 12, e0171264. [Google Scholar] [CrossRef]
- Song, Y.; Chen, Y.; Lv, J.; Xu, J.; Zhu, S.; Li, M.F. Comparative chloroplast genomes of Sorghum species: Sequence divergence and phylogenetic relationships. BioMed Res. Int. 2019, 2019, 5046958. [Google Scholar] [CrossRef]
- Zhou, T.; Ruhsam, M.; Wang, J.; Zhu, H.; Li, W.; Zhang, X.; Xu, Y.; Xu, F.; Wang, X. The complete chloroplast genome of Euphrasia regelii, pseudogenization of ndh genes and the phylogenetic relationships within orobanchaceae. Front. Genet. 2019, 10, 444. [Google Scholar] [CrossRef]
- Li, D.M.; Zhao, C.Y.; Liu, X.F. Complete chloroplast genome sequences of Kaempferia galanga and Kaempferia elegans: Molecular structures and comparative analysis. Molecules 2018, 24, 474. [Google Scholar] [CrossRef]
- He, L.; Qian, J.; Li, X.; Sun, Z.; Xu, X.; Chen, S. Complete chloroplast genome of medicinal plant Lonicera japonica: Genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules 2017, 22, 249. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, A.L.; Khan, M.A.; Waqas, M.; Kang, S.M.; Yun, B.W.; Lee, I.J. Chloroplast genomes of Arabidopsis halleri ssp. gemmifera and Arabidopsis lyrata ssp. petraea: Structures and comparative analysis. Sci. Rep. 2017, 7, 7556. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Dong, W.; Li, W.; Lu, Y.; Xie, X.; Jin, X.; Shi, J.; He, K.; Suo, Z. Comparative analysis of six Lagerstroemia complete chloroplast genomes. Front. Plant Sci. 2017, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Guo, L.; Zhao, W.; Xu, J.; Li, Y.; Zhang, X.; Shen, X.; Wu, M.; Hou, X. Complete chloroplast genome sequence and phylogenetic analysis of Paeonia ostii. Molecules 2018, 23, 246. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xiao, H.; Deng, C.; Xiong, L.; Yang, J.; Peng, C. The complete chloroplast genome sequences of the medicinal plant Pogostemon cablin. Int. J. Mol. Sci. 2016, 17, 820. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Wu, M.; Liao, B.; Liu, Z.; Bai, R.; Xiao, S.; Li, X.; Zhang, B.; Xu, J.; Chen, S. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules 2017, 22, 1330. [Google Scholar] [CrossRef]
- Naver, H.; Boudreau, E.; Rochaix, J.D. Functional studies of Ycf3: Its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell 2001, 13, 2731–2745. [Google Scholar] [CrossRef]
- Freeland, S.J.; Hurst, L.D. Load minimization of the genetic code: History does not explain the pattern. Proc. R. Soc. Lond. B 1998, 265, 2111–2119. [Google Scholar] [CrossRef]
- Błażej, P.; Wnętrzak, M.; Mackiewicz, D.; Mackiewicz, P. Optimization of the standard genetic code according to three codon positions using an evolutionary algorithm. PLoS ONE 2018, 13, e0201715. [Google Scholar] [CrossRef]
- Gonzalez, D.L.; Giannerini, S.; Rosa, R. On the origin of degeneracy in the genetic code. Interface Focus 2019, 9, 20190038. [Google Scholar] [CrossRef]
- Zhao, J.J.; Qi, B.; Ding, L.J.; Tang, X.Q. Based on RSCU and QRSCU research codon bias of F/10 and G/11 Xylanase. J. Food Sci. Biotechnol. 2010, 29, 755–764. [Google Scholar]
- Cavalier-Smith, T. Chloroplast evolution: Secondary symbiogenesis and multiple losses. Curr. Biol. 2002, 12, R62–R64. [Google Scholar] [CrossRef]
- Weng, M.L.; Blazier, J.C.; Govindu, M.; Jansen, R.K. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats and nucleotide substitution rates. Mol. Biol. Evol. 2013, 31, 645–659. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Zarei, A.; Zamani Faradonbeh, M.; Lawson, S. Evaluation of genetic variability among “Early Mature” Juglans regia using microsatellite markers and morphological traits. PeerJ 2017, 5, e3834. [Google Scholar] [CrossRef]
- Hu, Y.; Woeste, K.E.; Zhao, P. Completion of the chloroplast genomes of five Chinese Juglans and their contribution to chloroplast phylogeny. Front. Plant Sci. 2016, 7, 1955. [Google Scholar] [CrossRef]
- Zarei, A.; Sahraroo, A. Molecular characterization of Punica granatum l. accessions from Fars province of Iran using microsatellite markers. Hort Environ. Biotech. 2018, 59, 239–249. [Google Scholar] [CrossRef]
- Eguiluz, M.; Rodrigues, N.F.; Guzman, F.; Yuyama, P.; Margis, R. The chloroplast genome sequence from Eugenia uniflora, a myrtaceae from neotropics. Plant Sys. Evol. 2017, 303, 1199–1212. [Google Scholar] [CrossRef]
- AL-Saghir, M.G.; Porter, D.M. Taxonomic revision of the genus Pistacia L. (anacardiaceae). Am. J. Plant Sci. 2012, 3, 12–32. [Google Scholar] [CrossRef]
- Yi, T.; Wen, J.; Golan-Goldhirsh, A.; Parfitt, D.E. Phylogenetics and reticulate evolution in Pistacia (Anacardiaceae). Am. J. Bot. 2008, 95, 241–251. [Google Scholar] [CrossRef]
- Ibrahim Basha, A.; Padulosi, S.; Chabane, K.; Hadj-Hassan, A.; Dulloo, E.; Pagnotta, M.A.; Porceddu, E. Genetic diversity of Syrian pistachio (Pistacia vera L.) varieties evaluated by AFLP markers. Genet. Resour. Crop. Evol. 2007, 54, 1807–1816. [Google Scholar] [CrossRef]
- Ruhsam, M.; Rai, H.S.; Mathews, S.; Ross, T.G.; Graham, S.W.; Raubeson, L.A.; Mei, W.; Thomas, P.I.; Gardner, M.F.; Ennos, R.A.; et al. Does complete plastid genome sequencing improve species discrimination and phylogenetic resolution in Araucaria? Mol. Ecol. Res. 2015, 15, 1067–1078. [Google Scholar] [CrossRef]

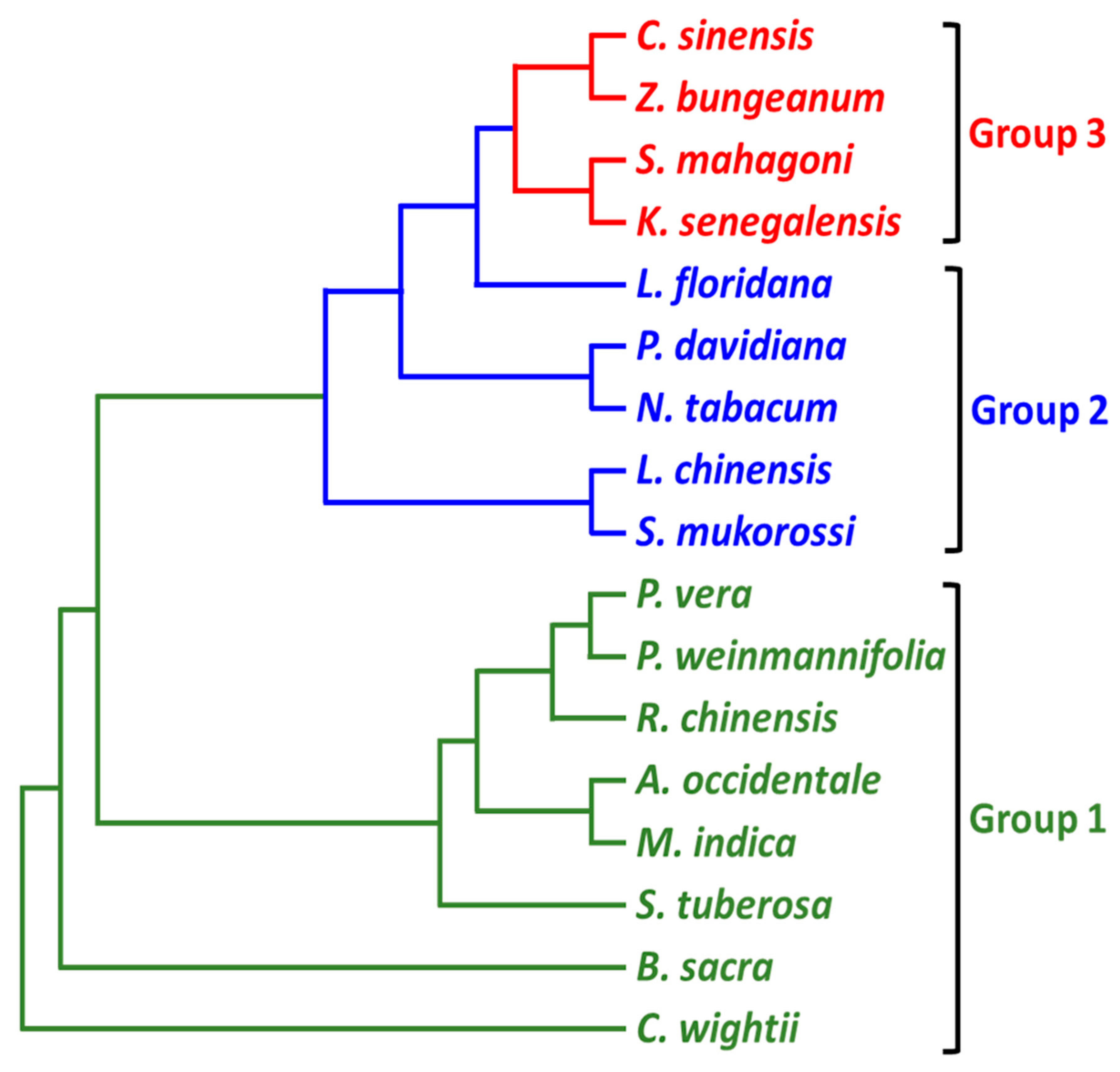
| Order | Family | Tribe | Genus | Species | NCBI Accession No. | Reference |
|---|---|---|---|---|---|---|
| Sapindales | Anacardiaceae | Rhoeae | Pistacia | P. vera | - | Current study |
| Sapindales | Anacardiaceae | Rhoeae | Pistacia | P. weinmannifolia | NC_037471.1 | [35] |
| Sapindales | Anacardiaceae | Rhoeae | Rhus | R. chinensis | NC_033535.1 | [36] |
| Sapindales | Anacardiaceae | Anacardieae | Anacardium | A. occidentale | NC_035235.1 | [37] |
| Sapindales | Anacardiaceae | Anacardieae | Mangifera | M. indica | NC_035239.1 | [37] |
| Sapindales | Anacardiaceae | Spondieae | Spondias | S. tuberosa | NC_030527.1 | [38] |
| Sapindales | Burseraceae | Bursereae | Commiphora | C. wightii | NC_036978.1 | unpublished |
| Sapindales | Burseraceae | Bursereae | Boswellia | B. sacra | NC_029420.1 | [39] |
| Sapindales | Sapindaceae | Nephelieae | Litchi | L. chinensis | NC_035238.1 | [37] |
| Sapindales | Sapindaceae | Sapindeae | Sapindus | S. mukorossi | KM454982.1 | [40] |
| Sapindales | Simaroubaceae | - | Leitneria | L. floridana | NC_030482.1 | [41] |
| Sapindales | Meliaceae | Swietenieae | Swietenia | S. mahagoni | NC_040009.1 | unpublished |
| Sapindales | Meliaceae | Swietenieae | Khaya | K. senegalensis | NC_037362.1 | [42] |
| Sapindales | Rutaceae | Aurantieae | Citrus | C. sinensis | DQ864733.1 | [43] |
| Sapindales | Rutaceae | Zanthoxyleae | Zanthoxylum | Z. bungeanum | NC_031386.1 | [44] |
| Rosales | Rosaceae | Pruneae | Prunus | P. davidiana | NC_039735.1 | [45] |
| Solanales | Solanaceae | Nicotianeae | Nicotiana | N. tabacum | Z00044.2 | [46] |
| Category | Gene Group | Gene Names |
|---|---|---|
| Self replication | Large subunit of ribosomal protein genes | rpl32, rpl23 a, rpl2 *,a, rpl33, rpl20, rpl36, rpl14, rpl16, rpl22 |
| Small subunit of ribosomal protein genes | rps7 a, rps12 *,a, rps15, rps19 a, rps16 *, rps2, rps4, rps14, rps18, rps11, rps8, rps3 | |
| DNA-dependent RNA polymerase genes | rpoA, rpoB, rpoC1 *,rpoC2 | |
| Ribosomal rRNA genes | CGW73_pgr008 a, (16S); CGW73_pgr007 a, (23S); CGW73_pgr008 a,(4.5S); CGW73_pgr0085, (5S) | |
| tRNA genes | trnM-CAU a, trnI-CAU a, trnL-CAA a, trnV-GAC a, trnI-GAU *,a, trnA-UGC *,a, trnR-ACG a, trnR-ACG a, trnR-ACG a, trnN-GUU a, trnL-UAG, trnH-GUG, trnH-GUG, trn-H-GUG, trnK-UUU *, trnQ-UUG, trnS-GCU, trnT-CGU *, trnG-UCC, trnR-UCU, trnD-GUC, trnD-GUC, trnY-GUA, trnE-UUC *, trnE-UUC *, trnT-UGU, trnS-GGA, trnM-CAU, trnM-CAU, trnS-UGA, trnG-GCC, trnG-GCC, trnT-GGU, trnL-UAA *, trnF-GAA, trnV-UAC *, trnM-CAU, trnW-CCA, trnP-UGG, trnP-UGG, trnE-UUC * | |
| Photosynthesis | Photosystem I | psaA, psaB, psaJ, psaC |
| Photosystem II | psbA, psbk, psbI, psbM, psbZ, psbC, psbD, psbJ, psbL, psbF, psbE, psbB, psbT, psbN, psbH | |
| Cytochrome b/f complex | petN, petA, petL, petG, petB, petD | |
| ATP synthase | atpA, atpF *, atpH, atpI, atpE, atpB | |
| NADH dehydrogenase | ndhB *,a, ndhF, ndhD, ndhE, ndhG, ndhI, ndhA *, ndhH, ndhJ, ndhK, ndhC | |
| RubisCo large subunit | rbcL | |
| Other genes | Protease | clpP * |
| Translation initiation factor | infA | |
| Maturase | matK | |
| Acetyl-CoA-carboxylase (subunit) | accD | |
| Envelope membrane protein | cemA | |
| Cytochrome synthesis (C-Type) | ccsA | |
| Chloroplast reading frames (hypothetical) | ycf2 a, ycf1, ycf3 *,ycf4 |
| T(U) | A | C | G | GC Content | SSRs | Length (bp) | % | |
|---|---|---|---|---|---|---|---|---|
| Whole Genome | 31.37 | 30.76 | 19.26 | 18.61 | 37.87 | 91 | 160,598 | 100 |
| LSC | 30.67 | 36.20 | 16.89 | 16.24 | 33.13 | 64 (70.33%) | 88,174 | 54.9 |
| SSC | 33.72 | 33.92 | 16.81 | 15.55 | 32.36 | 15 (16.48%) | 19,330 | 12.04 |
| IR | 28.38 | 28.67 | 20.67 | 22.27 | 42.94 | 12 (13.19%) | 26,547 | 16.53 |
| tRNA | 26.64 | 23.23 | 22.25 | 27.88 | 50.13 | 0 | 2346 | 1.46% |
| rRNA | 21.20 | 24.28 | 26.14 | 28.38 | 54.52 | 0 | 10,642 | 6.63% |
| Intron | 33.02 | 30.30 | 19.42 | 17.26 | 36.68 | 4 (4.4%) | 9099 | 5.65% |
| Intergenic space | 33.16 | 32.77 | 17.28 | 16.79 | 34.07 | 78 (85.71%) | 67,561 | 42.06 |
| Gene | Location | Strand | Start | End | Exon 1 | Intron 1 | Exon II | Intron II | Exon III |
|---|---|---|---|---|---|---|---|---|---|
| Base Pair (bp) | |||||||||
| trnR-TCT | LSC | - | 127 | 351 | 35 | 152 | 38 | ||
| atpF | LSC | - | 13,218 | 14,526 | 148 | 694 | 467 | ||
| rpoC1 | LSC | - | 22,314 | 25,133 | 435 | 777 | 1608 | ||
| trn-CTA | LSC | - | 39,132 | 39,262 | 38 | 56 | 37 | ||
| psaA | LSC | - | 42,601 | 44,742 | 1788 | 30 | 324 | ||
| ycf3 | LSC | - | 45,534 | 47,564 | 126 | 724 | 230 | 801 | 150 |
| clpP | LSC | - | 73,656 | 75,702 | 74 | 818 | 290 | 641 | 224 |
| rpl2 | IR | - | 88,274 | 89,760 | 394 | 626 | 467 | ||
| ycf2 | IR | + | 92,992 | 97,260 | 2404 | 57 | 1808 | ||
| ycf15 | IR | + | 97,351 | 97,948 | 155 | 292 | 151 | ||
| ndhB | IR | - | 98,944 | 101,154 | 870 | 588 | 753 | ||
| rrn23S | IR | + | 108,235 | 111,706 | 160 | 38 | 3274 | ||
| ndhA | SSC | - | 125,134 | 127,349 | 556 | 1124 | 536 | ||
| rrn23S | IR | - | 136,889 | 140,360 | 160 | 38 | 3274 | ||
| ndhB | IR | + | 147,441 | 149,654 | 777 | 681 | 756 | ||
| ycf15 | IR | - | 150,650 | 151,244 | 158 | 289 | 148 | ||
| ycf2 | IR | - | 151,338 | 155,633 | 2428 | 33 | 1835 | ||
| rpl2 | IR | + | 158,853 | 160,342 | 391 | 629 | 470 | ||
| Amino acid | Genome | Codon | No. | RSCU * | tRNA | Amino Acid | Genome | Codon | No. | RSCU * | tRNA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| (%) | (%) | ||||||||||
| Ala | 5.6 | GCA | 399 | 1.13 | trnA-UGC | Ile | 8.53 | ATA | 673 | 0.94 | trnI-UAU |
| GCC | 235 | 0.66 | ATC | 450 | 0.62 | trnI-GAU | |||||
| GCG | 174 | 0.49 | ATT | 1072 | 1.47 | ||||||
| GCT | 633 | 1.79 | Lys | 4.91 | AAA | 946 | 1.5 | trnK-UUU | |||
| Cys | 1.3 | TGT | 228 | 1.43 | trnC-ACA | AAG | 319 | 0.5 | |||
| TGC | 107 | 0.67 | Leu | 10.69 | CTA | 409 | 0.89 | trnL-UAG | |||
| Asp | 3.86 | GAT | 769 | 1.55 | CTC | 202 | 0.44 | ||||
| GAC | 226 | 0.45 | trnD-GUC | CTG | 201 | 0.44 | |||||
| Glu | 4.79 | GAA | 917 | 1.49 | trnE-UUC | CTT | 557 | 1.21 | trnL-UAA | ||
| GAG | 317 | 0.51 | TTA | 841 | 1.83 | ||||||
| Phe | 5.71 | TTC | 519 | 0.71 | trnF-GAA | TTG | 541 | 1.18 | trnL-CAA | ||
| TTT | 952 | 1.29 | Met | 2.49 | ATG | 584 | 1 | trnM-CAU | |||
| Gly | 7.16 | GGA | 718 | 1.56 | trnG-UCC | Asn | 4.6 | AAC | 297 | 0.5 | trnN-GUU |
| GGC | 192 | 0.42 | trnG-GCC | AAT | 886 | 1.5 | |||||
| GGG | 329 | 0.71 | Pro | 4.08 | CCA | 311 | 1.18 | trnP-UGG | |||
| GGT | 603 | 1.31 | CCC | 205 | 0.78 | ||||||
| His | 2.56 | CAC | 172 | 0.52 | trnH-GUG | CCG | 143 | 0.54 | |||
| CAT | 487 | 1.48 | CCT | 392 | 1.49 | ||||||
| Gln | 3.34 | CAA | 642 | 1.49 | trnQ-UUG | ||||||
| Thr | 5.19 | ACA | 405 | 1.21 | trnT-UGU | CAG | 219 | 0.51 | |||
| ACC | 275 | 0.82 | trnT-GGU | Arg | 5.96 | AGA | 466 | 1.82 | trnR-UCU | ||
| ACG | 149 | 0.45 | trnT-CGU | AGG | 166 | 0.65 | |||||
| ACT | 507 | 1.52 | CGA | 342 | 1.34 | trnR-ACG | |||||
| Val | 5.49 | GTA | 519 | 1.47 | trnV-UAC | CGC | 114 | 0.45 | |||
| GTC | 192 | 0.54 | trnV-GAC | CGG | 131 | 0.51 | |||||
| GTG | 192 | 0.54 | CGT | 316 | 1.24 | ||||||
| GTT | 511 | 1.45 | Ser | 7.77 | AGC | 117 | 0.35 | trnS-GCU | |||
| Trp | 1.74 | TGG | 467 | 1 | trnW-CCA | AGT | 417 | 1.25 | |||
| Tyr | 3.65 | TAC | 209 | 0.44 | trnY-GUA | TCA | 389 | 1.17 | trnS-UGA | ||
| TAT | 731 | 1.56 | TCC | 345 | 1.03 | trnS-GGA | |||||
| Stop | 0.57 | TAA | 63 | 1.28 | TCG | 186 | 0.56 | trnS-CGA | |||
| TAG | 42 | 0.85 | TCT | 547 | 1.64 | ||||||
| TGA | 43 | 0.87 |
| Species | Pistacia vera | Pistacia weinmannifolia | Anacardium occidentale | Mangifera indica | Rhus chinensis | Spondias tuberosa | Variation | SD |
|---|---|---|---|---|---|---|---|---|
| Single-copy region (large) | ||||||||
| Length (bp) | 88,174 | 88,402 | 87,727 | 86,673 | 96,882 | 89,550 | 10,209 bp | 4.13 |
| G + C (%) | 33.13 | 36 | 35.7 | 36 | 36.2 | 35.8 | 3.0% | 3.27 |
| Length (%) | 54.9 | 54.98 | 50.94 | 54.93 | 65.02 | 55.26 | 14.0% | 8.41 |
| Single-copy region (small) | ||||||||
| Length (bp) | 19,330 | 19,129 | 19,046 | 18,349 | 18,647 | 18,399 | 981 bp | 2.17 |
| G + C (%) | 32.36 | 32.9 | 32 | 32.4 | 32.5 | 32.2 | 1.0% | 0.94 |
| Length (%) | 12.04 | 11.9 | 11.06 | 11.63 | 12.51 | 11.36 | 1.0% | 4.38 |
| Inverted repeat | ||||||||
| Length (bp) | 26,547 | 26,618 | 32,713 | 25,792 | 33,474 | 27,045 | 7,684 bp | 11.98 |
| G + C (%) | 42.94 | 42.9 | 43 | 43 | 45.4 | 42.7 | 3.0% | 2.36 |
| Length (%) | 16.53 | 16.56 | 19 | 16.34 | 22.47 | 16.69 | 6.1% | 6.53 |
| Total | ||||||||
| Length (bp) | 160,598 | 160,767 | 172,199 | 157,780 | 149,011 | 162,039 | 23,188 bp | 4.65 |
| G + C (%) | 37.87 | 36.8 | 38.1 | 37.9 | 37.7 | 37.7 | 1.0% | 1.21 |
| Length (%) | 100 | 100 | 100 | 100 | 100 | 100 | 0.0% | 0 |
| Total genes | 132 (136) (+121) | 131 | 126 | 112 | 120 | 117 | 20 | 6.5 |
| Protein-coding genes | 87 | 87 | 79 | 78 | 84 | 83 | 9 | 4.64 |
| tRNA | 30 | 37 | 29 | 30 | 32 | 30 | 3 | 9.4 |
| rRNA | 4 | 4 | 4 | 4 | 4 | 4 | 0 | 0 |
| Intron-containing genes (# with 2 introns) | 13 (2 with 2) | 16 (2 with 2) | 12 (3 with 2) | 16 (2 with 2) | 13 (2 with 2) | 14 (3 with 2) | - | 9.56 |
| SSRs/Compound SSRs | 91/12 | 86/9 | 78/8 | 73/8 | 76/8 | 14/3 | 19 | 11.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarei, A.; Ebrahimi, A.; Mathur, S.; Lawson, S. The First Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Pistachio (Pistacia vera). Diversity 2022, 14, 577. https://doi.org/10.3390/d14070577
Zarei A, Ebrahimi A, Mathur S, Lawson S. The First Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Pistachio (Pistacia vera). Diversity. 2022; 14(7):577. https://doi.org/10.3390/d14070577
Chicago/Turabian StyleZarei, Abdolkarim, Aziz Ebrahimi, Samarth Mathur, and Shaneka Lawson. 2022. "The First Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Pistachio (Pistacia vera)" Diversity 14, no. 7: 577. https://doi.org/10.3390/d14070577
APA StyleZarei, A., Ebrahimi, A., Mathur, S., & Lawson, S. (2022). The First Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Pistachio (Pistacia vera). Diversity, 14(7), 577. https://doi.org/10.3390/d14070577






