Abstract
In the present study, we used the integrative taxonomy approach to redescribe Minibiotus intermedius based on the newly found topotypic population in Marburg (Germany). As the original type material is not available, we designate a neotype to stabilize the taxonomy of the genus Minibiotus. Obtained mitochondrial COI barcode sequence and nuclear markers, i.e., 18S rRNA and 28S rRNA of M. intermedius from the neotype locality, were unique and distinct from those deposited in GenBank. In the first redescription of M. intermedius, only four specimens and no eggs from the neotype locality were analyzed. Moreover, genetic analyses were not conducted and barcodes were not available. Therefore, the present study, by establishing the neotype and providing integrative data on the neotype population, helps to better define the Minibiotus taxonomy and prevents further misunderstandings in the future.
1. Introduction
Tardigrades inhabit aquatic (freshwater and marine) and terrestrial habitats, from the highest mountain peaks to deepest oceans, from the polar regions to the tropics. They are found in lichens, mosses, leaf litter, soil, sediments, and on aquatic plants [1]. Up to now, about 1400 species of hetero and eutardigrades have been described throughout the world [2,3,4,5].
The genus Minibiotus R.O. Schuster [6] was established more than forty years ago by Schuster et al. [6]. However, at first, the new genus was not easily accepted by other researchers who questioned its validity due to an insufficiently clear diagnosis [7,8]. Later, Claxton [9] published the most comprehensive revision of this genus based on animal and egg morphology, strengthening the generic diagnosis and, at the same time, its validity. In that time, 22 species were included into the genus Minibiotus; however, until now, many new species have been described. According to the current tardigrade checklist, 48 species are known in this genus [5]. The genus is mainly characterized by their antero-ventral mouth surrounded by 10 papulae, a narrow buccal tube with either a single or a double curvature, short ventral support, two or three short macroplacoids, and short macroplacoid row length [10]. However, recently, Stec et al. [10,11] stated that genetic data for the genus are very scarce (i.e., only 39 Minibiotus DNA sequences deposited in GenBank and many of them are not identified to species level), and also that detailed high-resolution data on the genus morphology are extremely limited. Up to now, genetic sequences are available for four nominal Minibiotus taxa: M. furcatus (Ehrenberg [12]), M. gumersindoi (Guil and Guidetti [13]), M. ioculator (Stec, Kristensen, and Michalczyk [10]), and M. pentannulatus (Londoño, Daza, Lisi, and Quiroga [10,14,15,16]). The available DNA sequences and extreme morphological diversity suggest that Minibiotus is probably polyphyletic and some species are closely related to Paramacrobiotus Guidetti et al. [16,17,18]. The mentioned phenotypic diversity of the genus accounts for the presence or absence of pores in the animal cuticle, egg processes enclosed within or without membrane, and bucco-pharyngal apparatuses with two or three macroplacoids [18,19]. The hypothesis on Minibiotus polyphyly based on the morphological diversity present in the genus is further supported by the recent study by Stec and Morek [20], who found and clarified a similar situation within the genus Tenuibiotus (Pilato and Lisi [21]). This study demonstrated that such morphological characteristics might generally be considered conservative and stable at the genus level. Importantly, beside the paraphyly problem in the genus Minibiotus, the more pressing issue concerns the insufficient description and characterization of the type species for the genus—M. intermedius (Plate [22]). As shown by several recent studies, integrative redescriptions of such important taxa are crucial in opening the widow for more precise quantification and description of tardigrade species diversity (e.g., [23,24,25,26,27,28,29,30,31,32,33]). Therefore, the correct and detailed redescription of type taxa is drastically needed.
In this paper, we provide an integrative redescription of M. intermedius from its original type locality in Marburg (Germany). The redescription is based on detailed phenotypic data of animals and eggs collected with the use of light and scanning electron microscopy. This morphological information is tightly associated with genetic data in the form of DNA sequences of three molecular markers (18S rRNA, 28S rRNA and COI).
2. Materials and Methods
2.1. Sampling
A single sample of mosses and lichens from a tree was collected in a mixed forest in Marburg (Germany) in September 2019 (sample code (SC) GR2, for more details see below). The sample was then packed in a paper envelope; dried at a temperature of ca. 20 °C; and delivered to the Department of Animal Taxonomy and Ecology at the Faculty of Biology, Adam Mickiewicz University in Poznań, Poland. The tardigrade collection, extraction, and mounting techniques followed the protocol of Dastych [34].
2.2. Microscopy and Imaging
A total of 38 specimens and eight eggs were mounted on microscope slides in Hoyer’s medium, and then examined under an Olympus BX41 Phase Contrast light Microscope (PCM) associated with an Olympus SC50 digital camera (Olympus Corporation, Shinjuku-ku, Japan).
Three eggs were prepared for scanning electron microscope (SEM) analysis according to the protocol in Roszkowska et al. [35] and examined under a high vacuum in Hitachi S3000N SEM.
All figures were assembled in Corel Photo-Paint 2017. For deep structures that could not be fully focused in a single photograph, a series of 2–50 images was taken every ca. 0.5 μm depth and then manually assembled into a single deep-focus image in Corel Photo-Paint 2017.
2.3. Morphometrics and Morphological Nomenclature
All measurements are given in micrometres [μm]. The sample size was adjusted following recommendations by Stec et al. [36]. Structures were measured only if their orientation was suitable. Body length was measured from the anterior extremity to the end of the body, excluding the hind legs. The type of bucco-pharyngeal apparatus and claws was classified according to Pilato and Binda [37]. The terminology used to describe oral cavity armature and egg shell morphology follows Michalczyk and Kaczmarek [38] and Kaczmarek and Michalczyk [39]. Macroplacoid length sequence is given according to Kaczmarek et al. [40]. The buccal tube length and the level of the stylet support insertion point were measured according to Pilato [41]. The pt index is the ratio of the length of a given structure to the length of the buccal tube expressed as a percentage [41]. Cuticular bars and muscle attachments under claws were classified according to Kiosya et al. [42]. All other measurements and nomenclature follow Kaczmarek and Michalczyk [39]. Morphometric data were handled using the “Parachela” ver. 1.8 template available from the Tardigrada Register [43]. Raw morphometric data for each analyzed species are provided as Supplementary Materials (SM. 1). Tardigrade taxonomy follows Bertolani et al. [16] and Stec et al. [18].
2.4. Genotyping
Three specimens of M. intermedius (isolates numbers: Min3GR, Min4GR, and Min6GR) were preliminarily identified in vivo using light microscopy (LM) prior to DNA extraction for genotyping analysis. Genomic DNA was extracted from individual animals using a Chelex® 100 resin (Bio-Rad) method [44] with modifications described in detail in Kaczmarek et al. [45]. The tardigrade exoskeleton was extracted from a pellet containing Chelex beads on the bottom of each tube. Obtained exoskeletons were mounted on a microscope slide in Hoyer’s medium for further morphological analysis and deposited in the collection of the Department of Animal Taxonomy and Ecology, Faculty of Biology, Adam Mickiewicz University in Poznań (Poland).
We sequenced three DNA fragments with different mutation rates. A fragment of the cytochrome oxidase subunit I (COI, mtDNA) was amplified using universal primers: HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) and LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) [46]. In turn, the cytoplasmic ribosome small (18S rRNA, nDNA) and large (28S rRNA, nDNA) subunit components were amplified using the following primers: SSU01_F (5′-AACCTGGTTGATCCTGCCAGT-3′) and SSU82_R (5′-TGATCCTTCTGCAGGTTCACCTAC-3′) [47] for the 18S rRNA sequences and 28SF0001 (5′-ACCCvCynAATTTAAGCATAT-3′) and 28SR0990 (5′-CCTTGGTCCGTGTTTCAAGAC-3′) [48] for the 28S rRNA sequences. For every PCR reaction, the solution contained 7.5 μL of ddH2O, 0.5 μL of 5 μM forward primer, 0.5 μL of 5 μM reverse primer, 10 μL of JumpStart™ Taq ReadyMix™ DNA polymerase (Sigma-Aldrich™), and 1.5 μL of genomic DNA extract. The PCR protocols for amplification of the mitochondrial COI gene fragment and nuclear 18S rRNA sequences, agarose gel electrophoresis, and sequencing were performed according to Mioduchowska et al. [49]. In turn, the PCR cycling profile to amplify the 28S rRNA gene fragment was as follows: initial denaturation at 95 °C for 5 min followed by 40 cycles of 95 °C for 30 s, 50 °C for 1 min, and 72 °C for 90 s, ending with 72 °C for 5 min.
2.5. Comparative Molecular Analysis
Obtained mitochondrial and nuclear sequences were checked for quality and were manually aligned in BioEdit v. 7.2.5 [50]. Basic local alignment search tool (BLAST) [51] searches were performed to verify the identity and homology of the obtained gene fragments with sequences deposited in the NCBI database. The COI sequence was translated into amino acid sequences with the invertebrate mitochondrial codon table and the 2th reading frame using the EMBOSS-TRANSEQ application [52,53].
For molecular comparisons, all sequences of species belonging to the genus Minibiotus were downloaded from the GenBank database and aligned using the ClustalW multiple alignment tool [54], implemented in BioEdit v. 7.2.5. Only the available GenBank sequences that represented a homologous fragment with nrDNA and mtDNA sequences obtained in our study were applied. Alignment sequences were trimmed to 472, 1138, and 678 bp for COI (13 sequences), 18S rRNA (6 sequences), and 28S rRNA (10 sequences) molecular markers, respectively. The uncorrected p-distances were calculated using pairwise deletion for the gap/missing data treatment option and the software MEGA X [55]. Detailed p-distance tables are presented as Supplementary Materials (SM.02).
Phylogenetic trees were computed using the software MEGA X by applying maximum likelihood (ML) analysis under the general settings of selected models with 1000 bootstraps. The best-fit substitution models were determined using jModelTest v. 2.1.4 [56] with the assumptions of both the Bayesian inference criterion (BIC) and the Akaike information criterion (AIC) [57]: the Hasegawa–Kishino–Yano with gamma (HKY + G) distribution (G parameter = 0.1899) [58] for COI sequences, the Kimura 2-parameter (K2) model [59] for 18S rRNA sequences, and the Kimura 2-parameter (K2 + G) model with gamma distribution (G parameter = 0.1709) for 28S rRNA sequences. The molecular markers of Macrobiotus porifini (GenBank accession numbers: COI–MT246659, 18S rRNA–MT241900, 28S rRNA–MT241897 [60]) were used as outgroups. Initial evolutionary trees for the heuristic search were generated by applying BioNJ and neighbor join algorithms to a matrix of pairwise distances (which was estimated using the maximum composite likelihood (MCL) approach). Finally, the topology of phylogenetic trees was selected with superior log likelihood value and visualized by FigTree v.1.4.3 and Inkscape 0.92.
All obtained sequences were deposited in GenBank under the following accession numbers: COI–ON005160, 18S rRNA–ON005188-8,9 and 28S rRNA–ON005193-95 (see also SM.02).
3. Results
3.1. Taxonomic Account
Phylum: Tardigrada Doyère [61]
Class: Eutardigrada Richters [62]
Order: Parachela Schuster, Nelson, Grigarick, and Christenberry [6]
Superfamily: Macrobiotoidea Thulin [63] (in [64])
Family: Macrobiotidae Thulin [63]
Genus: Minibiotus R.O. Schuster [6] (in [6])
Minibiotus intermedius (Plate [22])

Table 1.
Measurements [in μm] of selected morphological structures of individuals of Minibiotus intermedius mounted in Hoyer’s medium (N—number of specimens/structures measured, RANGE refers to the smallest and the largest structure among all measured specimens; SD—standard deviation).

Table 2.
Measurements [in μm] of selected morphological structures of the eggs of Minibiotus intermedius mounted in Hoyer’s medium (N–number of eggs/structures measured, RANGE refers to the smallest and the largest structure among all measured specimens; SD—standard deviation).
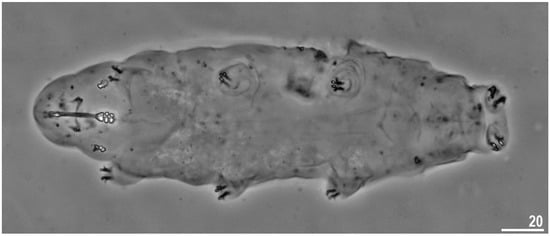
Figure 1.
Minibiotus intermedius: adult specimen, dorso-ventral projection (neotype, PCM). Scale bar in µm.
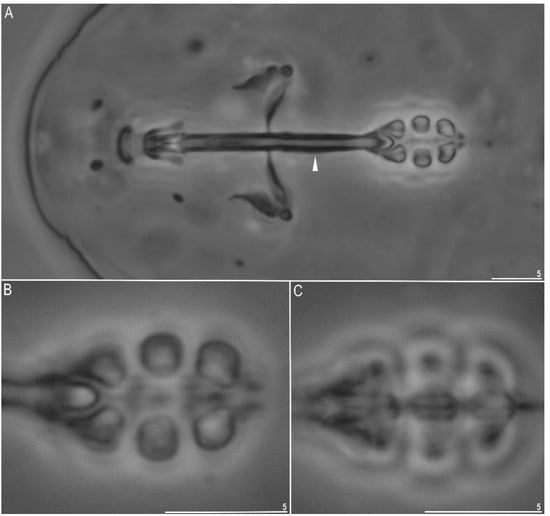
Figure 2.
Minibiotus intermedius: (A) bucco-pharyngeal apparatus, general view (neoparatype) (filled arrowhead indicates a thickening below stylet insertion point; (B) lateral rows of placoids (neoparatype); (C) central row of placoids (neoparatype). All in PCM. Scale bars in µm.
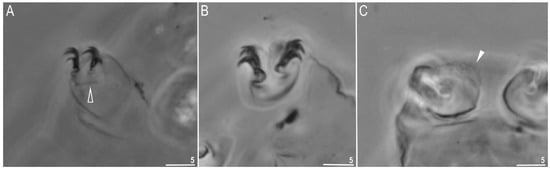
Figure 3.
Minibiotus intermedius: (A) claws I, empty arrowhead indicates double muscle attachments under the claws. (paratype); (B) claws IV (neotype); (C) filled arrowhead indicates granulation on leg IV (neotype). All in PCM. Scale bars in µm.
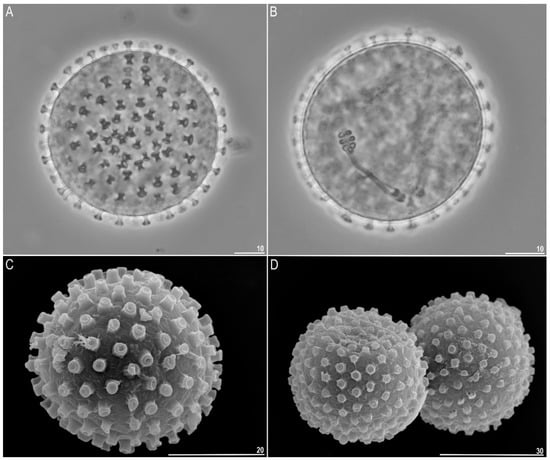
Figure 4.
Minibiotus intermedius: (A) egg chorion (PCM); (B) embryo in the egg (PCM); (C,D) egg chorion (SEM). Scale bars in µm.
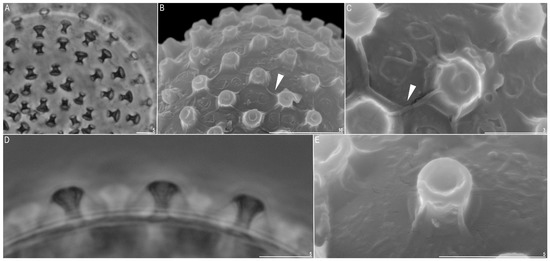
Figure 5.
Minibiotus intermedius: (A) egg surface with processes visible (PCM); (B,C) egg surface with processes (SEM) (filled arrowheads indicate striae); (D) egg processes, lateral view (PCM); (E) egg process (SEM). Scale bars in µm.
Neotype locality: Germany, 50°47′49″ N, 08°46′45″ E, 247 m asl, State of Hessen, Marburg, mosses and lichens from tree, mixed forest, 2 September 2019, coll. Johenn Sholl.
Material examined: 38 specimens and 8 eggs, i.e., neotype + 45 neoparatypes (37 specimens and 8 eggs) mounted on microscope slides in Hoyer’s medium, 3 eggs prepared for SEM, and 10 specimens prepared for molecular analyses. However, DNA sequences were obtained only from three specimens (isolates numbers Min3GR, Min4GR, and Min6GR).
Type depositories: Neotype (slide GR2/12), 35 neoparatypes (slides GR 2/2, and GR2/4–2/14) are deposited at the Department of Animal Taxonomy and Ecology, Faculty of Biology, Adam Mickiewicz University in Poznań, Uniwersystetu Poznańskiego 6, 61-614 Poznań (Poland). Ten neoparatypes (slides GR22/14 and GR2/16–2/18) are deposited at the Institute of Systematics and Evolution of Animals, Polish Academy of Sciences, Sławkowska 17, 31-016 Kraków (Poland).
3.1.1. Short Diagnosis
The body cuticle was smooth without pores and granulation, but very poorly visible granulation on the IV pair of legs was present. Large eye spots were also present. The buccal tube with an anterior and a posterior bend and slightly thicker below the stylet insertion point. The pharyngeal bulb had apophyses, three granular macroplacoids, and a small microplacoid (length sequence 2 ≤ 3 < 1). Eggs with processes (28–32 around the circumference) in the shape of a screw’s head were covered by a separate membrane.
3.1.2. Description of the Neotypic Population
Animals (measurements and statistics in Table 1). Body white in live specimens and transparent after fixation in Hoyer’s medium (Figure 1). Eyes were present in all specimens after mounting in the Hoyer’s medium (Figure 1). The body cuticle was smooth without pores and granulation (Figure 1).
Mouth antero-ventral surrounded by ten peribuccal papulae or shortened lamellae. The bucco-pharyngeal apparatus of the Minibiotus type (Figure 2A) had an anterior and a posterior bend (clearly visible in lateral view). The buccal tube was slightly thicker below the stylet insertion point (Figure 2A, arrow). Under PCM, only a third band of teeth was faintly visible (Figure 2A).
The pharyngeal bulb was spherical, with large triangular apophyses, three granular macroplacoids, and very small granular microplacoid placed very close to the third macroplacoid (Figure 2A,B). The macroplacoid length sequence was 2 ≤ 3 < 1. The first macroplacoid narrowed anteriorly (Figure 2A,B). All macroplacoids without constrictions (Figure 2A–C).
Claws stout, of the hufelandi type (Figure 3A,B). The primary branches had very large and distinct accessory points, a common tract, and an evident stalk (Figure 3A,B). Under PCM, very poorly visible granulation was present on the IV pair of legs (visible mainly in larger specimens) (Figure 3C, filled arrowhead). The lunulae was smooth on all legs. The cuticular bars under the claws were absent. Double muscle attachments were faintly marked under PCM (Figure 3A, empty arrowhead).
Eggs (measurements and statistics in Table 2) laid freely, white, spherical, or slightly ovoid (Figure 4A–D). Processes were nail-shaped (shaped like the head of a screw) (Figure 4A–D and Figure 5A–E). Each process was covered by a separate membrane (Figure 5A,B,D). The heads of the processes were always wider than the process bases. In SEM, a central and deep depression was present at the top of the processes (Figure 5B,C,E). This structure was also visible under PCM as a lighter circles at the processes tops (5D). Under PCM the egg surface between process smooth whereas under SEM usually striae extend from each process connecting it with the neighboring processes (Figure 5B,C, arrows). These striae (which are in fact formed by membrane) formed four quadratic areas that surrounded each egg process resembling poorly marked areolae. Within each of these areas’ wrinkles form 1–2 flat rose-like whorl structures (Figure 5B,C, arrows).
DNA sequences
We obtained good-quality sequences for the applied molecular markers:
- COI—GenBank: ON005160, 634 bp long;
- 18S rRNA—GenBank: ON005188-89, 1182 bp long;
- 28S rRNA—GenBank: ON005193-95, 753 bp long.
3.2. Comparisons with Other Genetic Sequences of Minibiotus Taxa
All the obtained sequences of M. intermedius were unique and distinct from those deposited in GenBank. The COI molecular marker exhibited a single sequence. In the conservative 18S rRNA gene fragment, we observed no differences between two sequences. In 28S rRNA, two haplotypes were found (three sequences), with a p-distance of 0.6%.
The ranges of uncorrected genetic p-distances between M. intermedius and other species/taxa belonging to the genus Minibiotus are as follows (please see SM2):
- (a)
- COI: 21.3–28.4% (23.7% on average), with the most similar being M. gumersindoi (FJ435803 [14]), M. furcatus (JX683828-29 [65]), and Minibiotus sp. (MW306857 [66]), and the least similar being M. ioculator (MT023412 [10]);
- (b)
- 18S rRNA: 0.4–1.0% (0.9% on average), with the most similar being M. gumersindoi (FJ435748 [15]), and the least similar being Minibiotus sp. (EU266934 [47]);
- (c)
- 28S rRNA: 4.8–13.7% (9.4% on average), with the most similar being M. gumersindoi (FJ435761 [15]), and the least similar being M. pentannulatus (MT024043 [10]).
3.3. Morphological Differential Diagnosis
Minibiotus intermedius, by the morphology of adults (smooth dorsal and ventral cuticle and absence of pores) and eggs (processes in shape of a screw’s head and covered by a separate membrane), is most similar to: M. continuus (Pilato and Lisi [67]), M. floriparus (Claxton [10]), and M. taiti (Claxton [10]). However it differs from:
- M. continuus by: the presence of eyes, a different macroplacoid length sequence (2 ≤ 3 < 1 in M. intermedius vs. 1 = 2 = 3 in M. continuus), and a higher number of processes on the egg circumference (28–32 in M. intermedius vs. 21–22 in M. continuus).
- M. floriparus by: a different macroplacoid length sequence (2 ≤ 3 < 1 in M. intermedius vs. 2 < 1 = 3 in M. floriparus), a lower pt of the stylet support insertion point (pt: 53.8–56.3 in M. intermedius vs. pt: ca. 64.4 in M. floriparus), the absence of granulation on legs I–III, the lack of pores on the distal tops of egg processes, larger eggs (egg bare diameter: 46.3–54.5 μm and full diameter: 52.1–61.4 μm in M. intermedius vs. ca. 62.0 μm and ca. 70.0 μm, respectively, in M. floriparus), lower egg processes (3.0–3.9 μm in M. intermedius vs. 5.5–6.0 μm in M. floriparus), narrower tops of processes (2.4–3.5 μm in M. intermedius vs. 6.0–7.0 μm in M. floriparus), and a higher number of processes on the egg circumference (28–32 in M. intermedius vs. 20–22 in M. floriparus).
- M. taiti by: lower pt of stylet support insertion point (pt: 53.8–56.3 in M. intermedius vs. pt: ca. 60.3 in M. taiti), the absence of granulation on legs I–III, and the lack of rings of small circles around the central pore on the top of egg processes.
3.4. Establishing of the New Neotype and Neoparatypes of M. intermedius
Taking into consideration that M. intermedius was described in 1888 by Plate, based on specimens from Chile and Germany, we can probably assume that the type material of M. intermedius no longer exists. What is more accurate diagnose of the species were poorly described in the past so that it was necessary to establish a neotype series of this species. Claxton [9] redescribed M. intermedius and established neotype altogether with three syntypes for four specimens collected in Marburg on 27 August 1994. Importantly, Claxton did not report any eggs from this locality, and she unjustifiably assigned eggs from other localities (Africa, Australia, Europe, New Zealand, and North and South America) for this redescription. This action should be criticized as it is commonly known that cryptic species or complexes of extremely similar species are often reported for tardigrades. In such a case, one should be extremely careful while assigning morphotype of eggs to the animal morphotype, especially in groups such as macrobiotids, in which egg ornaments hold a number of characters which are important in species identification. Moreover, the exact location (with geographic coordinates) in Marburg was also not reported by Claxton [9]. In addition, so far, DNA barcodes are unknown for M. intermedius, as confident species identification is currently impossible.
For this reason, considering all issues regarding the Claxton’s neotype, we decided that existing neotype and syntypes are invalid due to the lack of eggs from the locality, which makes the correct identification of the species impossible (see ICZN, article 75.3.4 and 75.3.5). In such a situation, we designate a new neotype (specimen) altogether with 45 neoparatypes (37 specimens and 8 eggs) of M. intermedius collected from the type locality in Marburg (Germany), which is in agreement with ICZN article 75.3.5. The detailed characterization of the neotype population by integrated analysis can stabilize the taxonomy of the genus and allow for more detailed exploration of its diversity. Specimens of the neotype series were deposited in the Department of Animal Taxonomy and Ecology of Adam Mickiewicz University in Poznań, Poland, as well as in the Institute of Systematics and Evolution of Animals of the Polish Academy of Sciences, Kraków, Poland. All the above-mentioned statements are in accordance with the International Commission on Zoological Nomenclature (ICZN) acts dedicated to establishing a neotype series.
4. Discussion
In our study, we used integrative taxonomy to describe M. intermedius specimens from one of reported terra typica—Marburg (Germany). Our analysis has shown that our specimens and their eggs are morphologically very similar to specimens and eggs used for the first redescription of this species by Claxton [9]. However, in the population studied by us, eggs were a little smaller than those described by Claxton [9], i.e. egg bare diameter: 40.0–45.0 μm (in [9]) vs. 46.3–54.5 μm (in studied population), egg full diameter: 45.0–52.0 μm vs. 52.1–61.4 μm. Moreover, a ring of tiny pores on the top of egg processes was reported by Claxton [9], though these pores were absent in the population in our study. However, as noted previously, eggs described by Claxton [9] were not collected in type locality in Marburg, but in many different localities. Therefore, in this situation, they cannot be confidently assigned to the true M. intermedius and it is very likely that they may belong to a completely different Minibiotus species. The detailed redescription presented by us in this study with detailed phenotypic and genetic data for the neotype population will effectively prevent similar misunderstandings in the future. This will directly contribute to the knowledge of the true distribution range of this species, which are most probably overestimated at present due to the mentioned identification problem [68,69,70,71,72,73,74].
Molecular markers (i.e., COI, 18S rRNA, and 28S rRNA sequences) were only available for four Minibiotus species and seven taxa belonging to an undefined species of the genus Minibiotus. The ML phylogenetic reconstructions based on the limited molecular data sets yielded a congruent topology (Figure 6) with two main clades: the first clade comprised M. intermedius (without pores) and M. gumersindoi (with pores) and the second clade contained M. furcatus (with pores). For M. pentannulatus (with pores) and M. ioculator (without pores), only COI and 28S rRNA sequences were available which were clustered together with the second clade. It is obvious that the taxonomic position of some sequences deposited in GenBank, described as M. furcatus, still needs further verification. We are not convinced which of these sequences have been correctly flagged, and all of them need revision. However, we assumed that the two COI sequences, i.e., deposited in GenBank as JX683828 and JX683829, seem to be questionable because they were clustered with the first clade.
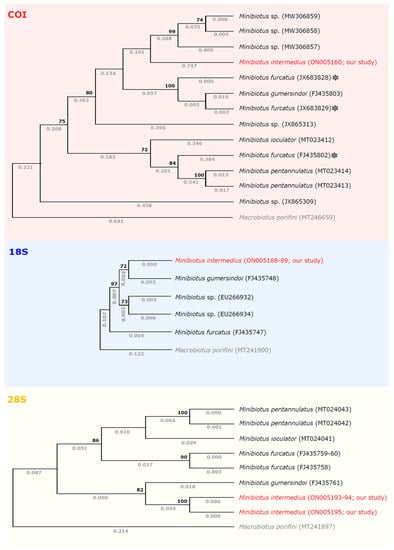
Figure 6.
The phylogenetic position (maximum likelihood analyses) of M. intermedius (marked in red) generated as follows: COI datasets under the HKY + G model, 18S rRNA datasets under the K2 model, and 28S rRNA datasets under the K2 + G model. The tree with the highest log likelihood is shown. Supporting bootstrap values are given above the branches (nodes with bootstrap <70 were collapsed) and the number of substitution events per site is given below the branches. The GenBank accession numbers of all the sequences applied are given in SM02. Species with a questionable position on the phylogenetic tree were marked in gray stars.
In summary, we propose a new population of the M. intermedius from Marburg as a type population of this species. Re-establishing the type species of the genus Minibiotus with provided integrative description should facilitate the description of new taxa within this genus.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d14050356/s1, SM1: Raw morphometric data of Minibiotus intermedius; SM2: Detailed p-distance between analyzed taxa.
Author Contributions
Conceptualization, Ł.K.; methodology, Ł.K., M.R., M.G., P.K. and M.M.; validation, Ł.K., M.R., M.G., P.K. and M.M.; formal analysis, Ł.K., M.R., M.G., P.K. and M.M.; investigation, Ł.K., M.R., P.K. and M.M.; resources, Ł.K.; writing—original draft preparation, Ł.K., M.R., P.K. and M.M.; writing—review and editing, Ł.K., M.R., M.G., P.K. and M.M.; visualization, M.R., M.G. and P.K.; supervision, Ł.K.; project administration, Ł.K.; funding acquisition, M.R., M.M. and P.K. All authors have read and agreed to the published version of the manuscript.
Funding
Milena Roszkowska and Pushpalata Kayastha are scholarship passport holders of the future interdisciplinary doctoral studies at the Faculty of Biology, Adam Mickiewicz University, Poznań POWR.03.02.00-00-I006/17. The work of Pushpalata Kayastha was also supported by grant UNIVERSYTET JUTRA No. POWR.03.05.00-00-Z303/17. The work of Monika Mioduchowska was supported by grant no. 2021/43/D/NZ8/00344.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Studies have been partially conducted in the framework of activities of the Biodiversity and Astrobiology Research group (BARG).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nelson, D.R.; Guidetti, R.; Rebecchi, L.; Kaczmarek, Ł.; McInnes, S.J. Phylum Tardigrada. In Thorp and Covich’s Freswater Invertebrates. Keys to Neotropical and Antarctic Fauna; Damborenea, C., Rogers, D.C., Thorp, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 5, pp. 505–522. [Google Scholar]
- Guidetti, R.G.; Bertolani, R.B. Tardigrade taxonomy: An updated check list of the taxa and a list of characters for their identification. Zootaxa 2005, 845, 1. [Google Scholar] [CrossRef]
- Degma, P.; Guidetti, R. Notes to the current checklist of Tardigrada. Zootaxa 2007, 1579, 41–53. [Google Scholar] [CrossRef]
- Vicente, F.; Bertolani, R. Considerations on the taxonomy of the Phylum Tardigrada. Zootaxa 2013, 3626, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Degma, P.; Bertolani, R.; Guidetti, R. Actual Checklist of Tardigrada Species. (2009–2021, 40th Edition: 19 July 2021). Available online: https://iris.unimore.it/retrieve/358743/Actual%20checklist%20of%20Tardigrada%2040th%20Edition%2019-07-21.pdf (accessed on 25 April 2022).
- Schuster, R.O.; Nelson, D.R.; Grigarick, A.A.; Christenberry, D. Systematic criteria of Eutardigrada. Trans. Am. Microsc. Soc. 1980, 99, 284–303. [Google Scholar] [CrossRef]
- Pilato, G. The systematics of Eutardigrada. J. Zool. Syst. Evol. Res. 1982, 20, 271–284. [Google Scholar] [CrossRef]
- Ramazzotti, G.; Maucci, W. Il Phylum Tardigrada. Mem. Ist. Ital. Idrobiol. 1983, 41, 1–1012. [Google Scholar]
- Claxton, S.K. A revision of the genus Minibiotus (Tardigrada: Macrobiotidae) with descriptions of eleven new species from Australia. Rec. Aust. Mus. 1998, 50, 125–160. [Google Scholar] [CrossRef] [Green Version]
- Stec, D.; Kristensen, R.M.; Michalczyk, Ł. An integrative description of Minibiotus ioculator sp. nov. from the Republic of South Africa with notes on Minibiotus pentannulatus Londoño et al., 2017 (Tardigrada: Macrobiotidae). Zool. Anz. A J. Comp. Zool. 2020, 286, 117–134. [Google Scholar] [CrossRef]
- Guidetti, R.; Bertolani, R.; Degma, P. New taxonomic position of several Macrobiotus species (Eutardigrada: Macrobiotidae). Zootaxa 2007, 1471, 61–68. [Google Scholar] [CrossRef]
- Ehrenberg, C.G. Beitrag zur Bestimmung des stationären mikroscopischen Lebens in bis 20,000 Fuss Alpenhöhe. Abhand. K. Akad. Wiss. 1859, 429–456. [Google Scholar]
- Guil, N.; Guidetti, R. A new species of Tardigrada (Eutardigrada: Macrobiotidae) from Iberian Peninsula and Canary Islands (Spain). Zootaxa 2005, 889, 1–11. [Google Scholar] [CrossRef]
- Londoño, R.; Daza, A.; Lisi, O.; Quiroga, S. New species of waterbear Minibiotus pentannulatus (Tardigrada: Macrobiotidae) from Colombia. Rev. Mex. Biodivers. 2017, 88, 807–814. [Google Scholar] [CrossRef]
- Guil, N.; Giribet, G. A comprehensive molecular phylogeny of tardigrades-adding genes and taxa to a poorly resolved phylum-level phylogeny. Cladistics 2012, 28, 21–49. [Google Scholar] [CrossRef] [PubMed]
- Bertolani, R.; Guidetti, R.; Marchioro, T.; Altiero, T.; Rebecchi, L.; Cesari, M. Phylogeny of Eutardigrada: New molecular data and their morphological support lead to the identification of new evolutionary lineages. Mol. Phylogenet. Evol. 2014, 76, 110–126. [Google Scholar] [CrossRef]
- Guidetti, R.; Schill, R.O.; Bertolani, R.; Dandekar, T.; Wolf, M. New molecular data for tardigrade phylogeny, with the erection of Paramacrobiotus gen. nov. J. Zool. Syst. Evol. Res. 2009, 47, 315–321. [Google Scholar] [CrossRef]
- Stec, D.; Vecchi, M.; Calhim, S.; Michalczyk, Ł. New multilocus phylogeny reorganises the family Macrobiotidae (Eutardigrada) and unveils complex morphological evolution of the Macrobiotus hufelandi group. Mol. Phylogenet. Evol. 2021, 160, 106987. [Google Scholar] [CrossRef]
- Stec, D.; Smolak, R.; Kaczmarek, Ł.; Michalczyk, Ł. An integrative description of Macrobiotus paulinae sp. nov. (Tardigrada: Eutardigrada: Macrobiotidae: Hufelandi group) from Kenya. Zootaxa 2015, 4052, 501–526. [Google Scholar] [CrossRef] [Green Version]
- Stec, D.; Morek, W. Reaching the Monophyly: Re-Evaluation of the Enigmatic Species Tenuibiotus hyperonyx (Maucci, 1983) and the Genus Tenuibiotus (Eutardigrada). Animals 2022, 12, 404. [Google Scholar] [CrossRef]
- Pilato, G.; Lisi, O.P.V. Tenuibiotus, a new genus of Macrobiotidae (Eutardigrada). Zootaxa 2011, 2761, 34–40. [Google Scholar] [CrossRef]
- Plate, L.H. Beiträge zur Naturgeschichte der Tardigraden. Zool. Jahrb. Anat. 1889, 3, 487–550. [Google Scholar] [CrossRef]
- Kaczmarek, Ł.; Zawierucha, K.; Buda, J.; Stec, D.; Gawlak, M.; Michalczyk, Ł.; Roszkowska, M. An integrative redescription of the nominal taxon for the Mesobiotus harmsworthi group (Tardigrada: Macrobiotidae) leads to descriptions of two new Mesobiotus species from Arctic. PLoS ONE 2018, 13, e0204756. [Google Scholar] [CrossRef]
- Stec, D.; Morek, W.; Gąsiorek, P.; Michalczyk, Ł. Unmasking hidden species diversity within the Ramazzottius oberhaeuseri complex, with an integrative redescription of the nominal species for the family Ramazzottiidae (Tardigrada: Eutardigrada: Parachela). Syst. Biodivers. 2018, 16, 357–376. [Google Scholar] [CrossRef]
- Guidetti, R.; Cesari, M.; Bertolani, R.; Altiero, T.; Rebecchi, L. High diversity in species, reproductive modes and distribution within the Paramacrobiotus richtersi complex (Eutardigrada, Macrobiotidae). Zool. Lett. 2019, 5, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Stec, D.; Krzywański, Ł.; Arakawa, K.; Michalczyk, Ł. A new redescription of Richtersius coronifer, supported by transcriptome, provides resources for describing concealed species diversity within the monotypic genus Richtersius (Eutardigrada). Zool. Lett. 2020, 6, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Stec, D.; Krzywański, Ł.; Zawierucha, K.; Michalczyk, Ł. Untangling systematics of the Paramacrobiotus areolatus species complex by an integrative redescription of the nominal species for the group, with multilocus phylogeny and species delineation in the genus Paramacrobiotus. Zool. J. Linn. Soc. 2020, 188, 694–716. [Google Scholar] [CrossRef]
- Stec, D.; Vecchi, M.; Maciejowski, W.; Michalczyk, Ł. Resolving the systematics of Richtersiidae by multilocus phylogeny and an integrative redescription of the nominal species for the genus Crenubiotus (Tardigrada). Sci. Rep. 2020, 10, 19418. [Google Scholar] [CrossRef]
- Roszkowska, M.; Grobys, D.; Bartylak, T.; Gawlak, M.; Kmita, H.; Kepel, A.; Kepel, M.; Parnikoza, I.; Kaczmarek, Ł. Integrative description of five Pseudechiniscus species (Heterotardigrada: Echiniscidae: The suillus-facettalis complex). Zootaxa 2020, 4763, 451–484. [Google Scholar] [CrossRef]
- Kaczmarek, Ł.; Kayastha, P.; Gawlak, M.; Mioduchowska, M.; Roszkowska, M. An integrative description of Diploechiniscus oihonnae (Richters, 1903) population from near the original type locality in Merok (Norway). Zootaxa 2021, 4964, 83–102. [Google Scholar] [CrossRef]
- Stec, D.; Vecchi, M.; Dudziak, M.; Bartels, P.J.; Calhim, S.; Michalczyk, Ł. Integrative taxonomy resolves species identities within the Macrobiotus pallarii complex (Eutardigrada: Macrobiotidae). Zool. Lett. 2021, 7, 9. [Google Scholar] [CrossRef]
- Stec, D.; Vončina, K.; Kristensen, R.M.; Michalczyk, Ł. The Macrobiotus ariekammensis species complex provides evidence for parallel evolution of claw elongation in macrobiotid tardigrades. Zool. J. Linn. Soc. 2021; in press. [Google Scholar] [CrossRef]
- Kaczmarek, Ł.; Kayastha, P.; Gawlak, M.; Mioduchowska, M.; Roszkowska, M. An integrative redescription of Echiniscus quadrispinosus quadrispinosus Richters, 1902 (Heterotardigrada; Echiniscidae) from the terra typica in Taunus Mountain Range (Europe; Germany). Eur. Zool. J. 2022; in press. [Google Scholar]
- Dastych, H. Niesporczaki (Tardigrada) Tatrzańskiego Parku Narodowego; Monografie Fauny Polski; PWN: Warszawa, Poland, 1980; pp. 1–232. [Google Scholar]
- Roszkowska, M.; Stec, D.; Gawlak, M.; Kaczmarek, Ł. An integrative description of a new tardigrade species Mesobiotus romani sp. nov. (Macrobiotidae: Harmsworthi group) from the Ecuadorian Pacific coast. Zootaxa 2018, 4450, 550–564. [Google Scholar] [CrossRef]
- Stec, D.; Gąsiorek, P.; Morek, W.; Kosztyła, P.; Zawierucha, K.; Michno, K.; Kaczmarek, Ł.; Prokop, Z.M.; Michalczyk, Ł. Estimating optimal sample size for tardigrade morphometry. Zool. J. Linn. Soc. 2016, 178, 776–784. [Google Scholar] [CrossRef] [Green Version]
- Pilato, G.; Binda, M.G. Definition of families, subfamilies, genera and subgenera of the Eutardigrada, and keys to their identification. Zootaxa 2010, 2404, 1–54. [Google Scholar] [CrossRef] [Green Version]
- Michalczyk, Ł.; Kaczmarek, Ł. A description of the new tardigrade Macrobiotus reinhardti (Eutardigrada: Macrobiotidae, harmsworthi group) with some remarks on the oral cavity armature within the genus Macrobiotus Schultze. Zootaxa 2003, 331, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Kaczmarek, Ł.; Michalczyk, Ł. The Macrobiotus hufelandi group (Tardigrada) revisited. Zootaxa 2017, 4363, 101–123. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, Ł.; Cytan, J.; Zawierucha, K.; Diduszko, D.; Michalczyk, Ł. Tardigrades from Peru (South America), with descriptions of three new species of Parachela. Zootaxa 2014, 3790, 357–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilato, G. Analisi di nuovi caratteri nello studio degli Eutardigradi. Animalia 1981, 8, 51–57. [Google Scholar]
- Kiosya, Y.; Pogwizd, J.; Matsko, Y.; Vecchi, M.; Stec, D. Phylogenetic position of two Macrobiotus species with a revisional note on Macrobiotus sottilei Pilato, Kiosya, Lisi & Sabella, 2012 (Tardigrada: Eutardigrada: Macrobiotidae). Zootaxa 2021, 4933, 113–135. [Google Scholar] [CrossRef]
- Michalczyk, Ł.; Kaczmarek, Ł. The Tardigrada Register: A comprehensive online data repository for tardigrade taxonomy. J. Limnol. 2013, 72, e22. [Google Scholar] [CrossRef]
- Casquet, J.; Thébaud, C.; Gillespie, R.G. Chelex without boiling, a rapid and easy technique to obtain stable amplifiable DNA from small amounts of ethanol-stored spiders. Mol. Ecol. Resour. 2011, 12, 136–141. [Google Scholar] [CrossRef]
- Kaczmarek, Ł.; Grobys, D.; Kulpa, A.; Bartylak, T.; Kmita, H.; Kepel, M.; Kepel, A.; Roszkowska, M. Two new species of the genus Milnesium Doyère, 1840 (Tardigrada, Apochela, Milnesiidae) from Madagascar. ZooKeys 2019, 884, 1–22. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxi-dase subunit I from diverse metazoan invertebrates. Mol. Marine Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Sands, C.J.; McInnes, S.J.; Marley, N.J.; Goodall-Copestake, W.P.; Convey, P.; Linse, K. Phylum Tardigrada: An “individual” approach. Cladistics 2008, 24, 861–871. [Google Scholar] [CrossRef]
- Mironov, S.V.; Dabert, J.; Dabert, M. A new feather mite species of the genus Proctophyllodes Robin, 1877 (Astigmata: Proctophyllodidae) from the Long-tailed Tit Aegithalos caudatus (Passeriformes: Aegithalidae)—morphological description with DNA barcode data. Zootaxa 2012, 3253, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Mioduchowska, M.; Kačarević, U.; Miamin, V.; Giginiak, Y.; Parnikoza, I.; Roszkowska, M.; Kaczmarek, Ł. Redescription of Antarctic eutardigrade Dastychius improvisus (Dastych, 1984) and some remarks on phylogenetic relationships within Isohypsibioidea. Eur. Zool. J. 2021, 88, 117–131. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Goujon, M.; McWilliam, H.; Li, W.; Valentin, F.; Squizzato, S.; Paern, J.; López, R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010, 38, W695–W699. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Posada, D.; Buckley, T.R. Model Selection and Model Averaging in Phylogenetics: Advantages of Akaike Information Criterion and Bayesian Approaches Over Likelihood Ratio Tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Kishino, H.; Yano, T.-A. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Kuzdrowska, K.A.; Mioduchowska, M.; Gawlak, M.; Bartylak, T.; Kepel, A.; Kepel, M.; Kaczmarek, Ł. Integrative description of Macrobiotus porifini sp. nov. (Macrobiotidae) from Madagascar and its phylogenetic position within the hufelandi group. Eur. Zool. J. 2021, 88, 375–389. [Google Scholar] [CrossRef]
- Doyère, P.L.N. Memoire sur les Tardigrades. Ann. Sci. Nat. Ser. Zool. 1840, 14, 269–362. [Google Scholar]
- Richters, F. Tardigrada. In Handbuch der Zoologie; Kükenthal, W., Krumbach, T., Eds.; Walter de Gruyter & Co.: Berlin, Germany; Leipzig, Germany, 1926; Volume 3, pp. 58–61. [Google Scholar]
- Thulin, G. Über die phylogenie und das system der tardigraden. Hereditas 2010, 11, 207–266. [Google Scholar] [CrossRef]
- Marley, N.; McInnes, S.J.; Sands, C. Phylum Tardigrada: A re-evaluation of the Parachela. Zootaxa 2011, 2819, 51–64. [Google Scholar] [CrossRef]
- Vicente, F.; Cesari, M.; Serrano, A.; Bertolani, R. The impact of fire on terrestrial tardigrade biodiversity: A first case-study from Portugal. J. Limnol. 2013, 72, e19. [Google Scholar] [CrossRef] [Green Version]
- Vecchi, M.; Ferrari, C.; Stec, D.; Calhim, S. Desiccation risk favours prevalence and diversity of tardigrade communities and influences their trophic structure in alpine ephemeral rock pools. Hydrobiologia, 2022; in press. [Google Scholar] [CrossRef]
- Pilato, G.; Lisi, O.P.V. Notes on some tardigrades from southern Mexico with description of three new species. Zootaxa 2006, 1236, 53–68. [Google Scholar] [CrossRef]
- McInnes, S.J. Zoogeographic distribution of terrestrial/freshwater tardigrades from current literature. Ann. Mag. Nat. Hist. 1994, 28, 257–352. [Google Scholar] [CrossRef]
- Meyer, H.A. Terrestrial and freshwater Tardigrada of the Americas. Zootaxa 2013, 3747, 1–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaczmarek, Ł.; Michalczyk, Ł.; McInnes, S.J. Annotated zoogeography of non-marine Tardigrada. Part I: Central America. Zootaxa 2014, 3763, 1–62. [Google Scholar] [CrossRef] [Green Version]
- Kaczmarek, Ł.; Michalczyk, Ł.; Mcinnes, S.J. Annotated zoogeography of non-marine Tardigrada. Part II: South America. Zootaxa 2015, 3923, 1–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaczmarek, Ł.; Michalczyk, Ł.; McInnes, S.J. Annotated zoogeography of non-marine Tardigrada. Part III: North America and Greenland. Zootaxa 2016, 4203, 1–249. [Google Scholar] [CrossRef]
- McInnes, S.J.; Michalczyk, Ł.; Kaczmarek, Ł. Annotated zoogeography of non-marine Tardigrada. Part IV: Africa. Zootaxa 2017, 4284, 1–74. [Google Scholar] [CrossRef] [Green Version]
- Michalczyk, Ł.; Kaczmarek, Ł.; Mcinnes, S.J. Annotated zoogeography of non-marine Tardigrada. Part V: Australasia. Zootaxa 2022, 5107, 1–119. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).