Single-Island Endemism despite Repeated Dispersal in Caribbean Micrathena (Araneae: Araneidae): An Updated Phylogeographic Analysis
Abstract
:1. Introduction
2. Materials and Methods
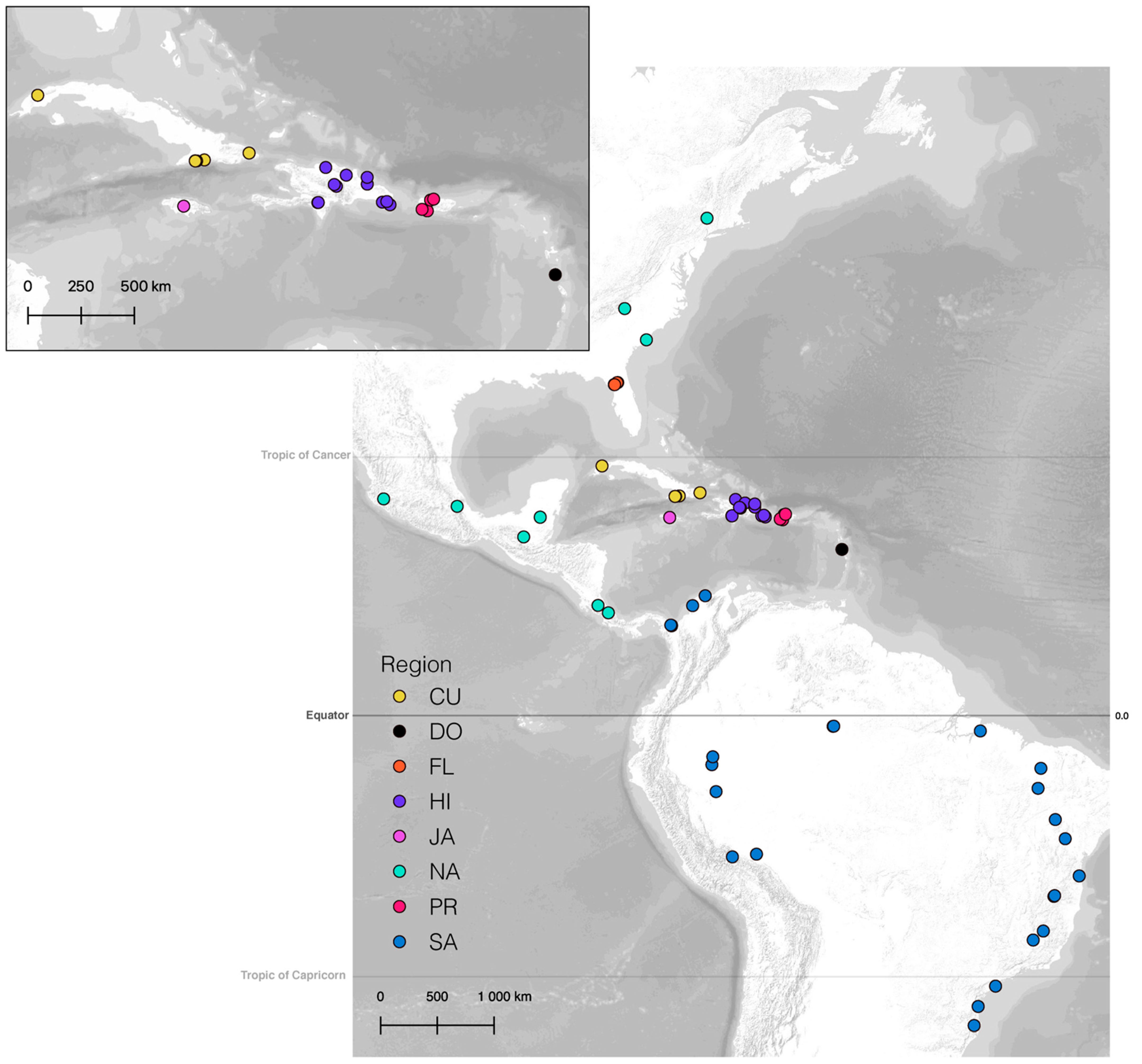
2.1. Specimen and Taxon Sampling
2.2. Tissue Extraction and PCR
2.3. Alignment and Phylogeny Building
2.4. Divergence Time Estimation and Biogeographic Modeling
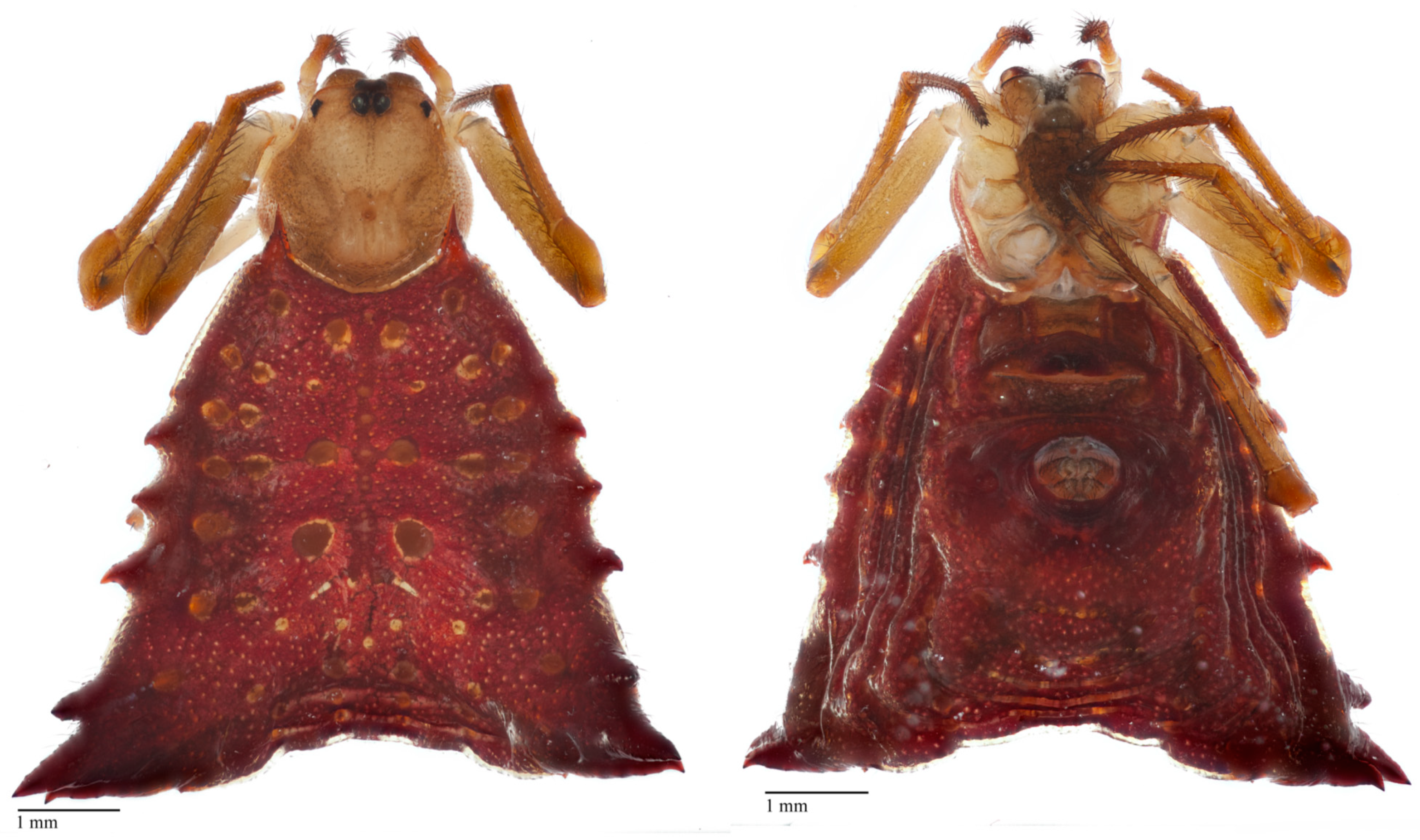
2.5. Specimen Photography
3. Results
3.1. Sequence Alignment
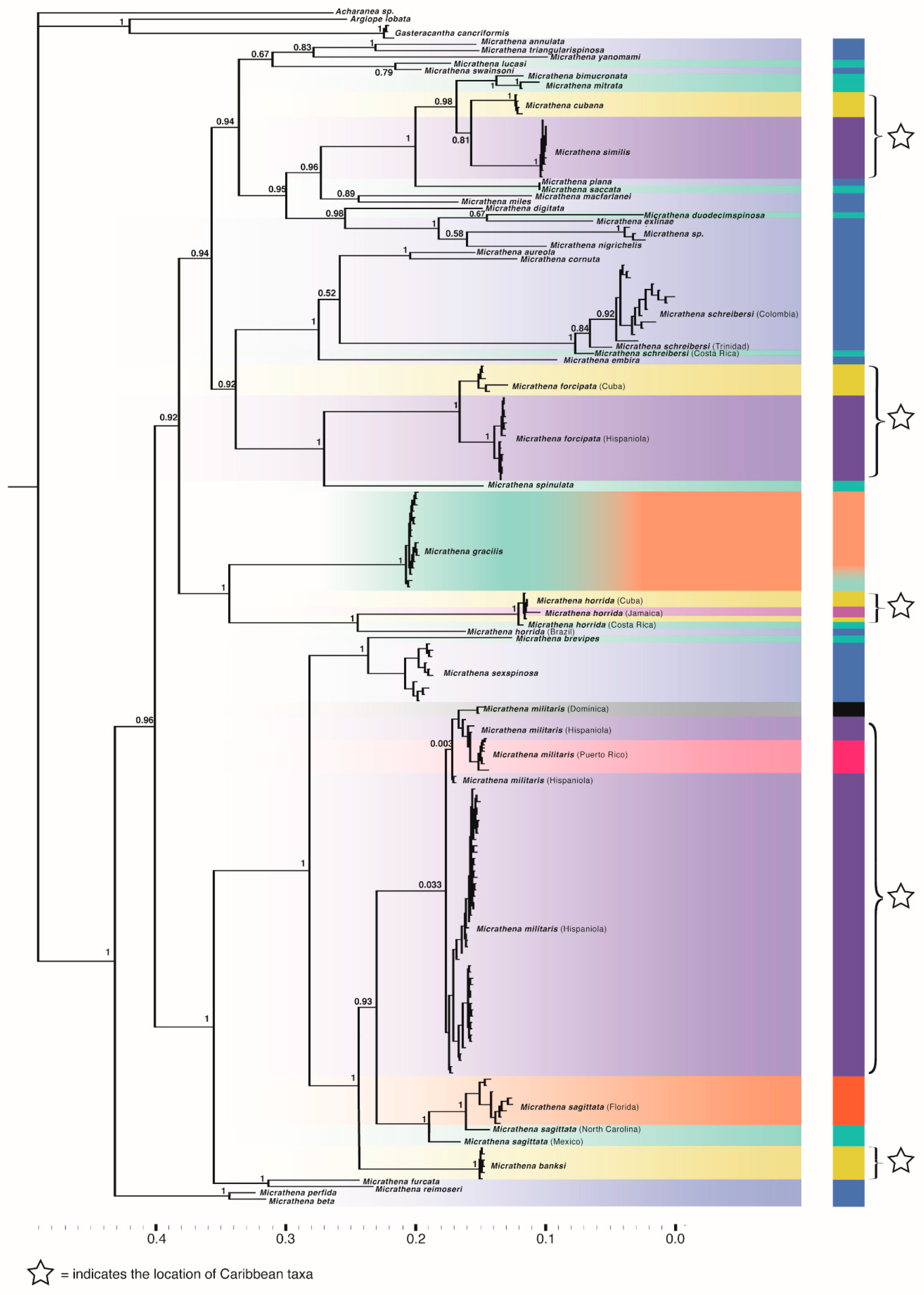
3.2. Phylogenetics
3.3. Divergence Times
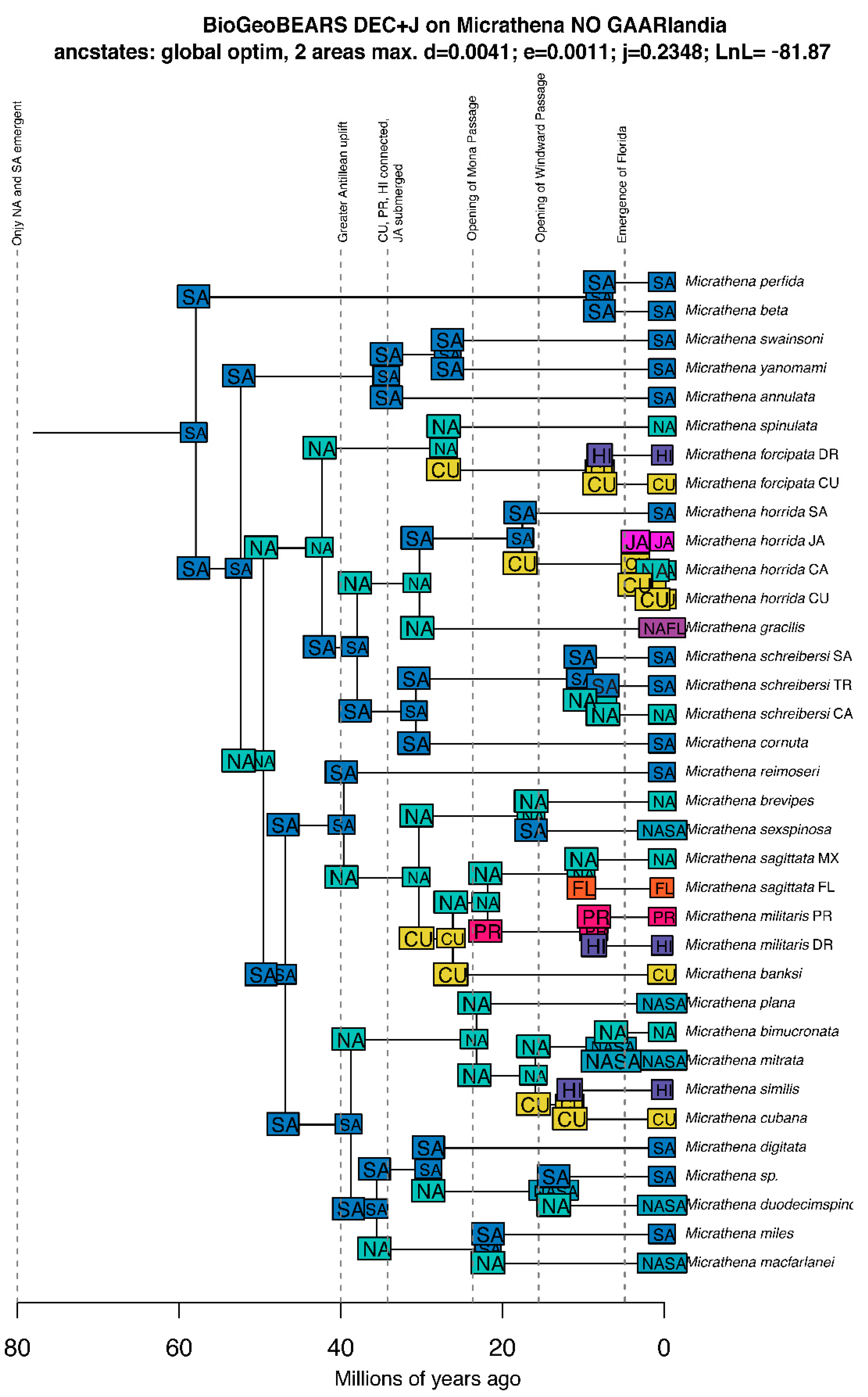
3.4. Biogeographic Patterns
3.4.1. Overview
3.4.2. Vicariance vs. Long Distance Dispersal
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilson, E.O.; MacArthur, R.H. The Theory of Island Biogeography; Princeton University Press: Princeston, NJ, USA, 1967. [Google Scholar]
- Carlquist, S.J. Island Biology; Columbia University Press: New York, NY, USA, 1974. [Google Scholar]
- Whittaker, R.J. Island Biogeography; Oxford University Press: Oxford, UK, 1998. [Google Scholar]
- Whittaker, R.J.; Fernández-Palacios, J.M. Island Biogeography: Ecology, Evolution, and Conservation; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- Gillespie, R. Community Assembly Through Adaptive Radiation in Hawaiian Spiders. Science 2004, 303, 356–359. [Google Scholar] [CrossRef] [Green Version]
- Craig, D.A.; Currie, D.C.; Joy, D.A. Geographical history of the central-western Pacific black fly subgenus Inseliellum (Diptera: Simuliidae: Simulium) based on a reconstructed phylogeny of the species, hot-spot archipelagoes and hydrological considerations. J. Biogeogr. 2001, 28, 1101–1127. [Google Scholar] [CrossRef]
- Ricklefs, R.; Bermingham, E. The West Indies as a laboratory of biogeography and evolution. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 2393–2413. [Google Scholar] [CrossRef] [Green Version]
- Myers, N.; Mittermeler, R.A.; Mittermeler, C.G.; Da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Rosen, D.E. A Vicariance Model of Caribbean Biogeography. Syst. Biol. 1975, 24, 431–464. [Google Scholar] [CrossRef]
- Pindell, J.L.; Barrett, S.F. Geologic evolution of the Caribbean region: A plate-tectonic persepective. In Decade of American Geology; Case, J.E., Dengo, G., Eds.; Geological Society of America: Boulder, CO, USA, 1990; The Geology of North America v. H; pp. 405–432. [Google Scholar]
- Pindell, J.L. Evolution of the Gulf of Mexico and the Caribbean. In Caribbean Geology, an Introduction; Donovan, S.K., Jackson, T.A., Eds.; The University of West Indies Publishers’ Association: Kingston, Jamaica, 1994; pp. 13–39. [Google Scholar]
- Burke, K. Tectonic evolution of the Caribbean. Annu. Rev. Earth Planet. Sci. 1988, 16, 201–230. [Google Scholar] [CrossRef]
- Donnelly, T.W. Geologic constraints on Caribbean biogeography. In Zoogeography of Caribbean Insects; Liebherr, J.K., Ed.; Cornell University Press: Ithaca, NY, USA, 1988; pp. 15–37. [Google Scholar]
- Alonso, R.; Crawford, A.J.; Bermingham, E. Molecular phylogeny of an endemic radiation of Cuban toads (Bufonidae: Peltophryne) based on mitochondrial and nuclear genes. J. Biogeogr. 2012, 39, 434–451. [Google Scholar] [CrossRef]
- Keppel, G.; Lowe, A.J.; Possingham, H.P. Changing Perspectives on the Biogeography of the Tropical South Pacific: Influences of Dispersal, Vicariance and Extinction. J. Biogeogr. 2009, 36, 1035–1054. [Google Scholar] [CrossRef]
- Crews, S.C.; Gillespie, R.G. Molecular systematics of Selenops spiders (Araneae: Selenopidae) from North and Central America: Implications for Caribbean biogeography. Biol. J. Linn. Soc. 2010, 101, 288–322. [Google Scholar] [CrossRef]
- Heinicke, M.P.; Duellman, W.E.; Hedges, S.B. Major Caribbean and Central American frog faunas originated by ancient oceanic dispersal. Proc. Natl. Acad. Sci. USA 2007, 104, 10092–10097. [Google Scholar] [CrossRef] [Green Version]
- Toussaint, E.F.A.; Balke, M. Historical biogeography of Polyura butterflies in the oriental Palaeotropics: Trans-archipelagic routes and South Pacific island hopping. J. Biogeogr. 2016, 43, 1560–1572. [Google Scholar] [CrossRef]
- Agnarsson, I.; Cheng, R.-C.; Kuntner, M. A multi-clade test supports the intermediate dispersal model of biogeography. PLoS ONE 2014, 9, e86780. [Google Scholar] [CrossRef]
- Iturralde-Vinent, M.A. Meso-Cenozoic Caribbean paleogeography: Implications for the historical biogeography of the region. Int. Geol. Rev. 2006, 48, 791–827. [Google Scholar] [CrossRef] [Green Version]
- Iturralde-Vinent, M.A.; MacPhee, R.D.E. Paleogeography of the Caribbean region: Implications for Cenozoic biogeography. Bull. Am. Museum Nat. Hist. 1999, 238, 1–95. [Google Scholar]
- Fabre, P.-H.; Vilstrup, J.T.; Raghavan, M.; Der Sarkissian, C.; Willerslev, E.; Douzery, E.J.P.; Orlando, L. Rodents of the Caribbean: Origin and diversification of hutias unravelled by next-generation museomics. Biol. Lett. 2014, 10, 20140266. [Google Scholar] [CrossRef] [PubMed]
- Klemen, Č.; Ingi, A.; Binford, G.J.; Matjaž, K. Biogeography of the Caribbean Cyrtognatha spiders. Sci. Reports (Nature Publ. Group) 2019, 9, 1–14. [Google Scholar]
- Esposito, L.A.; Bloom, T.; Caicedo-Quiroga, L.; Alicea-Serrano, A.M.; Sánchez-Ruíz, J.A.; May-Collado, L.J.; Binford, G.J.; Agnarsson, I. Islands within islands: Diversification of tailless whip spiders (Amblypygi, Phrynus) in Caribbean caves. Mol. Phylogenet. Evol. 2015, 93, 107–117. [Google Scholar] [CrossRef]
- Condamine, F.L.; Silva-Brandão, K.L.; Kergoat, G.J.; Sperling, F.A.H. Biogeographic and diversification patterns of Neotropical Troidini butterflies (Papilionidae) support a museum model of diversity dynamics for Amazonia. BMC Evol. Biol. 2012, 12, 82. [Google Scholar] [CrossRef] [Green Version]
- Dziki, A.; Binford, G.J.; Coddington, J.A.; Agnarsson, I. Spintharus flavidus in the Caribbean—A 30 million year biogeographical history and radiation of a ‘widespread species’. PeerJ 2015, 3, e1422. [Google Scholar] [CrossRef] [Green Version]
- Pfingstl, T.; Lienhard, A.; Baumann, J.; Koblmüller, S. A taxonomist’s nightmare--Cryptic diversity in Caribbean intertidal arthropods (Arachnida, Acari, Oribatida). Mol. Phylogenet. Evol. 2021, 163, 107240. [Google Scholar] [CrossRef]
- Esposito, L.A.; Prendini, L. Island ancestors and New World biogeography: A case study from the scorpions (Buthidae: Centruroidinae). Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Chamberland, L.; Salgado-Roa, F.C.; Basco, A.; Crastz-Flores, A.; Binford, G.J.; Agnarsson, I. Phylogeography of the widespread Caribbean spiny orb weaver Gasteracantha cancriformis. PeerJ 2020, 8, e8976. [Google Scholar] [CrossRef]
- Říčan, O.; Piálek, L.; Zardoya, R.; Doadrio, I.; Zrzavý, J. Biogeography of the Mesoamerican Cichlidae (Teleostei: Heroini): Colonization through the GAARlandia land bridge and early diversification. J. Biogeogr. 2013, 40, 579–593. [Google Scholar] [CrossRef] [Green Version]
- Tong, Y.; Binford, G.; Rheims, C.A.; Kuntner, M.; Liu, J.; Agnarsson, I. Huntsmen of the Caribbean: Multiple tests of the GAARlandia hypothesis. Mol. Phylogenet. Evol. 2019, 130, 259–268. [Google Scholar] [CrossRef]
- Agnarsson, I.; LeQuier, S.M.; Kuntner, M.; Cheng, R.-C.; Coddington, J.A.; Binford, G. Phylogeography of a good Caribbean disperser: Argiope argentata (Araneae, Araneidae) and a new ‘cryptic’species from Cuba. Zookeys 2016, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nieto-Blázquez, M.E.; Antonelli, A.; Roncal, J. Historical Biogeography of endemic seed plant genera in the Caribbean: Did GAAR landia play a role? Ecol. Evol. 2017, 7, 10158–10174. [Google Scholar] [CrossRef] [PubMed]
- Dávalos, L.M. Phylogeny and biogeography of Caribbean mammals. Biol. J. Linn. Soc. 2004, 81, 373–394. [Google Scholar] [CrossRef]
- Dávalos, L.M.; Turvey, S.T. West Indian mammals: The old, the new, and the recently extinct. In Bones, Clones, and Biomes: The History and Geography of Recent Neotropical Mammals; Patterson, B.D., Costa, L.P., Eds.; University of Chicago Press: Chicago, IL, USA, 2012; pp. 157–202. [Google Scholar]
- Crews, S.C.; Esposito, L.A. Towards a synthesis of the Caribbean biogeography of terrestrial arthropods. BMC Evol. Biol. 2020, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Philippon, M.; Cornée, J.-J.; Münch, P.; Van Hinsbergen, D.J.J.; Boudagher-Fadel, M.; Gailler, L.; Boschman, L.M.; Quillevere, F.; Montheil, L.; Gay, A.; et al. Eocene intra-plate shortening responsible for the rise of a faunal pathway in the northeastern Caribbean realm. PLoS ONE 2020, 15, e0241000. [Google Scholar]
- Ali, J.R. Colonizing the Caribbean: Is the GAARlandia land-bridge hypothesis gaining a foothold? J. Biogeogr. 2012, 39, 431–433. [Google Scholar] [CrossRef]
- Hedges, S.B. Paleogeography of the Antilles and Origin of West Indian Terrestrial Vertebrates. Ann. Missouri Bot. Gard. 2006, 93, 231–244. [Google Scholar] [CrossRef]
- Ali, J.R.; Hedges, S.B. Colonizing the Caribbean: New geological data and an updated land-vertebrate colonization record challenge the GAARlandia land-bridge hypothesis. J. Biogeogr. 2021, 48, 2699–2707. [Google Scholar] [CrossRef]
- Garrocq, C.; Lallemand, S.; Marcaillou, B.; Lebrun, J.; Padron, C.; Klingelhoefer, F.; Laigle, M.; Münch, P.; Gay, A.; Schenini, L.; et al. Genetic Relations Between the Aves Ridge and the Grenada Back-Arc Basin, East Caribbean Sea. J. Geophys. Res. Solid Earth 2021, 126, e2020JB020466. [Google Scholar] [CrossRef]
- Bond, J.E.; Hamilton, C.A.; Godwin, R.L.; Ledford, J.M.; Starrett, J. Phylogeny, evolution, and biogeography of the North American trapdoor spider family Euctenizidae (Araneae: Mygalomorphae) and the discovery of a new ‘endangered living fossil’along California’s central coast. Insect Syst. Divers. 2020, 4, 2. [Google Scholar] [CrossRef]
- Chousou-Polydouri, N.; Carmichael, A.; Szűts, T.; Saucedo, A.; Gillespie, R.; Griswold, C.; Wood, H.M. Giant Goblins above the waves at the southern end of the world: The biogeography of the spider family Orsolobidae (Araneae, Dysderoidea). J. Biogeogr. 2019, 46, 332–342. [Google Scholar] [CrossRef]
- Rix, M.G.; Wilson, J.D.; Huey, J.A.; Hillyer, M.J.; Gruber, K.; Harvey, M.S. Diversification of the mygalomorph spider genus Aname (Araneae: Anamidae) across the Australian arid zone: Tracing the evolution and biogeography of a continent-wide radiation. Mol. Phylogenet. Evol. 2021, 160, 107127. [Google Scholar] [CrossRef]
- Richardson, B.J. Evolutionary biogeography of Australian jumping spider genera (Araneae: Salticidae). Aust. J. Zool. 2020, 67, 162–172. [Google Scholar] [CrossRef]
- Chamberland, L.; McHugh, A.; Kechejian, S.; Binford, G.J.; Bond, J.E.; Coddington, J.; Dolman, G.; Hamilton, C.A.; Harvey, M.S.; Kuntner, M.; et al. From Gondwana to GAARlandia: Evolutionary history and biogeography of ogre-faced spiders (Deinopis). J. Biogeogr. 2018, 45, 2442–2457. [Google Scholar] [CrossRef] [Green Version]
- Hedges, S.B.; Hass, C.A.; Maxson, L.R. Caribbean biogeography: Molecular evidence for dispersal in West Indian terrestrial vertebrates. Proc. Natl. Acad. Sci. USA 1992, 89, 1909–1913. [Google Scholar] [CrossRef] [Green Version]
- Betancur-R, R.; Hines, A.; Acero, P.A.; Ortí, G.; Wilbur, A.E.; Freshwater, D.W. Reconstructing the lionfish invasion: Insights into Greater Caribbean biogeography. J. Biogeogr. 2011, 38, 1281–1293. [Google Scholar] [CrossRef]
- Klein, N.K.; Burns, K.J.; Hackett, S.J.; Griffiths, C.S. Molecular phylogenetic relationships among the wood warblers (Parulidae) and historical biogeography in the Caribbean basin. J. Caribb. Ornithol. 2004, 17, 3–17. [Google Scholar]
- Ferrier, S.; Powell, G.V.N.; Richardson, K.S.; Manion, G.; Overton, J.M.; Allnutt, T.F.; Cameron, S.E.; Mantle, K.; Burgess, N.D.; Faith, D.P.; et al. Mapping more of terrestrial biodiversity for global conservation assessment. Bioscience 2004, 54, 1101–1109. [Google Scholar] [CrossRef] [Green Version]
- McHugh, A.; Yablonsky, C.; Binford, G.; Agnarsson, I. Molecular phylogenetics of Caribbean Micrathena (Araneae:Araneidae) suggests multiple colonisation events and single island endemism. Invertebr. Syst. 2014, 28, 337–349. [Google Scholar] [CrossRef]
- Zhang, J.-X.; Maddison, W.P. Molecular phylogeny, divergence times and biogeography of spiders of the subfamily Euophryinae (Araneae: Salticidae). Mol. Phylogenet. Evol. 2013, 68, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, I.L.F.; Santos, A.J. Phylogenetic analysis of Micrathena and Chaetacis spiders (Araneae: Araneidae) reveals multiple origins of extreme sexual size dimorphism and long abdominal spines. Zool. J. Linn. Soc. 2012, 166, 14–53. [Google Scholar] [CrossRef] [Green Version]
- Levi, H.W. The spiny orb-weaver genera Micrathena and Chaetacis (Araneae: Araneidae). Los géneros de tejedoras de esferas espinosas Micrathena y Chaetacis (Araneae: Araneidae). Bull. Museum Comp. Zool. 1985, 150, 429–618. [Google Scholar]
- Bukowski, T.C.; Christenson, T.E. Natural history and copulatory behavior of the spiny orbweaving spider Micrathena gracilis (Araneae, Araneidae). J. Arachnol. 1997, 307–320. [Google Scholar]
- Bell, J.R.; Bohan, D.A.; Shaw, E.M.; Weyman, G.S. Ballooning dispersal using silk: World fauna, phylogenies, genetics and models. Bull. Entomol. Res. 2005, 95, 69–114. [Google Scholar] [CrossRef]
- World Spider Catalog. World Spider Catalog. Version 18.5. National History Museum Bern. Available online: http://ws.nmbe.ch (accessed on 1 March 2018).
- Scharff, N.; Coddington, J.A.; Blackledge, T.A.; Agnarsson, I.; Framenau, V.W.; Szűts, T.; Hayashi, C.Y.; Dimitrov, D. Phylogeny of the orb-weaving spider family Araneidae (Araneae: Araneoidea). Cladistics 2020, 36, 1–21. [Google Scholar] [CrossRef]
- Agnarsson, I.; Maddison, W.P.; Avilés, L. The phylogeny of the social Anelosimus spiders (Araneae: Theridiidae) inferred from six molecular loci and morphology. Mol. Phylogenet. Evol. 2007, 43, 833–851. [Google Scholar] [CrossRef]
- Kuntner, M.; Agnarsson, I. Biogeography and diversification of hermit spiders on Indian Ocean islands (Nephilidae: Nephilengys). Mol. Phylogenet. Evol. 2011, 59, 477–488. [Google Scholar] [CrossRef]
- Simon, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994, 87, 651–701. [Google Scholar] [CrossRef]
- Hedin, M.C.; Maddison, W.P. A combined molecular approach to phylogeny of the jumping spider subfamily Dendryphantinae (Araneae: Salticidae). Mol. Phylogenet. Evol. 2001, 18, 386–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guid. Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Green, P. Phrap. Version 1.090518. Available online: http://phrap.org (accessed on 19 June 2019).
- Green, P.; Ewing, B. Phred. Version 0.020425c. Available online: http://phrap.org/othersoftware.html (accessed on 19 June 2019).
- Maddison, D.R.; Maddison, W.P. Chromaseq: A Mesquite Package for Analyzing Sequence Chromatograms. Version 1.0 2011. Available online: http://mesquiteproject.org/packages/chromaseq (accessed on 19 June 2019).
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.6. Available online: http://mesquiteproject.org (accessed on 19 June 2019).
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Akaike, H. Information theory and an extension of the maximum likelihood principle. In Selected Papers of Hirotugu Akaike; Springer: Berlin/Heidelberg, Germany, 1998; pp. 199–213. [Google Scholar]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES science gateway: A community resource for phylogenetic analyses. In Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery, Salt Lake City, UT, USA, 18–21 July 2011; pp. 1–8. [Google Scholar]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [Green Version]
- Kuntner, M.; Arnedo, M.A.; Trontelj, P.; Lokovšek, T.; Agnarsson, I. A molecular phylogeny of nephilid spiders: Evolutionary history of a model lineage. Mol. Phylogenet. Evol. 2013, 69, 961–979. [Google Scholar] [CrossRef] [PubMed]
- Bidegaray-Batista, L.; Arnedo, M.A. Gone with the plate: The opening of the Western Mediterranean basin drove the diversification of ground-dweller spiders. BMC Evol. Biol. 2011, 11, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matzke, N.J. BioGeoBEARS: BioGeography with Bayesian (and likelihood) evolutionary analysis in R Scripts. R Packag. 2013, 1, 2013. [Google Scholar]
- Bartolini, C.; Buffler, R.T.; Blickwede, J.F. The Circum-Gulf of Mexico and the Caribbean: Hydrocarbon Habitats, Basin Formation, and Plate Tectonics, AAPG Memoir 79; American Association of Petroleum Geologists: Tulsa, OK, USA, 2003. [Google Scholar]
- Marshall, J.S. The geomorphology and physiographic provinces of Central America. Cent. Am. Geol. Resour. Hazards 2007, 1, 75–121. [Google Scholar]
- Mann, P. Overview of the tectonic history of northern Central America. Geol. Soc. Am. Spec. Pap. 2007, 428, 1–19. [Google Scholar]
- Randazzo, A.F.; Jones, D.S. The Geology of Florida; University Press of Florida: Gainesville, FL, USA, 1997. [Google Scholar]
- Ree, R.H.; Sanmartín, I. Conceptual and statistical problems with the DEC+ J model of founder-event speciation and its comparison with DEC via model selection. J. Biogeogr. 2018, 45, 741–749. [Google Scholar] [CrossRef]
- Ree, R.H.; Smith, S.A. Maximum Likelihood Inference of Geographic Range Evolution by Dispersal, Local Extinction, and Cladogenesis. Syst. Biol. 2008, 57, 4–14. [Google Scholar] [CrossRef] [Green Version]
- Kozub, D.; Khmelik, V.; Shapoval, Y.; Chentsov, V.; Yatsenko, S.; Litovchenko, B.; Starykh, V. Helicon Focus Software. Available online: http://heliconsoft.com (accessed on 26 January 2017).
- Garrison, N.L.; Rodriguez, J.; Agnarsson, I.; Coddington, J.A.; Griswold, C.E.; Hamilton, C.A.; Hedin, M.; Kocot, K.M.; Ledford, J.M.; Bond, J.E. Spider phylogenomics: Untangling the Spider Tree of Life. PeerJ 2016, 4, e1719. [Google Scholar] [CrossRef] [Green Version]
- Čandek, K.; Agnarsson, I.; Binford, G.J.; Kuntner, M. Global biogeography of Tetragnatha spiders reveals multiple colonization of the Caribbean. BioRxiv 2018. [Google Scholar] [CrossRef]
- Peck, S.B. Aerial dispersal of insects between and to islands in the Galapagos Archipelago, Ecuador. Ann. Entomol. Soc. Am. 1994, 87, 218–224. [Google Scholar] [CrossRef]
- Aldrich, J.R. Others Dispersal of the southern green stink bug, Nezara viridula (L.) (Heteroptera: Pentatomidae), by hurricane Hugo. Proc. Entomol. Soc. Washingt. 1990, 92, 757–759. [Google Scholar]
- Zimmerman, E.C. Insects of Hawaii; University of Hawaii Press: Honolulu, HI, USA, 1948; Volume 7. [Google Scholar]
- Oey, L.; Ezer, T.; Lee, H. Loop Current, rings and related circulation in the Gulf of Mexico: A review of numerical models and future challenges. Geophys. Monogr. Geophys. Union 2005, 161, 31. [Google Scholar]
- Harrison, S.E.; Harvey, M.S.; Cooper, S.J.B.; Austin, A.D.; Rix, M.G. Across the Indian Ocean: A remarkable example of trans-oceanic dispersal in an austral mygalomorph spider. PLoS ONE 2017, 12, e0180139. [Google Scholar] [CrossRef] [Green Version]
- Ceccarelli, F.S.; Opell, B.D.; Haddad, C.R.; Raven, R.J.; Soto, E.M.; Ramírez, M.J. Around the world in eight million years: Historical biogeography and evolution of the spray zone spider Amaurobioides (Araneae: Anyphaenidae). PLoS ONE 2016, 11, e0163740. [Google Scholar] [CrossRef] [PubMed]
- Duque-Caro, H. Neogene stratigraphy, paleoceanography and paleobiogeography in northwest South America and the evolution of the Panama Seaway. Palaeogeogr. Palaeoclimatol. Palaeoecol. 1990, 77, 203–234. [Google Scholar] [CrossRef]
- Coates, A.G.; Jackson, J.B.C.; Collins, L.S.; Cronin, T.M.; Dowsett, H.J.; Bybell, L.M.; Jung, P.; Obando, J.A. Closure of the Isthmus of Panama: The near-shore marine record of Costa Rica and western Panama. Geol. Soc. Am. Bull. 1992, 104, 814–828. [Google Scholar] [CrossRef]
- Holcombe, T.L.; Moore, W.S. Paleocurrents in the eastern Caribbean: Geologic evidence and implications. Mar. Geol. 1977, 23, 35–56. [Google Scholar] [CrossRef]
- Houle, A. Floating islands: A mode of long-distance dispersal for small and medium-sized terrestrial vertebrates. Divers. Distrib. 1998, 4, 201–216. [Google Scholar]
- Lee, V.M.J.; Kuntner, M.; Li, D. Ballooning behavior in the golden orbweb spider Nephila pilipes (Araneae: Nephilidae). Front. Ecol. Evol. 2015, 3, 2. [Google Scholar] [CrossRef] [Green Version]



| Genus | Species | Barcode | Country/Region | Latitude | Longitude | 16S | CO1 | ITS2 |
|---|---|---|---|---|---|---|---|---|
| Micrathena | annulata | MIC007 | Brazil | 26.08933S | 48.64006W | KJ157272 | ||
| Micrathena | aureola | MIC009 | Brazil | 4.904167S | 42.79083W | KJ157249 | ||
| Micrathena | banksi | 784750 | Cuba | 20.05269N | 76.50296W | KJ156991 | KJ157215 | KJ157104 |
| Micrathena | banksi | 784760 | Cuba | 20.0107N | 76.8843W | KJ156992 | KJ157216 | |
| Micrathena | banksi | 784976 | Cuba | 20.00939N | 76.89402W | KJ156993 | KJ157217 | KJ157105 |
| Micrathena | banksi | 785101 | Cuba | 20.00939N | 76.89402W | KJ156994 | KJ157220 | KJ157106 |
| Micrathena | banksi | 785175 | Cuba | 20.33178N | 74.56919W | KJ156995 | KJ157219 | KJ157107 |
| Micrathena | banksi | 787933 | Cuba | 20.01742N | 76.89781W | KJ156996 | KJ157218 | KJ157108 |
| Micrathena | beta | MIC238 | Peru | 4.5674444S | 73.45925W | KX687306 | ||
| Micrathena | bimucronata | MIC123 | Costa Rica | 10.233518N | 84.075411W | KJ157236 | ||
| Micrathena | brevipes | MIC121 | Costa Rica | 9.552960N | 83.112910W | KJ157223 | ||
| Micrathena | cornuta | MIC199 | Peru | 12.8088056S | 69.30175W | KX687309 | ||
| Micrathena | cubana | 784355 | Cuba | 20.01309N | 76.83400W | KJ156997 | KJ157224 | KJ157109 |
| Micrathena | cubana | 784820 | Cuba | 20.00874N | 76.88777W | KJ156998 | KJ157225 | KJ157110 |
| Micrathena | cubana | 785048 | Cuba | 22.65707N | 83.70161W | KJ156999 | KJ157226 | KJ157111 |
| Micrathena | cubana | 787840 | Cuba | 20.33178N | 74.56919W | KJ157000 | KJ157227 | |
| Micrathena | digitata | MIC017 | Brazil | 11.39983S | 40.52206W | KJ157238 | ||
| Micrathena | duodecimspinosa | 00004833A | Costa Rica | San Antonio de Escazú | x | x | ||
| Micrathena | embira | MIC182 | Brazil | 9.642419S | 41.446727W | KX687311 | ||
| Micrathena | exlinae | MIC147 | Brazil | 0.99185S | 62.15915W | KX687313 | ||
| Micrathena | forcipata | 00002846A | Cuba | Juan Gonzalez, Guamá | x | x | ||
| Micrathena | forcipata | 00002848A | Cuba | 20.01309N | 76.83400W | x | x | |
| Micrathena | forcipata | 00002845A | Cuba | 20.01309N | 76.83400W | x | x | |
| Micrathena | forcipata | 784425 | Cuba | 20.00939N | 76.89402W | KJ157002 | KJ157256 | KJ157113 |
| Micrathena | forcipata | 787842 | Cuba | 20.33178N | 74.56919W | KJ157003 | KJ157257 | |
| Micrathena | forcipata | 782311 | Hispaniola | 18.355536N | 68.61825W | KJ157004 | KJ157258 | |
| Micrathena | forcipata | 782434 | Hispaniola | 19.34405N | 69.46635W | KJ157005 | KJ157260 | KJ157114 |
| Micrathena | forcipata | 784362 | Hispaniola | 18.32902N | 68.80995W | KJ157006 | KJ157264 | KJ157115 |
| Micrathena | forcipata | 784366 | Hispaniola | 18.32902N | 68.80995W | KJ157271 | KJ157116 | |
| Micrathena | forcipata | 784447 | Hispaniola | 18.2205360N | 68.480607W | KJ157007 | KJ157261 | KJ157117 |
| Micrathena | forcipata | 785054 | Hispaniola | 19.746175N | 71.257726W | KJ157008 | KJ157263 | KJ157118 |
| Micrathena | forcipata | 785282 | Hispaniola | 18.355536N | 68.6185W | KJ157009 | KJ157259 | KJ157119 |
| Micrathena | forcipata | 785682 | Hispaniola | 18.2205360N | 68.480607W | KJ157010 | KJ157 | |
| Micrathena | forcipata | 787132 | Hispaniola | 18.310010 N | 71.6000 W | KJ157265 | ||
| Micrathena | forcipata | 787135 | Hispaniola | 18.310010 N | 71.6000 W | KJ157011 | KJ157266 | |
| Micrathena | forcipata | 787150 | Hispaniola | 18.310010 N | 71.6000 W | KJ157012 | KJ157267 | KJ157121 |
| Micrathena | forcipata | 787153 | Hispaniola | 18.310010 N | 71.6000 W | KJ157013 | KJ157269 | KJ157122 |
| Micrathena | forcipata | 787210 | Hispaniola | 18.310010 N | 71.6000 W | KJ157014 | KJ157268 | KJ157123 |
| Micrathena | forcipata | 787243 | Hispaniola | 18.310010 N | 71.6000 W | KJ157015 | KJ157270 | KJ157124 |
| Micrathena | furcata | MIC037 | Brazil | 27.66667 S | 49.01667W | KJ157242 | ||
| Micrathena | gracilis | 10000619A | FL, USA | 29.4776N | 82.5627W | x | x | |
| Micrathena | gracilis | 10000629A | FL, USA | 29.62986N | 82.29880W | x | ||
| Micrathena | gracilis | 10000627A | FL, USA | 29.62986N | 82.29880W | x | ||
| Micrathena | gracilis | 10000638A | FL, USA | 29.63680N | 82.23961W | x | x | |
| Micrathena | gracilis | 10000644A | FL, USA | 29.46368N | 82.52898W | x | ||
| Micrathena | gracilis | 10000642A | FL, USA | 29.62688N | 82.29878W | x | ||
| Micrathena | gracilis | 10000643A | FL, USA | 29.62688N | 82.29878W | x | ||
| Micrathena | gracilis | 00000804A | NC, USA | 35.44842N | 81.58694W | KJ157250 | KJ157188 | |
| Micrathena | gracilis | 00000954A | SC, USA | 33.03913N | 79.56459W | KJ157084 | KJ157252 | KJ157192 |
| Micrathena | gracilis | 00000935A | SC, USA | 33.03913N | 79.56459W | KJ157083 | KJ157254 | KJ157191 |
| Micrathena | gracilis | 00000889A | SC, USA | 33.03913N | 79.56459W | KJ157082 | KJ157251 | KJ157190 |
| Micrathena | gracilis | 00000984A | SC, USA | 33.03913N | 79.56459W | KJ157086 | KJ157253 | KJ157194 |
| Micrathena | gracilis | 00000988A | SC, USA | 33.03913N | 79.56459W | KJ157087 | KJ157255 | KJ157195 |
| Micrathena | gracilis | 00002487A | NY, USA | 42.01807N | 73.91707W | KJ157088 | KJ157196 | |
| Micrathena | gracilis | 00002501A | NY, USA | 42.01807N | 73.91707W | KJ157089 | KJ157197 | |
| Micrathena | gracilis | 00000976A | SC, USA | 33.03913N | 79.56459W | KJ157085 | KJ157193 | |
| Micrathena | horrida | MIC042 | Brazil | 16.59553S | 41.57925W | KJ157248 | ||
| Micrathena | horrida | MIC122 | Costa Rica | 10.233518N | 84.075411W | KJ157245 | ||
| Micrathena | horrida | 00003552A | Jamaica | 18.1635N | 77.39410W | x | x | |
| Micrathena | horrida | 784351 | Cuba | 20.00939N | 76.89402W | KJ157016 | KJ157243 | KJ157125 |
| Micrathena | horrida | 784751 | Cuba | 20.00939N | 76.89402W | KJ157017 | KJ157246 | KJ157126 |
| Micrathena | horrida | 787913 | Cuba | 20.00939N | 76.89402W | KJ157018 | KJ157247 | KJ157127 |
| Micrathena | horrida | 787919 | Cuba | 20.00939N | 76.89402W | KJ157019 | KJ157244 | KJ157128 |
| Micrathena | lucasi | 00004785A | Costa Rica | San Antonio de Escazú | ||||
| Micrathena | macfarlanei | MIC054 | Brazil | 19.65000S | 42.56667W | KJ157241 | ||
| Micrathena | miles | MIC142 | Peru | 3.82975S | 73.375333W | KX687317 | ||
| Micrathena | militaris | 10000526A | Dominica | 15.32710N | 61.3381W | x | x | |
| Micrathena | militaris | 10000528A | Dominica | 15.32710N | 61.3381W | x | x | |
| Micrathena | militaris | 782365 | Hispaniola | 18.355536N | 068.61825W | KJ157020 | KJ157129 | |
| Micrathena | militaris | 784338 | Hispaniola | 18.32902N | 068.80995W | KJ157021 | KJ157273 | |
| Micrathena | militaris | 784363 | Hispaniola | 18.32902N | 068.80995W | KJ157022 | KJ157293 | KJ157130 |
| Micrathena | militaris | 784403 | Hispaniola | 18.32902N | 068.80995W | KJ157023 | KJ157298 | KJ157131 |
| Micrathena | militaris | 784430 | Hispaniola | 18.32902N | 068.80995W | KJ157024 | KJ157132 | |
| Micrathena | militaris | 784448 | Hispaniola | 18.32902N | 068.80995W | KJ157025 | KJ157294 | KJ157133 |
| Micrathena | militaris | 784458 | Hispaniola | 18.32902N | 068.80995W | KJ157026 | KJ157134 | |
| Micrathena | militaris | 784503 | Hispaniola | 18.3150011N | 71.580556W | KJ157027 | KJ157300 | KJ157135 |
| Micrathena | militaris | 784531 | Hispaniola | 18.355536N | 068.61825W | KJ157028 | KJ157136 | |
| Micrathena | militaris | 784566 | Hispaniola | 18.32902N | 068.80995W | KJ157029 | KJ157296 | KJ157137 |
| Micrathena | militaris | 784671 | Hispaniola | 19.06707N | 069.46355W | KJ157030 | KJ157138 | |
| Micrathena | militaris | 784721 | Hispaniola | 18.32902N | 068.80995W | KJ157031 | KJ157310 | KJ157139 |
| Micrathena | militaris | 784759 | Hispaniola | 18.355536N | 068.61825W | KJ157032 | KJ157277 | KJ157140 |
| Micrathena | militaris | 784762 | Hispaniola | 18.2205360N | 68.4806070W | KJ157033 | KJ157141 | |
| Micrathena | militaris | 784772 | Hispaniola | 18.32902N | 068.80995W | KJ157034 | KJ157287 | KJ157142 |
| Micrathena | militaris | 784806 | Hispaniola | KJ157035 | KJ157143 | |||
| Micrathena | militaris | 784926 | Hispaniola | KJ157036 | KJ157144 | |||
| Micrathena | militaris | 785066 | Hispaniola | 19.06707N | 069.46355W | KJ157037 | KJ157145 | |
| Micrathena | militaris | 785080 | Hispaniola | 18.32902N | 068.80995W | KJ157038 | KJ157274 | KJ157146 |
| Micrathena | militaris | 785099 | Hispaniola | 18.32902N | 068.80995W | KJ157313 | ||
| Micrathena | militaris | 785128 | Hispaniola | 18.355536N | 068.61825W | KJ157039 | KJ157147 | |
| Micrathena | militaris | 785144 | Hispaniola | 19.746175N | 71.257726W | KJ157040 | KJ157148 | |
| Micrathena | militaris | 785169 | Hispaniola | 18.355536N | 068.61825W | KJ157041 | KJ157290 | KJ157149 |
| Micrathena | militaris | 785173 | Hispaniola | 19.06707N | 069.46355W | KJ157042 | KJ157314 | KJ157150 |
| Micrathena | militaris | 785174 | Hispaniola | 19.06707N | 069.46355W | KJ157043 | KJ157292 | KJ157151 |
| Micrathena | militaris | 785194 | Hispaniola | 18.355536N | 068.61825W | KJ157044 | ||
| Micrathena | militaris | 785208 | Hispaniola | 18.2205360N | 68.4806070W | KJ157045 | KJ157297 | KJ157152 |
| Micrathena | militaris | 785219 | Hispaniola | 18.355536N | 068.61825W | KJ157046 | KJ157286 | KJ157153 |
| Micrathena | militaris | 785263 | Hispaniola | 18.355536N | 068.61825W | KJ157047 | KJ157154 | |
| Micrathena | militaris | 785273 | Hispaniola | 19.432213N | 070.371412W | KJ157048 | KJ157275 | KJ157155 |
| Micrathena | militaris | 785280 | Hispaniola | 18.32902N | 068.80995W | KJ157049 | KJ157315 | KJ157156 |
| Micrathena | militaris | 785312 | Hispaniola | 19.34405N | 069.46635W | KJ157050 | KJ157280 | KJ157157 |
| Micrathena | militaris | 785401 | Hispaniola | 19.06707N | 069.46355W | KJ157051 | KJ157276 | KJ157158 |
| Micrathena | militaris | 785402 | Hispaniola | 19.34405N | 069.46635W | KJ157052 | KJ157285 | KJ157159 |
| Micrathena | militaris | 785423 | Hispaniola | 18.355536N | 068.61825W | KJ157053 | KJ157160 | |
| Micrathena | militaris | 785461 | Hispaniola | 19.06707N | 069.46355W | KJ157054 | KJ157281 | |
| Micrathena | militaris | 785502 | Hispaniola | 19.06707N | 069.46355W | KJ157055 | KJ157301 | KJ157161 |
| Micrathena | militaris | 785512 | Hispaniola | 19.06707N | 069.46355W | KJ157056 | KJ157316 | KJ157162 |
| Micrathena | militaris | 785524 | Hispaniola | 18.355536N | 068.61825W | KJ157057 | KJ157311 | KJ157163 |
| Micrathena | militaris | 785527 | Hispaniola | 19.34405N | 069.46635W | KJ157058 | KJ157279 | KJ157164 |
| Micrathena | militaris | 785563 | Hispaniola | 19.06707N | 069.46355W | KJ157059 | KJ157295 | KJ157165 |
| Micrathena | militaris | 785604 | Hispaniola | 19.06707N | 069.46355W | KJ157060 | KJ157288 | KJ157166 |
| Micrathena | militaris | 785706 | Hispaniola | 19.06707N | 069.46355W | KJ157061 | KJ157278 | KJ157167 |
| Micrathena | militaris | 785709 | Hispaniola | 19.06707N | 069.46355W | KJ157312 | KJ157168 | |
| Micrathena | militaris | 785722 | Hispaniola | 19.06707N | 069.46355W | KJ157062 | KJ157283 | KJ157169 |
| Micrathena | militaris | 785729 | Hispaniola | 19.34405N | 069.46635W | KJ157063 | KJ157284 | KJ157170 |
| Micrathena | militaris | 785743 | Hispaniola | 19.06707N | 069.46355W | KJ157064 | KJ157282 | KJ157171 |
| Micrathena | militaris | 785769 | Hispaniola | 19.06707N | 069.46355W | KJ157065 | KJ157172 | |
| Micrathena | militaris | 787068 | Hispaniola | 18.980122N | 70.798425W | KJ157066 | KJ157299 | KJ157173 |
| Micrathena | militaris | 787106 | Hispaniola | 18.980122N | 70.798425W | KJ157067 | KJ157289 | KJ157174 |
| Micrathena | militaris | 787148 | Hispaniola | 18.3150011N | 71.580556W | KJ157068 | KJ157291 | KJ157175 |
| Micrathena | militaris | 787152 | Hispaniola | 18.3150011N | 71.580556W | KJ157069 | KJ157176 | |
| Micrathena | militaris | 787166 | Hispaniola | 18.3150011N | 71.580556W | KJ157070 | KJ157177 | |
| Micrathena | militaris | 787190 | Hispaniola | 18.3150011N | 71.580556W | KJ157071 | KJ157178 | |
| Micrathena | militaris | 787208 | Hispaniola | 18.3150011N | 71.580556W | KJ157072 | KJ157179 | |
| Micrathena | militaris | 787212 | Hispaniola | 18.3150011N | 71.580556W | KJ157073 | KJ157180 | |
| Micrathena | militaris | 787214 | Hispaniola | 18.3150011N | 71.580556W | KJ157001 | KJ157112 | |
| Micrathena | militaris | 392672 | Puerto Rico | 17.971472N | 66.867958W | KJ157074 | KJ157302 | KJ157181 |
| Micrathena | militaris | 392677 | Puerto Rico | 17.971472N | 66.867958W | KJ157075 | KJ157303 | KJ157182 |
| Micrathena | militaris | 782048 | Puerto Rico | 18.414373N | 66.728722W | KJ157076 | KJ157307 | KJ157183 |
| Micrathena | militaris | 782126 | Puerto Rico | 18.173264N | 66.590149W | KJ157077 | KJ157308 | KJ157184 |
| Micrathena | militaris | 782153 | Puerto Rico | 18.414373N | 66.728722W | KJ157078 | KJ157306 | KJ157185 |
| Micrathena | militaris | 782174 | Puerto Rico | 18.414373N | 66.728722W | KJ157079 | KJ157304 | KJ157186 |
| Micrathena | militaris | 782201 | Puerto Rico | 18.032518N | 67.094653W | KJ157080 | KJ157305 | KJ157187 |
| Micrathena | militaris | 783400 | Puerto Rico | 18.45226N | 66.59711W | KJ157309 | ||
| Micrathena | mitrata | 10000679A | Mexico | 19.79357N | 104.0554W | x | x | |
| Micrathena | mitrata | 00002849A | Mexico | 19.79357N | 104.0554W | x | x | |
| Micrathena | nigrichelis | MIC056 | Brazil | 20.43481S | 43.50906W | KJ157239 | ||
| Micrathena | perfida | MIC026 | Brazil | 24.387111S | 47.017583W | KX687318 | ||
| Micrathena | plana | MIC062 | Brazil | 16.53294S | 41.51042W | KJ157240 | ||
| Micrathena | reimoseri | MIC072 | Brazil | 11.399833S | 40.522056W | KX687321 | ||
| Micrathena | saccata | MIC076 | Brazil | 1.424828S | 48.43802W | KJ157237 | ||
| Micrathena | sagittata | 10000618A | FL, USA | 29.4776N | 082.5627W | x | ||
| Micrathena | sagittata | 10000621A | FL, USA | 29.63703N | 082.23976W | x | ||
| Micrathena | sagittata | 10000631A | FL, USA | 29.62986N | 082.29880W | x | x | |
| Micrathena | sagittata | 10000633A | FL, USA | 29.62986N | 082.29880W | x | ||
| Micrathena | sagittata | 10000636A | FL, USA | 29.63680N | 082.23961W | x | x | |
| Micrathena | sagittata | 10000634A | FL, USA | 29.46397N | 082.55285W | x | x | |
| Micrathena | sagittata | 10000639A | FL, USA | 29.63680N | 082.23961W | x | ||
| Micrathena | sagittata | 10000640A | FL, USA | 29.62688N | 082.29878W | x | ||
| Micrathena | sagittata | 00002847A | Mexico | 18.18963N | 89.46333W | x | ||
| Micrathena | sagittata | 00000833A | SC, USA | 33.03913 N | 79.56459W | KJ157081 | KJ157221 | KJ157189 |
| Micrathena | schreibersi | 00002357A | Colombia | Bucaramanga | x | |||
| Micrathena | schreibersi | 10000650A | Colombia | 8.39104N | 77.21548W | x | ||
| Micrathena | schreibersi | 10000652A | Colombia | 8.39104N | 77.21548W | x | ||
| Micrathena | schreibersi | 10000653A | Colombia | 8.39104N | 77.21548W | x | x | |
| Micrathena | schreibersi | 10000664A | Colombia | 8.424N | 77.29216W | x | ||
| Micrathena | schreibersi | 10000673A | Colombia | 8.39104N | 77.21548W | x | ||
| Micrathena | schreibersi | 10000658A | Colombia | 8.39104N | 77.21548W | x | ||
| Micrathena | schreibersi | 10000651A | Colombia | 8.39104N | 77.21548W | x | x | |
| Micrathena | schreibersi | 10000663A | Colombia | 8.424N | 77.29216W | x | ||
| Micrathena | schreibersi | 10000665A | Colombia | 8.424N | 77.29216W | x | x | |
| Micrathena | schreibersi | 00004787A | Colombia | 10.21192N | 75.25403W | x | x | |
| Micrathena | schreibersi | 00004818A | Trinidad | x | x | |||
| Micrathena | schreibersi | 00002900A | Costa Rica | 10.430686N | 84.007089W | x | x | |
| Micrathena | schreibersi | 00000936A | Colombia | 7.062695N | 73.073058W | KJ157090 | KJ157318 | KJ157198 |
| Micrathena | schreibersi | 00002357A | Colombia | 7.062695N | 73.073058W | KJ157092 | KJ157319 | KJ157199 |
| Micrathena | sexspinosa | 10000690A | Colombia | 8.35249N | 77.22118W | x | ||
| Micrathena | sexspinosa | 10000659A | Colombia | 8.35249N | 77.22118W | x | ||
| Micrathena | sexspinosa | 10000674A | Colombia | 8.35249N | 77.22118W | x | x | |
| Micrathena | sexspinosa | 10000677A | Colombia | 11.120083N | 74.082805W | x | ||
| Micrathena | sexspinosa | 10000683A | Colombia | 11.120083N | 74.082805W | x | ||
| Micrathena | sexspinosa | 10000669A | Colombia | 8.39104N | 77.21548W | x | x | |
| Micrathena | sexspinosa | 10000670A | Colombia | 8.39104N | 77.21548W | x | x | |
| Micrathena | sexspinosa | 10000681A | Colombia | 8.35249N | 77.22118W | x | ||
| Micrathena | sexspinosa | 10000678A | Colombia | 8.35249N | 77.22118W | x | ||
| Micrathena | sexspinosa | 00000987A | Colombia | 7.062695N | 73.073058W | KJ157091 | KJ157222 | |
| Micrathena | similis | 785024 | Hispaniola | 19.34405N | 69.46635W | KJ157093 | KJ157228 | KJ157200 |
| Micrathena | similis | 785496 | Hispaniola | 19.34405N | 69.46635W | KJ157094 | KJ157232 | KJ157201 |
| Micrathena | similis | 787265 | Hispaniola | 19.05116N | 70.88866W | KJ157095 | KJ157233 | KJ157202 |
| Micrathena | similis | 787297 | Hispaniola | 19.05116N | 70.88866W | KJ157096 | KJ157203 | |
| Micrathena | similis | 787308 | Hispaniola | 19.03627N | 70.54337W | KJ157097 | KJ157229 | KJ157204 |
| Micrathena | similis | 787309 | Hispaniola | 19.05116N | 70.88866W | KJ157098 | KJ157205 | |
| Micrathena | similis | 787311 | Hispaniola | 19.05116N | 70.88866W | KJ157235 | KJ157206 | |
| Micrathena | similis | 787318 | Hispaniola | 19.03627N | 70.54337W | KJ157099 | KJ157234 | KJ157207 |
| Micrathena | similis | 787320 | Hispaniola | 19.05116N | 70.88866W | KJ157100 | KJ157230 | KJ157208 |
| Micrathena | similis | 787322 | Hispaniola | 19.05116N | 70.88866W | KJ157101 | KJ157231 | KJ157209 |
| Micrathena | sp. | 10000656A | Colombia | 11.120083N | 74.082805W | x | ||
| Micrathena | sp. | 10000671A | Colombia | 11.120083N | 74.082805W | x | x | |
| Micrathena | sp. | 00006693A | Colombia | 11.120083N | 74.082805W | x | x | |
| Micrathena | spinulata | MIC205 | Mexico | 19.1381667N | 97.2045W | KX687324 | ||
| Micrathena | triangularispinosa | MIC156 | Brazil | 0.97799S | 62.10292W | KX687327 | ||
| Micrathena | yanomami | MIC193 | Peru | 13.055639S | 71.546194W | KX687332 | ||
| Outgroups | ||||||||
| Achaearanea | sp. | 784841 | Cuba | 21.59166N | 77.78822W | KJ157211 | ||
| Argiope | lobata | Arg0160 | Spain | Missing GPS data | KJ156988 | KJ157103 | ||
| Gasteracantha | cancriformis | 787198 | Hispaniola | 18.3150011N | 71.580556W | KJ156989 | KJ157212 | |
| Gasteracantha | cancriformis | 784515 | Hispaniola | 18.2205260N | 68.480607W | KJ157213 | ||
| Gasteracantha | cancriformis | 782149 | Puerto Rico | 18. 172979N | 66.491798W | KJ156990 | KJ157214 |
| Polymerase Chain Reaction (PCR) Conditions | ||||
|---|---|---|---|---|
| Gene | Forward Primer | Reverse Primer | Annealing Temp. (°C) | Fragment Length (bp) |
| Internal transcribed spacer 2 (ITS-2) | ITS4 | ITS5.8 | 47 | 350–500 |
| Jerry | C1-N-2776 | 46 | ~1250 | |
| Cytochrome oxidase subunit 1 (CO1) | LCO11490 | C1-N-2776 | 48 | ~1250 |
| Model | LnL | Number of Parameters | d | e | j | AICc | AICc Weight | ΔAICc |
|---|---|---|---|---|---|---|---|---|
| DEC + J no-GAARlandia | −81.87 | 3 | 0.0041 | 0.0011 | 0.2 | 170.5 | 0.56 | 0 |
| BAYAREALIKE + J no-GAARlandia | −82.46 | 3 | 0.0019 | 0.01 | 0.2 | 171.7 | 0.31 | 1.2 |
| DIVALIKE + J no-GAARlandia | −83.53 | 3 | 0.0048 | 0.001 | 0.2 | 173.8 | 0.11 | 3.3 |
| BAYAREALIKE + J GAARlandia | −85.26 | 3 | 0.023 | 0.011 | 0.8 | 177.3 | 0.019 | 6.8 |
| DIVALIKE no-GAARlandia | −95.23 | 2 | 0.013 | 0.0033 | 0 | 194.8 | 2.9 × 10−6 | 24.3 |
| DEC + J GAARlandia | −94.48 | 3 | 0.025 | 1.00 × 10−12 | 2.4 | 195.7 | 1.90 × 10−6 | 25.2 |
| DIVALIKE + J GAARlandia | −97.42 | 3 | 0.027 | 1.00 × 10−12 | 1.7 | 201.6 | 9.90 × 10−8 | 31.1 |
| DEC no-GAARlandia | −99.69 | 2 | 0.013 | 0.0063 | 0 | 203.8 | 3.40 × 10−8 | 33.3 |
| BAYAREALIKE no-GAARlandia | −107.9 | 2 | 0.017 | 0.025 | 0 | 220.2 | 8.90 × 10−12 | 49.7 |
| BAYAREALIKE GAARlandia | −112 | 2 | 0.24 | 0.025 | 0 | 228.4 | 1.50 × 10−13 | 57.9 |
| DIVALIKE GAARlandia | −112.8 | 2 | 0.11 | 0.0058 | 0 | 230 | 6.90 × 10−14 | 59.5 |
| DEC GAARlandia | −112.9 | 2 | 0.16 | 0.01 | 0 | 230.2 | 6.00 × 10−14 | 59.7 |
| Species-Group | Magalhaēs et al., 2012 | McHugh et al., 2014 | Current Micrathena Study |
|---|---|---|---|
| furcula | M. cubana, M. similis | M. cubana, M. similis | M. cubana, M. similis |
| militaris | M. banksi, M. militaris, M. sagittata, M. sexspinosa | M. banksi, M. militaris, M. sagittata, M. sexspinosa | M. banksi, M. militaris, M. sagittata, M. sexspinosa |
| gracilis | M. horrida, M. gracilis, M. forcipata | M. horrida, M. gracilis1 | M. horrida, M. gracilis2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shapiro, L.; Binford, G.J.; Agnarsson, I. Single-Island Endemism despite Repeated Dispersal in Caribbean Micrathena (Araneae: Araneidae): An Updated Phylogeographic Analysis. Diversity 2022, 14, 128. https://doi.org/10.3390/d14020128
Shapiro L, Binford GJ, Agnarsson I. Single-Island Endemism despite Repeated Dispersal in Caribbean Micrathena (Araneae: Araneidae): An Updated Phylogeographic Analysis. Diversity. 2022; 14(2):128. https://doi.org/10.3390/d14020128
Chicago/Turabian StyleShapiro, Lily, Greta J. Binford, and Ingi Agnarsson. 2022. "Single-Island Endemism despite Repeated Dispersal in Caribbean Micrathena (Araneae: Araneidae): An Updated Phylogeographic Analysis" Diversity 14, no. 2: 128. https://doi.org/10.3390/d14020128
APA StyleShapiro, L., Binford, G. J., & Agnarsson, I. (2022). Single-Island Endemism despite Repeated Dispersal in Caribbean Micrathena (Araneae: Araneidae): An Updated Phylogeographic Analysis. Diversity, 14(2), 128. https://doi.org/10.3390/d14020128







