Solenysa, a Cretaceous Relict Spider Group in East Asia
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling
2.2. Laboratory Protocols for Molecular Data
2.3. Phylogenetic Analyses
2.4. Estimation of Divergence Times
2.5. Lineages Tracing through Time
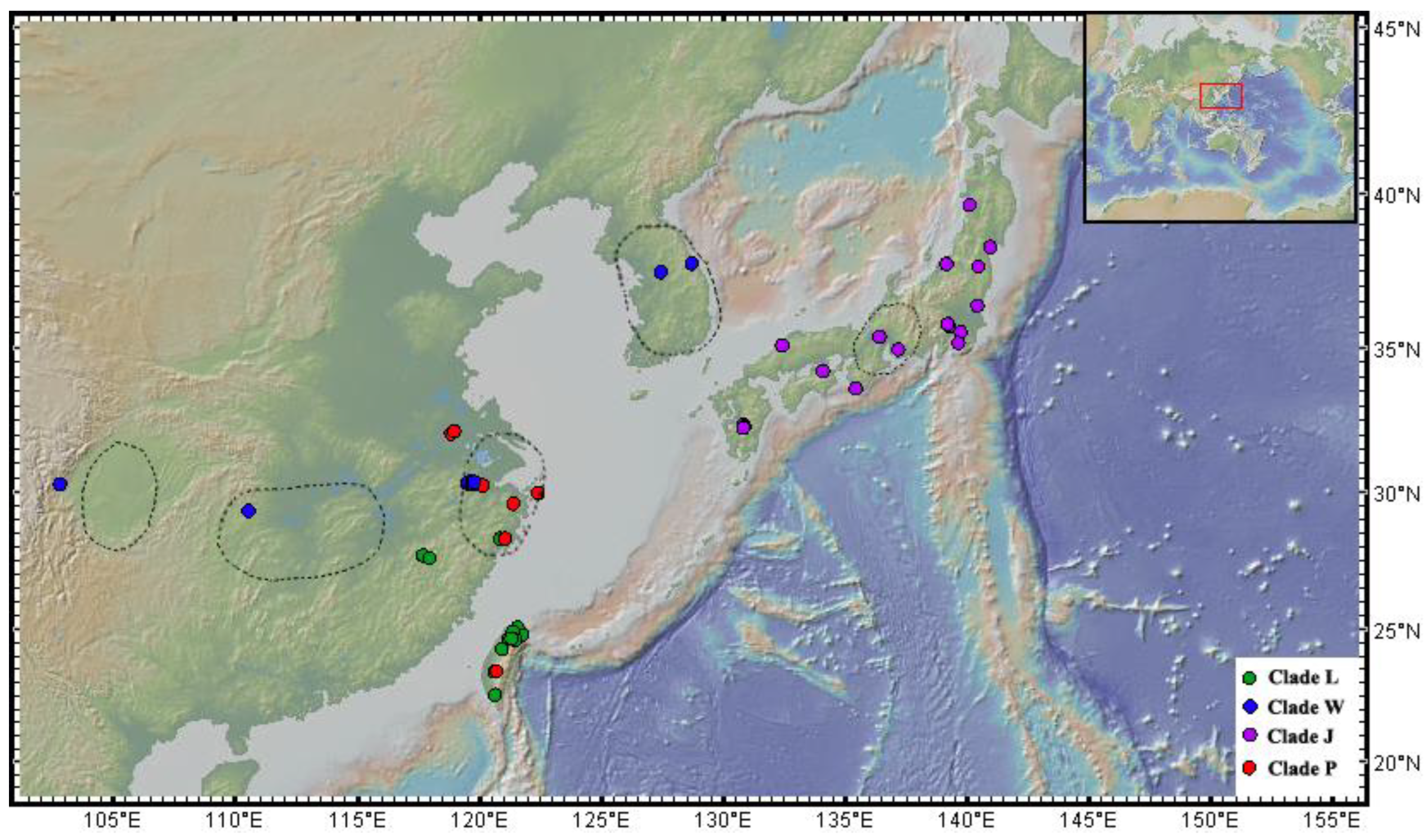
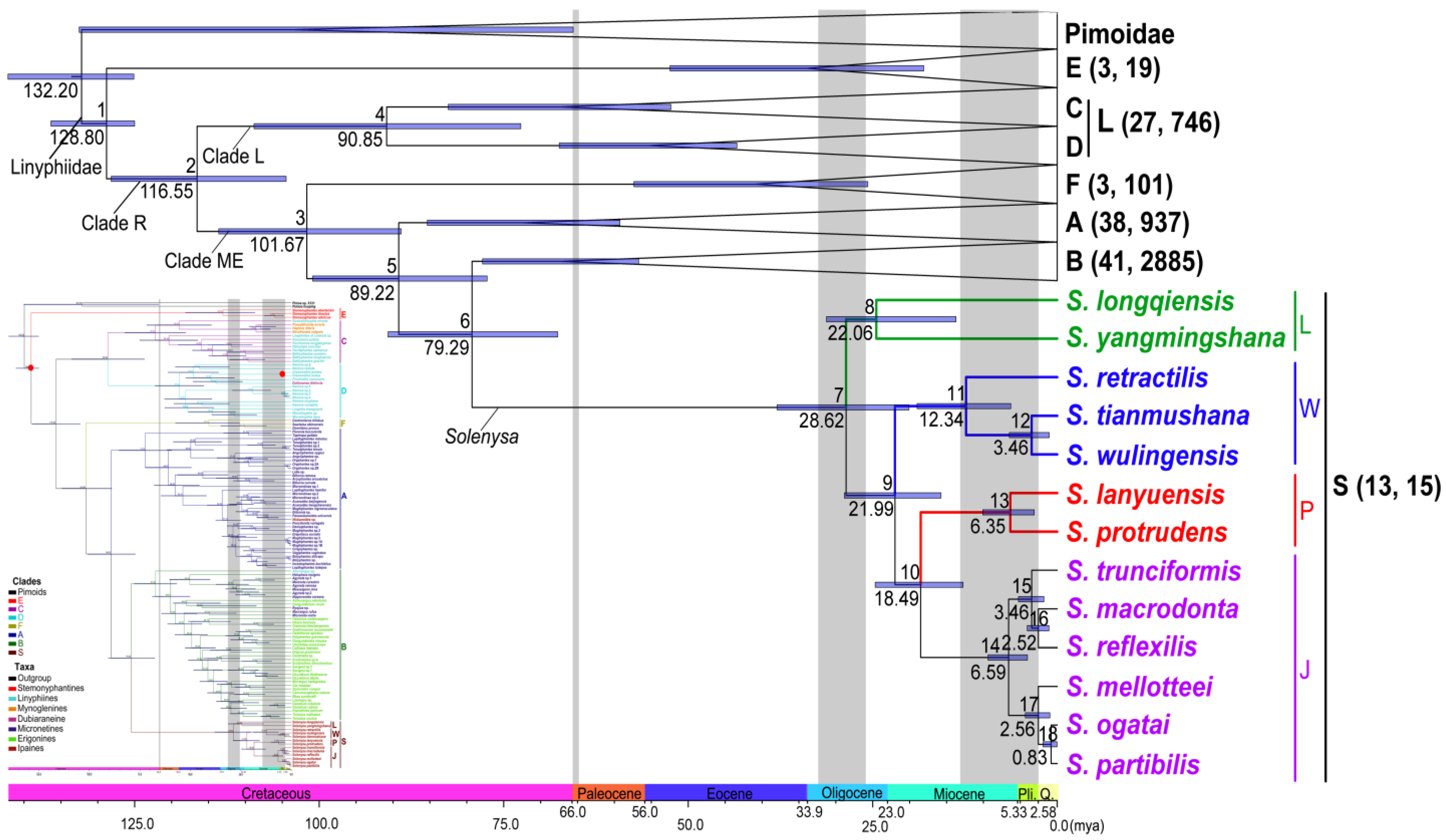
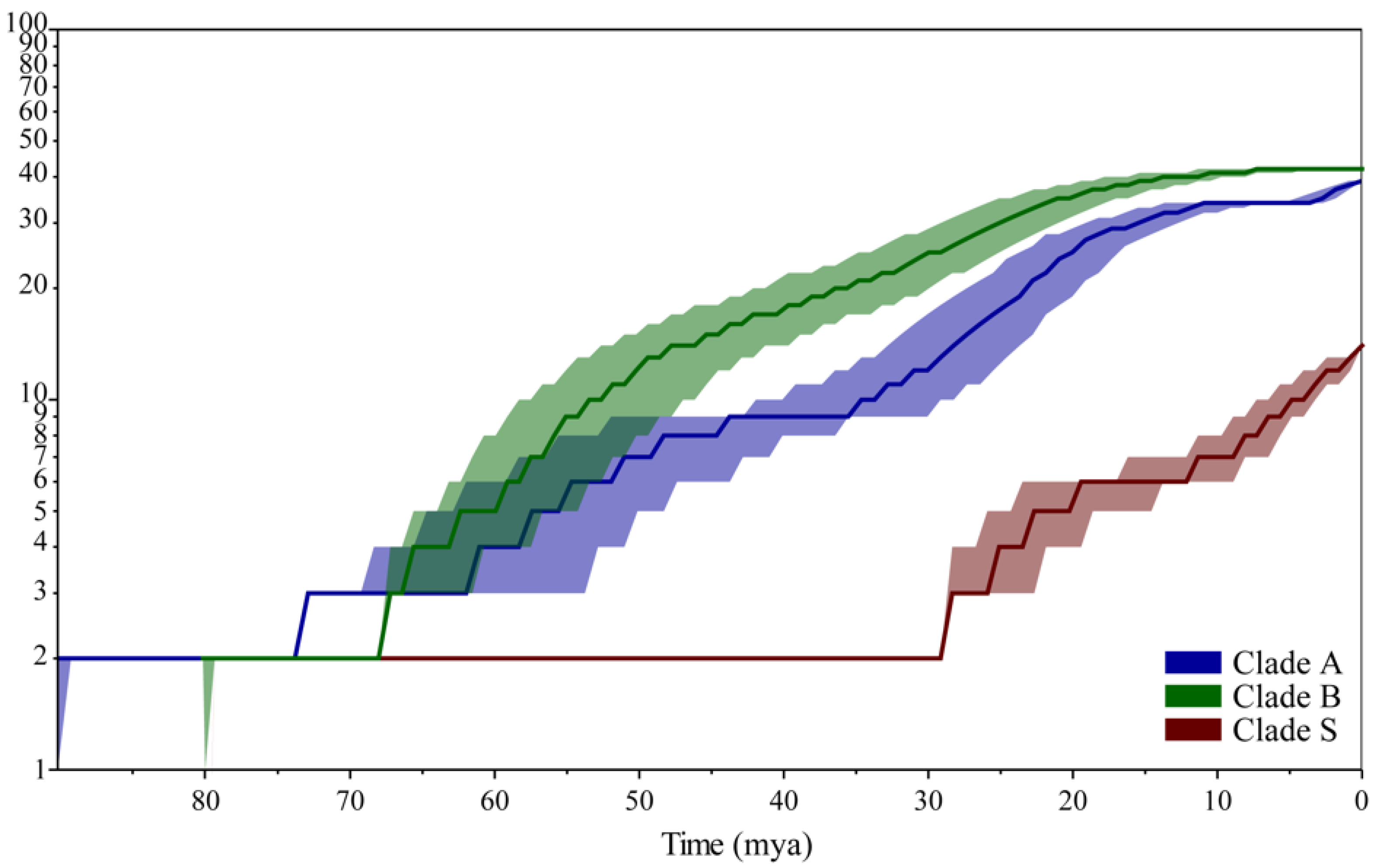
3. Results
3.1. DNA Sequence Data
3.2. Phylogeny of Solenysa
3.3. Divergence Times Estimation
3.4. Temporal Patterns of Linyphiids Diversification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnedo, M.A.; Hormiga, G.; Scharff, N. Higher-level phylogenetics of linyphiid spiders (Araneae, Linyphiidae) based on morphological and molecular evidence. Cladistics 2009, 25, 231–262. [Google Scholar] [CrossRef] [PubMed]
- Dabert, M.; Witalinski, W.; Kaźmierski, A.; Olszanowski, Z.; Dabert, J. Molecular phylogeny of acariform mites (Acari, Arachnida): Strong conflict between phylogenetic signal and long-branch attraction artifacts. Mol. Phylogenetics Evol. 2010, 56, 222–241. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.P.; Kaluziak, S.T.; Perez-Porro, A.; González, V.L.; Hormiga, G.; Wheeler, W.C.; Giribet, G. Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol. Biol. Evol. 2014, 31, 2963–2984. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ballesteros, J.A.; Hormiga, G.; Chesters, D.; Zhan, Y.; Sun, N.; Zhu, C.; Chen, W.; Tu, L. Resolving the phylogeny of a speciose spider group, the family Linyphiidae (Araneae). Mol. Phylogenetics Evol. 2015, 91, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Bininda-Emonds, O.R.P.; Cardillo, M.; Jones, K.E.; MacPhee, R.D.E.; Beck, R.M.D.; Grenyer, R.; Price, S.A.; Vos, R.A.; Gittleman, J.L.; Purvis, A. The delayed rise of present-day mammals. Nature 2007, 446, 507–512. [Google Scholar] [CrossRef]
- Near, T.J.; Dornburg, A.; Kuhn, K.L.; Eastman, J.T.; Pennington, J.N.; Patarnello, T.; Zane, L.; Fernández, D.A.; Jones, C.D. Ancient climate change, antifreeze, and the evolutionary diversification of Antarctic fishes. Proc. Natl. Acad. Sci. USA 2012, 109, 3434–3439. [Google Scholar] [CrossRef]
- Arnedo, M.A.; Hormiga, G. Repeated colonization, adaptive radiation and convergent evolution in the sheet-weaving spiders (Linyphiidae) of the south Pacific Archipelago of Juan Fernandez. Cladistics 2020, 37, 317–342. [Google Scholar] [CrossRef]
- Hormiga, G.; Kulkarni, S.; Moreira, T.D.S.; Dimitrov, D. Molecular phylogeny of pimoid spiders and the limits of Linyphiidae, with a reassessment of male palpal homologies (Araneae, Pimoidae). Zootaxa 2021, 5026, 71–101. [Google Scholar] [CrossRef]
- Dimitrov, D.; Hormiga, G. Spider Diversification Through Space and Time. Annu. Rev. Entomol. 2021, 66, 225–241. [Google Scholar] [CrossRef]
- Bond, J.E.; Opell, B.D. Testing adaptive radiation and key innovation hypotheses in spiders. Evolution 1998, 52, 403–414. [Google Scholar] [CrossRef]
- Penney, D. Does the fossil record of spiders track that of their principal prey, the insects? Trans. R. Soc. Edinburgh Earth Sci. 2003, 94, 275–281. [Google Scholar] [CrossRef]
- Penney, D.; Ortuño, V.M. Oldest true orb-weaving spider (Araneae: Araneidae). Biol. Lett. 2006, 2, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Selden, P.A.; Penney, D. Fossil spiders. Biol. Rev. 2010, 85, 171–206. [Google Scholar] [CrossRef]
- Lopardo, L.; Michalik, P.; Hormiga, G. Take a deep breath… The evolution of the respiratory system of symphytognathoid spiders (Araneae, Araneoidea). Org. Divers. Evol. 2021, 1–33. [Google Scholar] [CrossRef]
- Garrison, N.L.; Rodriguez, J.; Agnarsson, I.; Coddington, J.A.; Griswold, C.E.; Hamilton, C.A.; Hedin, M.; Kocot, K.M.; Ledford, J.M.; Bond, J.E. Spider phylogenomics: Untangling the Spider Tree of Life. PeerJ 2016, 4, e1719. [Google Scholar] [CrossRef] [PubMed]
- Penney, D.; Selden, P.A. The oldest linyphiid spider, in Lower Cretaceous Lebanese amber (Araneae, Linyphiidae, Linyphiinae). J. Arachnol. 2002, 30, 487–493. [Google Scholar] [CrossRef]
- World Spider Catalog. Version 22.5. Natural History Museum Bern. Available online: http://wsc.nmbe.ch (accessed on 20 November 2021). [CrossRef]
- Tanasevitch, A.V. Linyphiid Spiders of the World. Available online: http://www.andtan.newmail.ru/list (accessed on 20 November 2021).
- Dimitrov, D.; Lopardo, L.; Giribet, G.; Arnedo, M.; Álvarez-Padilla, F.; Hormiga, G. Tangled in a sparse spider web: Single origin of orb weavers and their spinning work unravelled by denser taxonomic sampling. Proc. R. Soc. B Boil. Sci. 2011, 279, 1341–1350. [Google Scholar] [CrossRef]
- Dimitrov, D.; Benavides, L.R.; Arnedo, M.; Giribet, G.; Griswold, C.E.; Scharff, N.; Hormiga, G. Rounding up the usual suspects: A standard target-gene approach for resolving the interfamilial phylogenetic relationships of ecribellate orb-weaving spiders with a new family-rank classification (Araneae, Araneoidea). Cladistics 2017, 33, 221–250. [Google Scholar] [CrossRef]
- Wheeler, W.C.; Coddington, J.A.; Crowley, L.M.; Dimitrov, D.; Goloboff, P.A.; Griswold, C.E.; Hormiga, G.; Prendini, L.; Ramírez, M.J.; Sierwald, P.; et al. The spider tree of life: Phylogeny of Araneae based on target-gene analyses from an extensive taxon sampling. Cladistics 2017, 33, 574–616. [Google Scholar] [CrossRef]
- Eberhard, W.G. Evolution of genitalia: Theories, evidence, and new directions. Genetica 2010, 138, 5–18. [Google Scholar] [CrossRef]
- Tu, L.; Li, S. A review of the linyphiid spider genus Solenysa (Araneae, Linyphiidae). J. Arachnol. 2006, 34, 87–97. [Google Scholar] [CrossRef]
- Tu, L.; Hormiga, G. Phylogenetic analysis and revision of the linyphiid spider genus Solenysa (Araneae: Linyphiidae: Erigoninae). Zool. J. Linn. Soc. 2011, 161, 484–530. [Google Scholar] [CrossRef]
- Saaristo, M.I. A new subfamily of linyphiid spiders based on a new genus created for the keyserlingi-group of the genus Lepthyphantes (Aranei: Linyphiidae). Arthropoda Sel. 2007, 16, 33–42. [Google Scholar]
- Namkung, J. Two unrecorded species of linyphiid spiders from Korea. Korean Arachnol. 1986, 2, 11–18. [Google Scholar]
- Li, S.; Song, D. On two new species of soil linyphiid spiders from China (Araneae: Linyphiidae: Erigoninae). Acta Arachnol. Sin. 1992, 1, 6–9. [Google Scholar]
- Gao, J.C.; Zhu, C.D.; Sha, Y.H. Two new species of the genus Solenysa from China (Araneae: Linyphiidae: Erigoninae). Acta Arachnol. Sin. 1993, 2, 65–68. [Google Scholar]
- Tu, L.; Ono, H.; Li, S. Two new species of Solenysa Simon, 1894 (Araneae: Linyphiidae) from Japan. Zootaxa 2007, 1426, 57–62. [Google Scholar] [CrossRef]
- Ono, H. Notes on Japanese spiders of the genera Paikiniana and Solenysa (Araneae, Linyphiidae). Bull. Natl. Mus. Nat. Sci. Ser. A 2011, 37, 121–129. [Google Scholar]
- Tu, L.; Wang, F.; Ono, H. A review of Solenysa spiders from Japan (Araneae, Linyphiidae), with a comment on the type species S. mellotteei Simon, 1894. ZooKeys 2015, 481, 39–56. [Google Scholar] [CrossRef]
- Tian, J.; Tu, L. A new species of the spider genus Solenysa from China (Araneae, Linyphiidae). Zootaxa 2018, 4531, 142–146. [Google Scholar] [CrossRef]
- Blest, A.D. The tracheal arrangement and the classification of linyphiid spiders. J. Zool. 1976, 180, 185–194. [Google Scholar] [CrossRef]
- Hormiga, G. Higher level phylogenetics of erigonine spiders (Araneae, Linyphiidae, Erigoninae). Smithson. Contrib. Zool. 2000, 1–160. [Google Scholar] [CrossRef]
- Schmitz, A. Metabolic rates during rest and activity in differently tracheated spiders (Arachnida, Araneae): Pardosa lugubris (Lycosidae) and Marpissa muscosa (Salticidae). J. Comp. Physiol. B 2004, 174, 519–526. [Google Scholar] [CrossRef]
- Spider Ecophysiology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2013; pp. 1–529.
- Levi, H.W. Adaptations of respiratory systems of spiders. Evolution 1967, 21, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.51. Available online: http://www.mesquiteproject.org (accessed on 20 November 2021).
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Shimodaira, H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002, 51, 492–508. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.R.; Drummond, A.J. bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol. Biol. 2017, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Fernández, R.; Kallal, R.J.; Dimitrov, D.; Ballesteros, J.A.; Arnedo, M.A.; Giribet, G.; Hormiga, G. Phylogenomics, diversification dynamics, and comparative transcriptomics across the Spider Tree of Life. Curr. Biol. 2018, 28, 1489–1497.e5. [Google Scholar] [CrossRef]
- Scharff, N.; Coddington, J.A.; Blackledge, T.A.; Agnarsson, I.; Framenau, V.; Szűts, T.; Hayashi, C.Y.; Dimitrov, D. Phylogeny of the orb-weaving spider family Araneidae (Araneae: Araneoidea). Cladistics 2020, 36, 1–21. [Google Scholar] [CrossRef]
- Magalhaes, I.L.F.; Azevedo, G.H.F.; Michalik, P.; Ramírez, M.J. The fossil record of spiders revisited: Implications for calibrating trees and evidence for a major faunal turnover since the Mesozoic. Biol. Rev. 2020, 95, 184–217. [Google Scholar] [CrossRef]
- Hormiga, G.; Arnedo, M.; Gillespie, R.G. Speciation on a conveyor belt: Sequential colonization of the Hawaiian Islands by Orsonwelles spiders (Araneae, Linyphiidae). Syst. Biol. 2003, 52, 70–88. [Google Scholar] [CrossRef]
- Hewitt, G.M. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Hewitt, G.M. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 183–195. [Google Scholar] [CrossRef]
- Blackledge, T.A.; Scharff, N.; Coddington, J.A.; Szüts, T.; Wenzel, J.W.; Hayashi, C.Y.; Agnarsson, I. Reconstructing web evolution and spider diversification in the molecular era. Proc. Natl. Acad. Sci. USA 2009, 106, 5229–5234. [Google Scholar] [CrossRef] [PubMed]
- Nayar, A. Asteroid impact may have gassed Earth. Nature 2009, 2009–2012. [Google Scholar] [CrossRef]
- Opell, B.D. The influence of web monitoring tactics on the tracheal systems of spiders in the family Uloboridae (Arachnida, Araneida). Zoomorphology 1987, 107, 255–259. [Google Scholar] [CrossRef]
- Lopardo, L.; Hormiga, G. On the synaphrid spider Cepheia longiseta (Simon 1881) (Araneae, Synaphridae). Am. Museum Novit. 2007, 20052, 18. [Google Scholar] [CrossRef]
- Lopardo, L.; Hormiga, G. Out of the twilight zone: Phylogeny and evolutionary morphology of the orb-weaving spider family Mysmenidae, with a focus on spinneret spigot morphology in symphytognathoids (Araneae, Araneoidea). Zool. J. Linn. Soc. 2015, 173, 527–786. [Google Scholar] [CrossRef]
- Foelix, R.F. Biology of Spiders, 3rd ed.; Oxford University Press: New York, NY, USA, 2011. [Google Scholar]
- Zachos, J.C.; Dickens, G.R.; Zeebe, R.E. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature 2008, 451, 279–283. [Google Scholar] [CrossRef]
- Strömberg, C.A. Evolution of grasses and grassland ecosystems. Annu. Rev. Earth Planet. Sci. 2011, 39, 517–544. [Google Scholar] [CrossRef]
- Guo, Z.T.; Sun, B.; Zhang, Z.S.; Peng, S.Z.; Xiao, G.Q.; Ge, J.Y.; Hao, Q.Z.; Qiao, Y.S.; Liang, M.Y.; Liu, J.F.; et al. A major reorganization of Asian climate by the Early Miocene. Clim. Past 2008, 4, 153–174. [Google Scholar] [CrossRef]
- Law, C.J. Evolutionary shifts in extant mustelid (Mustelidae: Carnivora) cranial shape, body size and body shape coincide with the Mid-Miocene Climate Transition. Biol. Lett. 2019, 15, 20190155. [Google Scholar] [CrossRef]
- Hampe, A.; Jump, A.S. Climate Relicts: Past, Present, Future. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 313–333. [Google Scholar] [CrossRef]
- Grandcolas, P.; Nattier, R.; Trewick, S. Relict species: A relict concept? Trends Ecol. Evol. 2014, 29, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Grandcolas, P.; Trewick, S. Biodiversity Conservation and Phylogenetic Systematics, Topics in Biodiversity and Conservation; Springer Nature: Basel, Switzerland, 2016. [Google Scholar]
- Fu, J.; Weadick, C.J.; Zeng, X.; Wang, Y.; Liu, Z.; Zheng, Y.; Li, C.; Hu, Y. Phylogeographic analysis of the Bufo gargarizans species complex: A revisit. Mol. Phylogenetics Evol. 2005, 37, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Borzée, A.; Santos, J.; Sanchez-Ramirez, S.; Bae, Y.; Heo, K.; Jang, Y.; Jowers, M.J. Phylogeographic and population insights of the Asian common toad (Bufo gargarizans) in Korea and China: Population isolation and expansions as response to the ice ages. PeerJ 2017, 5, e4044. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Cao, L.-J.; Li, B.-Y.; Gong, Y.-J.; Hoffmann, A.A.; Wei, S.-J. Multiple refugia from penultimate glaciations in East Asia demonstrated by phylogeography and ecological modelling of an insect pest. BMC Evol. Biol. 2018, 18, 152. [Google Scholar] [CrossRef]
- Du, Z.; Ishikawa, T.; Liu, H.; Kamitani, S.; Tadauchi, O.; Cai, W.; Li, H. Phylogeography of the assassin bug Sphedanolestes impressicollis in East Asia inferred from mitochondrial and nuclear gene sequences. Int. J. Mol. Sci. 2019, 20, 1234. [Google Scholar] [CrossRef]
- Kurata, S.; Sakaguchi, S.; Hirota, S.K.; Kurashima, O.; Suyama, Y.; Nishida, S.; Ito, M. Refugia within refugium of Geranium yesoense (Geraniaceae) in Japan were driven by recolonization into the southern interglacial refugium. Biol. J. Linn. Soc. 2021, 132, 552–572. [Google Scholar] [CrossRef]
- Habel, J.C.; Assmann, T. Relict Species: Phylogeography and Conservation Biology; Springer eBooks: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Shi, C.-M.; Ji, Y.-J.; Liu, L.; Wang, L.; Zhang, D.-X. Impact of climate changes from Middle Miocene onwards on evolutionary diversification in Eurasia: Insights from the mesobuthid scorpions. Mol. Ecol. 2013, 22, 1700–1716. [Google Scholar] [CrossRef]
- Mammola, S.; Hormiga, G.; Arnedo, M.A.; Isaia, M. Unexpected diversity in the relictual European spiders of the genus Pimoa (Araneae: Pimoidae). Invertebr. Syst. 2016, 30, 566–587. [Google Scholar] [CrossRef][Green Version]

| Gene | Primer | Sequence (5′–3′) | Annealing Temperature | Voucher |
|---|---|---|---|---|
| COI | C1-J-1718(F) | GGA GGA TTT GGA AAT TGA TTA GTT CC | 45 °C | Simon et al., 1994 |
| C1-N-219(R) | CCC GGT AAA ATT AAA ATA TAA ACT TC | Simon et al., 1994 | ||
| 16S | LR-N-13398 (F) | CGC CTG TTT AAC AAA AAC AT | 42–45 °C | Arnedo et al., 2004 |
| LR-J-12864 (R) | CTC CGG TTT GAA CTC AGA TCA | Simon et al., 1994 | ||
| 18S | 18Sa2.0 (F) | ATG GTT GCA AAG CTG AAA C | 58 °C | Giribet et al., 1999 |
| 5F (F) | GCG AAA GCA TTT GCC AAG AA | Giribet et al., 1999 | ||
| 9R (R) | GAT CCT TCC GCA GGT TCA CCT AC | Giribet et al., 1999 | ||
| 28S | 28Sa (F) | GAC CCG TCT TGA AAC ACG GA | 48 °C | Whiting et al., 1997 |
| 28Sb (R) | TCG GAA GGA ACC AGC TAC TA | Whiting et al., 1997 | ||
| H3 | H3a (F) | ATG GCT CGT ACC AAG CAG AC(ACG) GC | 48 °C | Colgan et al., 1998 |
| H3b (R) | ATA TCC TT(AG) GGC AT(AG) AT(AG) GTG AC | Colgan et al., 1998 |
| Family | Subfamily | Species | 16s | 18s | 28s | COI | H3 | Voucher |
|---|---|---|---|---|---|---|---|---|
| Pimoidae | Pimoa sp. X131 | AY230940 | AY230893 | AY231072 | AY231025 | AY230985 | Arnedo et al., 2004 | |
| Linyphiidae | Micronetinae | Acanoides beijingensis | KJ027589 | KJ027587 | KJ027580 | KJ027582 | KJ027583 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Acanoides hengshanensis | KJ027585 | KJ027588 | KJ027584 | KJ027586 | KJ027581 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Agyneta ramosa | FJ838670 | FJ838694 | FJ838648 | FJ838740 | Arnedo et al., 2009 | |
| Linyphiidae | Micronetinae | Agyneta sp.1 | KT003097 | KT002904 | KT003003 | KT002707 | KT002804 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Agyneta sp.2 | KT003098 | KT002905 | KT003004 | KT002708 | KT002805 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Allomengea sp. | KT003099 | KT002906 | KT002709 | KT002805 | Wang et al., 2015 | |
| Linyphiidae | Micronetinae | Anguliphantes sp. | KT002907 | KT003005 | KT002710 | KT002807 | Wang et al., 2015 | |
| Linyphiidae | Micronetinae | Anguliphantes zygius | KT003100 | KT002908 | KT003006 | KT002711 | KT002808 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Arcuphantes arcuatulus | KT003007 | KT002809 | Wang et al., 2015 | |||
| Linyphiidae | Erigoninae | Asthenargus edentulus | KT003101 | KT002909 | KT003008 | KT002712 | KT002810 | Wang et al., 2015 |
| Linyphiidae | Linyphiinae | Australolinyphia remota | FJ838671 | FJ838695 | FJ838718 | FJ838649 | FJ838741 | Arnedo et al., 2009 |
| Linyphiidae | Linyphiinae | Bathyphantes eumenis | KT003101 | KT002910 | KT003009 | KT002713 | KT002811 | Wang et al., 2015 |
| Linyphiidae | Linyphiinae | Bathyphantes gracilis | FJ838672 | FJ838696 | FJ838650 | FJ838742 | Arnedo et al., 2009 | |
| Linyphiidae | Linyphiinae | Bathyphantes tongluensis | KT003104 | KT002912 | KT003011 | KT002715 | KT002813 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Bifurcia curvata | KT003105 | KT002913 | KT003012 | KT002716 | KT002814 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Bifurcia ramosa | KT003106 | KT002914 | KT003013 | KT002717 | KT002815 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Bolyphantes alticeps | AY078660 | AY078667 | AY078678 | AY078691 | AY078700 | Hormiga et al., 2003 |
| Linyphiidae | Micronetinae | Bolyphantes sp. | KT003107 | KT002915 | KT002718 | KT002816 | Wang et al., 2015 | |
| Linyphiidae | Micronetinae | Centromerus trilobus | KT003108 | KT002916 | KT003014 | KT002718 | KT002817 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Ceratinella sp. | KT003109 | KT002917 | KT003015 | KT002818 | Wang et al., 2015 | |
| Linyphiidae | Erigoninae | Collinsia inerrans | KT003110 | KT002918 | KT003016 | KT002720 | KT002819 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Concavocephalus rubens | KT002919 | KT003017 | KT002721 | KT002820 | Wang et al., 2015 | |
| Linyphiidae | Micronetinae | Crispiphantes sp. | KT003111 | KT002920 | KT003018 | KT002722 | KT002821 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Denisiphantes sp. | KT003112 | KT002921 | KT003019 | KT002723 | KT002822 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Dicymbium libidinosum | KT003113 | KT002922 | KT003020 | KT002724 | KT002823 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Dicymbium tibiale | KT003114 | KT002923 | KT003021 | KT002725 | KT002824 | Wang et al., 2015 |
| Linyphiidae | Linyphiinae | Diplostyla concolor | FJ838673 | FJ838697 | FJ838651 | FJ838743 | Arnedo et al., 2009 | |
| Linyphiidae | Micronetinae | Doenitzius pruvus | KT003116 | KT002925 | KT003023 | KT002727 | KT002826 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Drapetisca socialis | FJ838674 | FJ838698 | FJ838652 | FJ838744 | Arnedo et al., 2009 | |
| Linyphiidae | Dubiaraneinae | Dubiaranea distincta | FJ838675 | FJ838699 | FJ838722 | FJ838653 | FJ838745 | Arnedo et al., 2009 |
| Linyphiidae | Micronetinae | Eldonia sp. | KT003117 | KT002926 | KT003024 | KT002728 | KT002827 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Erigone prominens | KT002927 | KT003025 | KT002729 | KT002828 | Wang et al., 2015 | |
| Linyphiidae | Micronetinae | Floronia bucculenta | FJ838676 | FJ838700 | FJ838654 | FJ838746 | Arnedo et al., 2009 | |
| Linyphiidae | Linyphiinae | Frontinella communis | FJ838677 | FJ838701 | FJ838724 | FJ838655 | FJ838747 | Arnedo et al., 2009 |
| Linyphiidae | Erigoninae | Gnathonarium taczanowskii | KT003119 | KT002929 | KT003027 | KT002730 | KT002830 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Gonatium rubellum | FJ838679 | FJ838703 | FJ838656 | FJ838749 | Arnedo et al., 2009 | |
| Linyphiidae | Erigoninae | Gonatium rubens | KT003120 | KT002930 | KT003028 | KT002732 | KT002831 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Gongylidiellum vivum | FJ838678 | FJ838702 | FJ838725 | FJ838748 | Arnedo et al., 2009 | |
| Linyphiidae | Erigoninae | Gongylidioides rimatus | KT003121 | KT002931 | KT003029 | KT002733 | KT002832 | Wang et al., 2015 |
| Linyphiidae | Mynogleninae | Haplinis diloris | FJ838680 | FJ838704 | FJ838657 | FJ838750 | Arnedo et al., 2009 | |
| Linyphiidae | Micronetinae | Helophora insignis | FJ838681 | FJ838705 | FJ838658 | FJ838751 | Arnedo et al., 2009 | |
| Linyphiidae | Erigoninae | Hilaira herniosa | KT003123 | KT002933 | KT003030 | KT002735 | KT002834 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Hylyphantes graminicola | KT003124 | KT002934 | KT003031 | KT002736 | KT002835 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Hypselistes jacksoni | KT002935 | KT003032 | KT002737 | KT002836 | Wang et al., 2015 | |
| Linyphiidae | Micronetinae | Incestophantes kochiellus | KT003125 | KT002936 | KT003033 | KT002738 | KT002836 | Wang et al., 2015 |
| Linyphiidae | Linyphiinae | Kaestneria pullata | KT003126 | KT002937 | KT003034 | KT002739 | KT002838 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Lasiargus sp. | KT003127 | KT002938 | KT003035 | KT002740 | KT002839 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Lepthyphantes hamifer | KT003128 | KT002939 | KT003036 | KT002741 | KT002840 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Lepthyphantes luteipes | KT003129 | KT002940 | KT002742 | KT002841 | Wang et al., 2015 | |
| Linyphiidae | Micronetinae | Lepthyphantes minutus | AY078663 | AY078673 | AY078681 | AY078689 | AY078705 | Hormiga et al., 2003 |
| Linyphiidae | Micronetinae | Lidia sp. | KT003130 | KT002941 | KT003037 | KT002841 | Wang et al., 2015 | |
| Linyphiidae | Linyphiinae | Linyphia triangularis | AY078664 | AY078668 | AY078682 | AY078693 | AY078702 | Hormiga et al., 2003 |
| Linyphiidae | Linyphiinae | Laetesia sp. | FJ838682 | FJ838706 | FJ838659 | FJ838752 | Arnedo et al., 2009 | |
| Linyphiidae | Micronetinae | Macrargus rufus | KT003133 | KT002944 | KT003040 | KT002745 | KT002845 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Maso sundevalli | KT002945 | KT003041 | KT002746 | KT002846 | Wang et al., 2015 | |
| Linyphiidae | Micronetinae | Meioneta rurestris | FJ838683 | FJ838707 | FJ838660 | FJ838753 | Arnedo et al., 2009 | |
| Linyphiidae | Micronetinae | Mesasigone mira | KT003134 | KT002946 | KT002746 | KT002847 | Wang et al., 2015 | |
| Linyphiidae | Erigoninae | Micrargus herbigradus | KT003135 | KT002947 | KT003042 | KT002748 | KT002848 | Wang et al., 2015 |
| Linyphiidae | Linyphiinae | Microlinyphia dana | AY078665 | AY078677 | AY078683 | AY078690 | Hormiga et al., 2003 | |
| Linyphiidae | Linyphiinae | Microlinyphia sp. | KT003136 | KT002948 | KT003043 | KT002749 | KT002849 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Microneta viaria | FJ838684 | FJ838708 | FJ838661 | FJ838754 | Arnedo et al., 2009 | |
| Linyphiidae | Micronetinae | Micronetine sp.1 | KT003138 | KT002950 | KT002751 | KT002851 | Wang et al., 2015 | |
| Linyphiidae | Micronetinae | Micronetine sp.2 | KT003139 | KT002951 | KT003045 | KT002752 | KT002851 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Micronetine sp.3 | KT003140 | KT002952 | KT003046 | KT002753 | KT002853 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Mughiphantes nigromaculatus | KT003187 | KT003001 | KT003095 | KT002802 | KT002902 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Mughiphantes sp.1A | KT003141 | KT002953 | KT003047 | KT002754 | KT002854 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Mughiphantes sp.1B | KT003142 | KT002954 | KT003048 | KT002755 | KT002855 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Mughiphantes sp.2 | KT003143 | KT002955 | KT002756 | KT002855 | Wang et al., 2015 | |
| Linyphiidae | Micronetinae | Mughiphantes sp.3 | KT003144 | KT002956 | KT003049 | KT002757 | KT002857 | Wang et al., 2015 |
| Linyphiidae | Linyphiinae | Neriene emphana | KT003145 | KT002957 | KT003050 | KT002758 | KT002858 | Wang et al., 2015 |
| Linyphiidae | Linyphiinae | Neriene radiata | AY078710 | AY078670 | AY078684 | AY078696 | AY078709 | Hormiga et al., 2003 |
| Linyphiidae | Linyphiinae | Neriene sp.2 | KT003146 | KT002958 | KT003051 | KT002759 | KT002859 | Wang et al., 2015 |
| Linyphiidae | Linyphiinae | Neriene sp.3 | KT003147 | KT002959 | KT003052 | KT002760 | KT002859 | Wang et al., 2015 |
| Linyphiidae | Linyphiinae | Neriene sp.4 | KT003148 | KT002960 | KT003053 | KT002761 | KT002861 | Wang et al., 2015 |
| Linyphiidae | Linyphiinae | Neriene sp.5 | KT003149 | KT002961 | KT003054 | KT002762 | KT002862 | Wang et al., 2015 |
| Linyphiidae | Linyphiinae | Neriene sp.6 | KT003150 | KT002962 | KT003055 | KT002763 | KT002863 | Wang et al., 2015 |
| Linyphiidae | Linyphiinae | Neriene variabilis | AY078711 | AY078669 | AY078685 | AY078699 | AY078706 | Hormiga et al., 2003 |
| Linyphiidae | Micronetinae | Nippononeta coreana | KT003151 | KT002963 | KT003056 | KT002764 | KT002864 | Wang et al., 2015 |
| Linyphiidae | Mynogleninae | Novafroneta vulgaris | FJ838710 | FJ838663 | FJ838756 | Arnedo et al., 2009 | ||
| Linyphiidae | Erigoninae | Oedothorax apicatus | KT003152 | KT002964 | KT003057 | KT002765 | KT002864 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Oia imadatei | KT003153 | KT002965 | KT003058 | KT002765 | KT002866 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Oreoneta tienshangensis | KT003154 | KT002966 | KT003059 | KT002765 | KT002867 | Wang et al., 2015 |
| Linyphiidae | Linyphiinae | Orsonwelles malus | AY078737 | AY078676 | AY078687 | AY078697 | AY078708 | Hormiga et al., 2003 |
| Linyphiidae | Linyphiinae | Orsonwelles polites | AY078725 | AY078671 | AY078686 | AY078755 | AY078701 | Hormiga et al., 2003 |
| Linyphiidae | Micronetinae | Oryphantes sp.1 | KT003155 | KT002967 | KT003060 | KT002768 | KT002868 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Oryphantes sp.2A | KT003156 | KT002968 | KT003061 | KT002769 | KT002869 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Oryphantes sp.2B | KT003157 | KT002969 | KT003062 | KT002801 | KT002870 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Ostearius melanopygius | FJ838688 | FJ838712 | FJ838758 | Arnedo et al., 2009 | ||
| Linyphiidae | Linyphiinae | Pacifiphantes zakharovi | KT003159 | KT002971 | KT003064 | KT002771 | KT002872 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Parawubanoides unicornis | KT003160 | KT002972 | KT003065 | KT002772 | KT002873 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Poeciloneta variegata | KT003161 | KT002973 | KT003066 | KT002772 | KT002874 | Wang et al., 2015 |
| Linyphiidae | Linyphiinae | Porrhomma longjiangense | KT003162 | KT002974 | KT003067 | KT002774 | KT002875 | Wang et al., 2015 |
| Linyphiidae | Mynogleninae | Pseudafroneta incerta | FJ838690 | FJ838714 | FJ838737 | FJ838666 | FJ838760 | Arnedo et al., 2009 |
| Linyphiidae | Stemonyphantinae | Putaoa huaping | KT003163 | KT002975 | KT003068 | KT002775 | KT002876 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Ryojius sp. | KT003069 | KT002776 | KT002877 | Wang et al., 2015 | ||
| Linyphiidae | Micronetinae | Saaristoa ebinoensis | KT003164 | KT002976 | KT003070 | KT002777 | KT002878 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Savignia sp.1 | KT003165 | KT002977 | KT003071 | KT002778 | KT002879 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Savignia sp.2 | KT003166 | KT002978 | KT003072 | KT002779 | KT002880 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Scotinotylus sp.A | KT003167 | KT002979 | KT003073 | KT002780 | KT002881 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Scotinotylus tianschanicus | KT003188 | KT003002 | KT003096 | KT002803 | KT002903 | Wang et al., 2015 |
| Linyphiidae | Ipainae | Solenysa lanyuensis * | OL691622 | OL691625 | OL691629 | OL693167 | OL702838 | CNU |
| Linyphiidae | Ipainae | Solenysa longqiensis | KT003169 | KT002981 | KT003075 | KT002782 | KT002883 | Wang et al., 2015 |
| Linyphiidae | Ipainae | Solenysa macrodonta * | OL691623 | OL691627 | OL691631 | OL693169 | OL702840 | CNU |
| Linyphiidae | Ipainae | Solenysa mellotteei | KT003168 | KT002980 | KT003074 | KT002780 | KT002882 | Wang et al., 2015 |
| Linyphiidae | Ipainae | Solenysa ogatai * | OL691626 | OL691630 | OL693168 | OL702839 | CNU | |
| Linyphiidae | Ipainae | Solenysa partibilis | KT003170 | KT002983 | KT003077 | KT002784 | KT002885 | Wang et al., 2015 |
| Linyphiidae | Ipainae | Solenysa protrudens | KT003171 | KT002984 | KT003078 | KT002785 | KT002886 | Wang et al., 2015 |
| Linyphiidae | Ipainae | Solenysa reflexilis | KT003172 | KT002985 | KT003079 | KT002786 | KT002887 | Wang et al., 2015 |
| Linyphiidae | Ipainae | Solenysa retractilis | KT003174 | KT002987 | KT003081 | KT002788 | KT002889 | Wang et al., 2015 |
| Linyphiidae | Ipainae | Solenysa tianmushana | KT003175 | KT002988 | KT003082 | KT002788 | KT002890 | Wang et al., 2015 |
| Linyphiidae | Ipainae | Solenysa trunciformis | KT002982 | KT003076 | KT002783 | KT002884 | Wang et al., 2015 | |
| Linyphiidae | Ipainae | Solenysa wulingensis | KT003176 | KT002989 | KT003083 | KT002790 | Wang et al., 2015 | |
| Linyphiidae | Ipainae | Solenysa yangmingshana * | OL691624 | OL691628 | OL693166 | OL702837 | CNU | |
| Linyphiidae | Stemonyphantinae | Stemonyphantes abantensis | KT003177 | KT002990 | KT003084 | KT002791 | KT002891 | Wang et al., 2015 |
| Linyphiidae | Stemonyphantinae | Stemonyphantes lineatus | FJ838691 | FJ838715 | FJ838667 | FJ838761 | Arnedo et al., 2009 | |
| Linyphiidae | Stemonyphantinae | Stemonyphantes sibiricus | FJ838692 | FJ838668 | FJ838762 | Arnedo et al., 2009 | ||
| Linyphiidae | Erigoninae | Styloctetor compar | KT003178 | KT002991 | KT003085 | KT002792 | KT002892 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Tapinopa guttata | KT003179 | KT002992 | KT003086 | KT002893 | Wang et al., 2015 | |
| Linyphiidae | Micronetinae | Tenuiphantes sp.1 | KT002993 | KT003087 | KT002793 | KT002894 | Wang et al., 2015 | |
| Linyphiidae | Micronetinae | Tenuiphantes sp.2 | KT003180 | KT002994 | KT003087 | KT002794 | KT002895 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Tenuiphantes tenuis | FJ838693 | FJ838716 | FJ838669 | FJ838763 | Arnedo et al., 2009 | |
| Linyphiidae | Erigoninae | Ternatus malleatus | KT003181 | KT002995 | KT003089 | KT002795 | KT002896 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Ternatus siculus | KT003182 | KT002996 | KT003090 | KT002795 | KT002897 | Wang et al., 2015 |
| Linyphiidae | Erigoninae | Ummeliata insecticeps | KT003184 | KT002998 | KT003092 | KT002798 | KT002899 | Wang et al., 2015 |
| Linyphiidae | Micronetinae | Vagiphantes vaginatus | KT003185 | KT002999 | KT003093 | KT002799 | KT002900 | Wang et al., 2015 |
| Linyphiidae | Ipainae | Wubanoides sp. | KT003186 | KT003000 | KT003094 | KT002800 | KT002901 | Wang et al., 2015 |
| Node | Mean | 95% HPD Lower | 95% HPD Upper |
|---|---|---|---|
| 1 | 128.80 | 125.14 | 136.35 |
| 2 | 116.55 | 104.48 | 128.18 |
| 3 | 101.67 | 88.88 | 113.61 |
| 4 | 90.85 | 72.69 | 108.83 |
| 5 | 89.22 | 77.24 | 100.88 |
| 6 | 79.29 | 67.69 | 90.67 |
| 7 | 28.62 | 20.09 | 37.98 |
| 8 | 22.06 | 13.71 | 31.27 |
| 9 | 21.99 | 15.74 | 28.85 |
| 10 | 18.49 | 12.75 | 24.66 |
| 11 | 12.34 | 6.29 | 18.98 |
| 12 | 3.46 | 1.09 | 6.50 |
| 13 | 6.35 | 3.10 | 10.10 |
| 14 | 6.59 | 4.03 | 9.39 |
| 15 | 3.46 | 1.80 | 5.27 |
| 16 | 2.52 | 1.12 | 4.07 |
| 17 | 2.56 | 0.99 | 4.33 |
| 18 | 0.83 | 0.02 | 1.94 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, J.; Zhan, Y.; Shi, C.; Ono, H.; Tu, L. Solenysa, a Cretaceous Relict Spider Group in East Asia. Diversity 2022, 14, 120. https://doi.org/10.3390/d14020120
Tian J, Zhan Y, Shi C, Ono H, Tu L. Solenysa, a Cretaceous Relict Spider Group in East Asia. Diversity. 2022; 14(2):120. https://doi.org/10.3390/d14020120
Chicago/Turabian StyleTian, Jiahui, Yongjia Zhan, Chengmin Shi, Hirotsugu Ono, and Lihong Tu. 2022. "Solenysa, a Cretaceous Relict Spider Group in East Asia" Diversity 14, no. 2: 120. https://doi.org/10.3390/d14020120
APA StyleTian, J., Zhan, Y., Shi, C., Ono, H., & Tu, L. (2022). Solenysa, a Cretaceous Relict Spider Group in East Asia. Diversity, 14(2), 120. https://doi.org/10.3390/d14020120







