Morphological and Molecular Evidence for Two New Species within Russula Subgenus Brevipes from China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Morphological Studies
2.2. Molecular Study and Phylogenetic Analysis
3. Results
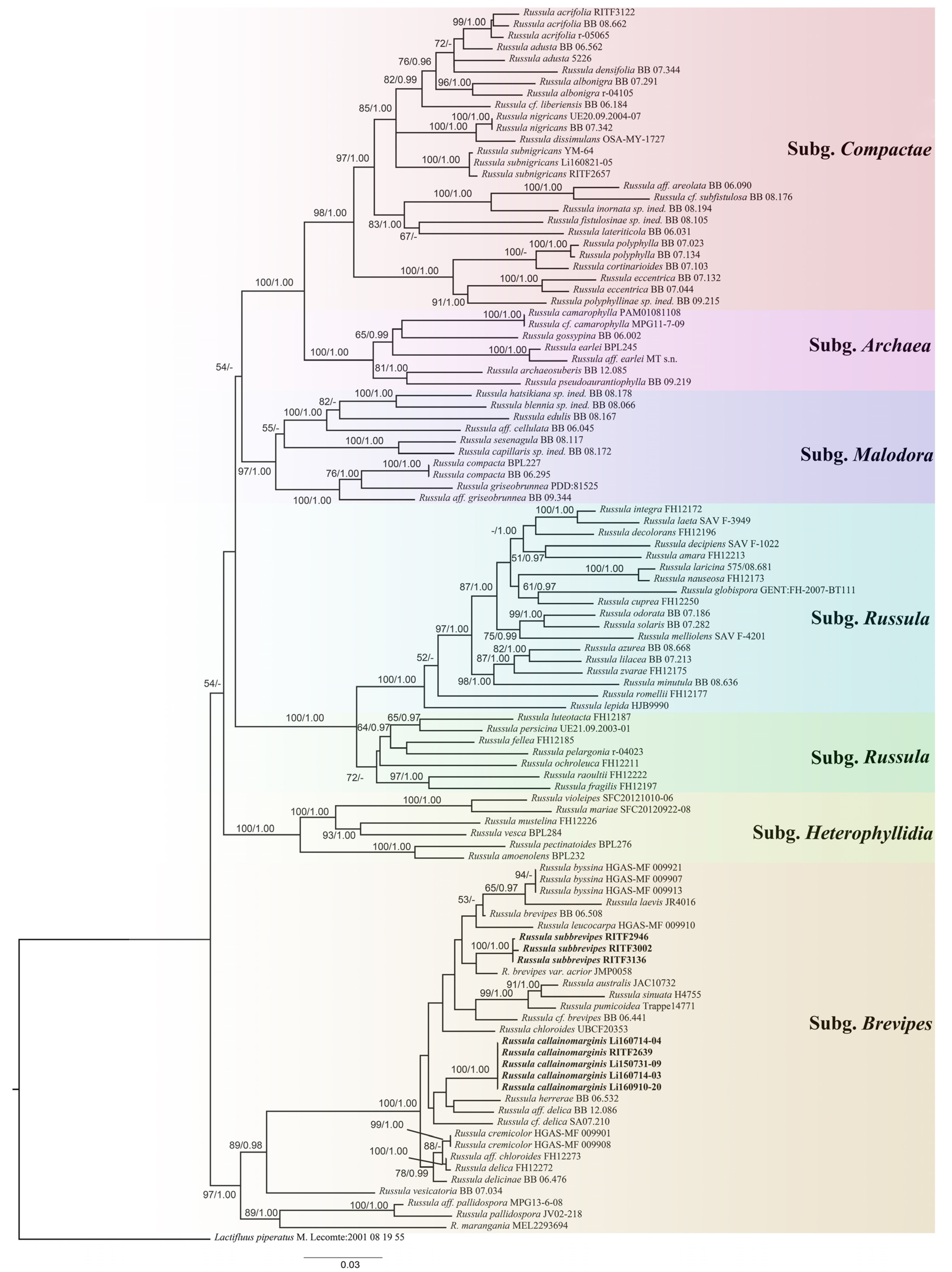
3.1. Molecular Phylogeny
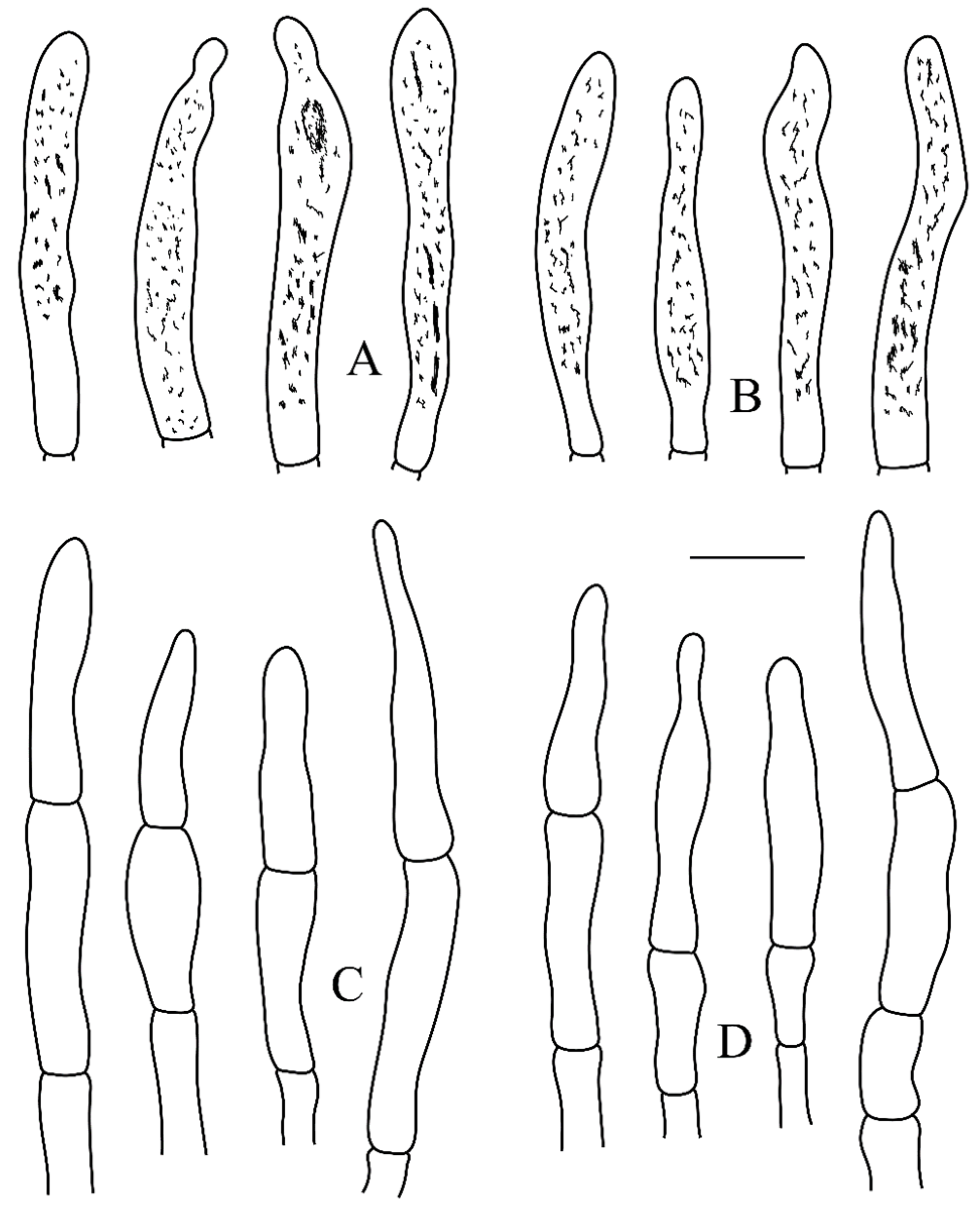
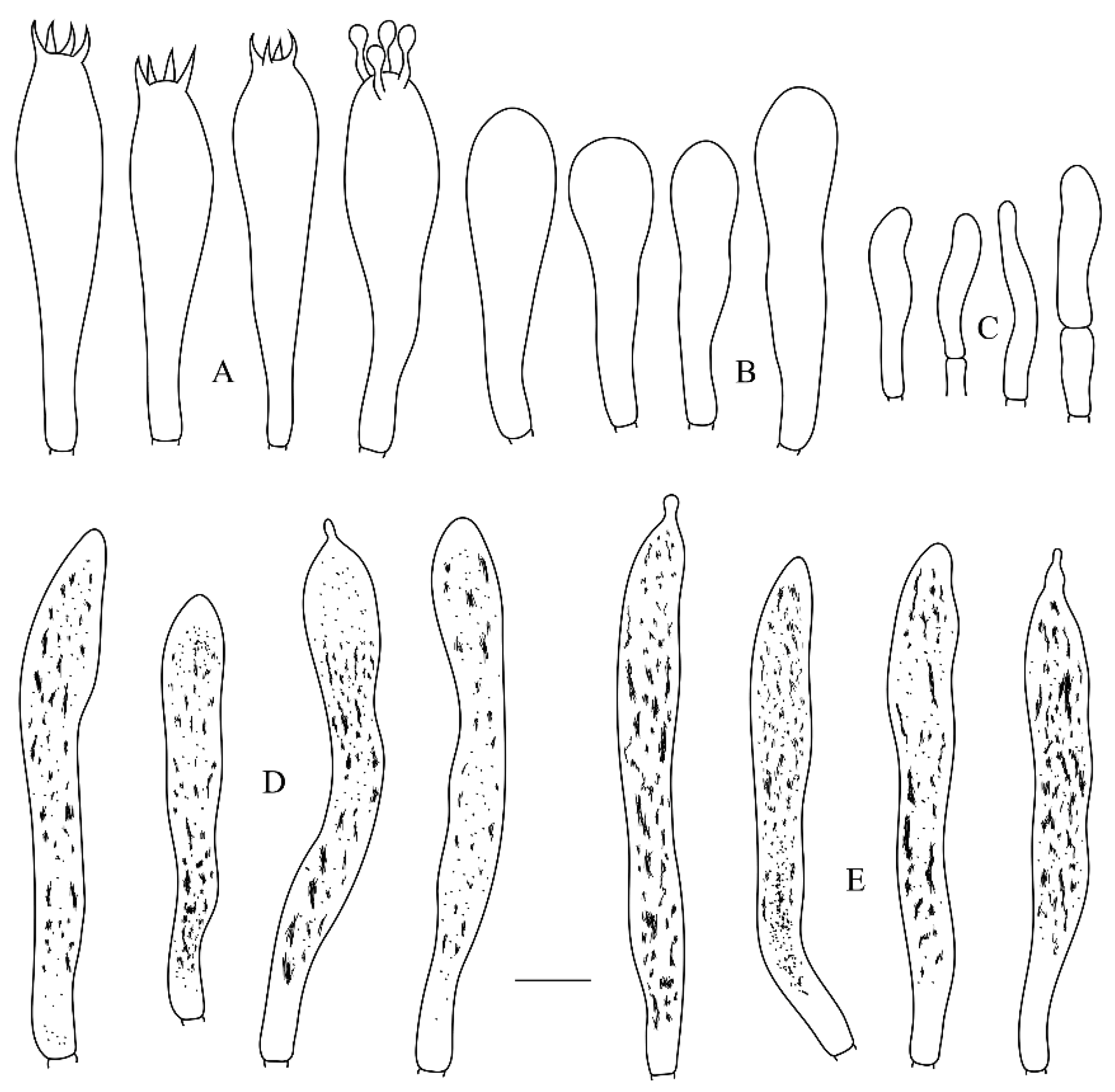
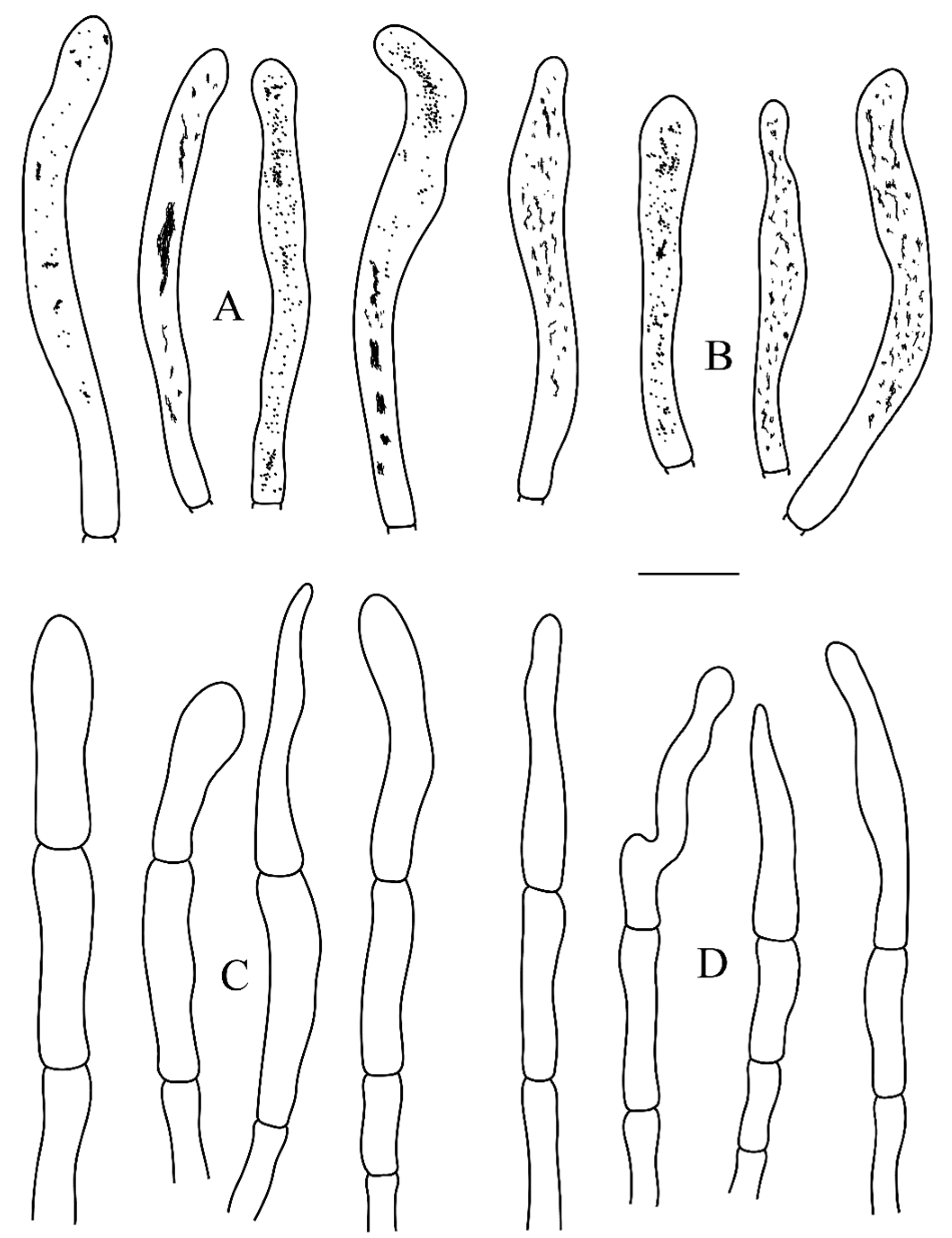
3.2. Taxonomy
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Persoon, C.H. Observations Mycologicae, Seu, Descriptiones Tam Novorum Quan Notabilium Fungorum; Apud Petrum P. Wolf: Lipsiae, Germany, 1796. [Google Scholar]
- Kirk, P.M.; Cannon, P.F.; David, J.C.; Minter, D.W.; Stalpers, J.A. Ainsworth and Bisby’s Dictionary of the Fungi, 10th ed.; CAB International Press: Wallingford, UK, 2008. [Google Scholar]
- Adamčík, S.; Looney, B.; Caboň, M.; Jančovičová, S.; Adamčíková, K.; Avis, P.G.; Barajas, M.; Bhatt, R.P.; Corrales, A.; Das, K.; et al. The quest for a globally comprehensible Russula language. Fungal Divers. 2019, 99, 369–449. [Google Scholar] [CrossRef]
- Buyck, B.; Hofstetter, V.; Eberhardt, U.; Verbeken, A.; Kauff, F. Walking the thin line between Russula and Lactarius: The dilemma of Russula subsect. Ochricompactae. Fungal Divers. 2008, 8, 15–40. [Google Scholar]
- Looney, B.P.; Meidl, P.; Piatek, M.J.; Miettinen, O.; Martin, F.M.; Matheny, P.B.; Labbé, J.L. Russulaceae: A new genomic dataset to study ecosystem function and evolutionary diversification of ectomycorrhizal fungi with their tree associates. New Phytol. 2018, 218, 54–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romagnesi, H. Statuts et noms nouveaux pour les taxa infragénériques dans le genre Russula. Doc. Mycol. 1987, 18, 39–40. [Google Scholar]
- Singer, R. The Agaricales in Modern Taxonmy, 4th ed.; Koeltz Scientific Books: Königstein im Taunus, Germany, 1986. [Google Scholar]
- Bon, M. Clé onographique des russules d’Europe. Doc. Mycol. 1988, 18, 1–120. [Google Scholar]
- Sarnari, M. Monografia Illustrate de Genere Russula in Europa. Tomo Primo; AMB, Centro Studi Micologici: Trento, Italy, 1998. [Google Scholar]
- Buyck, B.; Zoller, S.; Hofstetter, V. Walking the thin lin ten years later: The dilemma of above- versus below-ground features to support phylogenies in the Russulaceae (Basidiomycota). Fungal Divers. 2018, 89, 267–292. [Google Scholar] [CrossRef]
- Buyck, B.; Wang, X.H.; Adamčíková, K.; Caboň, M.; Jančovičová, S.; Hofstetter, V.; Adamčík, S. One step closer to unravelling the origin of Russula: Subgenus Glutinosae subg. nov. Mycosphere 2020, 11, 285–304. [Google Scholar] [CrossRef]
- Hongsanan, S.; Hyde, K.D.; Bahkali, A.H.; Camporesi, E.; Chomnunti, P.; Ekanayaka, H.; Gomes, A.A.M.; Hofstetter, V.; Jones, E.B.G.; Pinho, D.B.; et al. Fungal Biodiversity Profiles 11-20. Cryptog. Mycol. 2015, 36, 355–380. [Google Scholar] [CrossRef]
- Eberhardt, U. Molecular kinship analyses of the agaricoid Russulaceae: Correspondence with mycorrhizal anatomy and sporocarp features in the genus Russula. Mycol. Prog. 2002, 1, 201–223. [Google Scholar] [CrossRef]
- Miller, S.L.; Buyck, B. Molecular phylogeny of the genus Russula in Europe with a comparison of modern infrageneric classifications. Mycol. Res. 2002, 106, 259–276. [Google Scholar] [CrossRef]
- Li, G.J.; Deng, C.Y.; Shi, L.Y.; Wang, J.; Meng, Q.F.; Li, S.M. Three new species of Russula subsect. Lactarioideae from China. Mycosystema 2020, 39, 618–636. [Google Scholar]
- Jiang, X.M.; Li, Y.K.; Liang, J.F.; Wu, J.R. Russula brunneovinacea sp. nov., from north-eastern China. Mycotaxon 2018, 132, 789–797. [Google Scholar] [CrossRef]
- Chen, B.; Song, J.; Chen, Y.l.; Zhang, J.H.; Liang, J.F. Morphological and phylogenetic evidence for two new species of Russula subg. Heterophyllidia from Guangdong Province of China. MycoKeys 2021, 82, 139–157. [Google Scholar]
- Li, G.J.; Li, S.M.; Buyck, B.; Zhao, S.Y.; Xie, X.J.; Shi, L.Y.; Deng, C.Y.; Meng, Q.F.; Sun, Q.B.; Yan, J.Q.; et al. Three new Russula species in sect. Ingratae (Russulales, Basidiomycota) from southern China. MycoKeys 2021, 84, 103–139. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Xie, X.C.; Buyck, B. Two novel species of subgenus Russula crown clade (Russulales, Basidiomycota) from China. Eur. J. Taxon. 2021, 775, 15–33. [Google Scholar] [CrossRef]
- Vellinga, E.C.; Noordeloos, M.E. Glossary. In Flora Agaricina Neerlandica 5; Noordeloos, M.E., Kuyper, T.W., Vellinga, E.C., Eds.; A.A. Balkema Publishers: Rotterdam, The Netherlands, 1988; pp. 6–11. [Google Scholar]
- Kornerup, A.; Wanscher, J.H. Taschenlexikon der Farben, 3rd ed.; Muster-Schmidt Verlag: Göttingen, Germany, 1981. [Google Scholar]
- Buyck, B. Valeur taxonomique du bleu de crésyl pour le genra Russula. Bull. De La Société Mycol. Fr. 1989, 120, 1–6. [Google Scholar]
- Zhou, L.L.; Liang, J.F. An improved protocol for extraction of DNA from macrofungi. Guangdong Fore. Sci. Tech. 2011, 27, 13–16. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. PCR Protocols: A Guide to Methods and Applications; Academic Press: New York, NY, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.J.; Hall, B.D. Body plan evolution of ascomycetes, as inferred from an RNA polymerase II, phylogeny. Proc. Natl. Acad. Sci. USA 2004, 101, 4507–4512. [Google Scholar] [CrossRef] [Green Version]
- Matheny, P.B. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol. Phyl. Evol. 2005, 35, 1–20. [Google Scholar] [CrossRef]
- Liang, J.F.; Xu, J.; Yang, Z.L. Divergence, dispersal and recombination in Lepiota cristata from China. Fungal Divers. 2009, 38, 105–124. [Google Scholar]
- Nilsson, R.H.; Tedersoo, L.; Abarenkov, K.; Ryberg, M.; Kristiansson, E.; Hartmann, M.; Schoch, C.L.; Nylander, J.A.A.; Bergsten, J.; Porter, T.M.; et al. Five simple guidelines for establishing basic authenticity and reliability of newly generated fungal ITS sequences. Mycokeys 2012, 4, 37–63. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0b10; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Posada, D.; Crandall, K.A. Model test: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nylander, J.A.A. MrModeltest 2.3. Computer Program and Documentation Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Knudsen, H.; Borgen, T. Russulaceae in Greenland. Arctic and Alpine Mycology 1; University of Washington Press: Seattle, WA, USA, 1982. [Google Scholar]
- Li, G.J.; Li, S.F.; Zhao, D.; Wen, H.A. Recent research progress of Russula (Russulales, Agaricomycetes): A review. Mycosystema 2015, 34, 821–848. [Google Scholar]
- Roberts, C. Russulas of Southern Vancouver Island Coastal Forests. Doctoral Dissertation, University of Victoria, Victoria, BC, Canada, 2007. [Google Scholar]
- Shaffer, R.L. Notes on the subsection Crassotunicatinae and other species of Russula. Lloydia 1970, 33, 49–96. [Google Scholar]
- Buyck, B.; Adamčík, S. Type studies in Russula subsection Lactarioideae from North America and a tentative key to North American species. Cryptogam. Mycol. 2013, 34, 259–279. [Google Scholar] [CrossRef]



| Species | Collection No. | Location | GenBank Accession No. | |||
|---|---|---|---|---|---|---|
| ITS | LSU | RPB2 | mtSSU | |||
| Lactifluus piperatus | M. Lecomte:2001 08 19 55 | France | KF220121 | KF220214 | KF220287 | NC_038056 |
| Russula acrifolia | r-05065 | USA | JF834363 | JF834510 | JF834460 | - |
| R.acrifolia | BB 08.662 | Italy | - | KU237535 | KU237821 | KU237381 |
| R.acrifolia | RITF3122 | China | MH911600 a | MH911611 a | MH911626 a | - |
| R.adusta | 5226 | Italy | JF908669 | - | - | - |
| R.adusta | BB 06.562 | Canada | - | KU237476 | KU237762 | KU237320 |
| R.albonigra | r-04105 | USA | JF834355 | JF834503 | JF834452 | - |
| R.albonigra | BB 07.291 | Slovakia | KU237536 | KU237822 | KU237382 | |
| R.amara | FH12213 | Germany | KT933998 | KT933859 | KT933930 | - |
| R.amoenolens | BPL232 | USA | KT933954 | KT933813 | KT933884 | - |
| R.archaeosuberis | BB 12.085 | Italy | KY800355 | KU237593 | KU237878 | KU237441 |
| R. aff. areolata | BB 06.090 | Madagascar | - | KU237471 | KU237757 | KU237315 |
| R. australis | JAC10732 | New Zealand | MW683746 | MW683616 | ||
| R.azurea | BB 08.668 | Italy | JN944002 | KU237529 | KU237815 | KU237375 |
| R.blennia sp. ined. | BB 08.066 | Madagascar | - | KU237556 | KU237842 | KU237404 |
| R.brevipes | BB 06.508 | Mexico | - | KU237479 | KU237765 | KU237323 |
| R. cf. brevipes | BB 06.441 | Mexico | - | KU237483 | KU237769 | KU237327 |
| R. brevipes var. acrior | JMP0058 | USA | EU819422 | |||
| R. callainomarginis | RITF2639 | China | MH286463 a | MH286468 a | MH911624 a | MH911616 a |
| R. callainomarginis | Li160714-03 | China | MH911604 a | - | - | - |
| R. callainomarginis | Li150731-09 | China | MH911605 a | - | - | - |
| R. callainomarginis | Li160910-20 | China | MH911606 a | - | - | - |
| R. callainomarginis | Li160714-04 | China | MH911607 a | - | - | - |
| R.camarophylla | PAM01081108 | China | DQ421982 | DQ421982 | DQ421938 | - |
| R. cf. camarophylla | MPG11-7-09 | Spain | - | KU237579 | KU237865 | KU237427 |
| R.capillaris sp. ined. | BB 08.172 | Madagascar | - | KU237553 | KU237839 | KU237399 |
| R. aff. cellulata | BB 06.045 | Madagascar | - | KU237454 | KU237740 | KU237298 |
| R.chloroides | UBCF20353 | Canada | KC581331 | KC581331 | - | - |
| R. aff. chloroides | FH12273 | Belgium | KT934015 | KT933876 | KT933947 | |
| R.compacta | BPL227 | USA | KT933952 | KT933810 | KT933881 | - |
| R.compacta | BB 06.295 | USA | - | KU237480 | KU237766 | KU237324 |
| R.cortinarioides | BB 07.103 | USA | KP033480 | KP033491 | KP033502 | KU237402 |
| R.cuprea | FH12250 | Slovakia | KT934010 | KT933871 | KT933942 | - |
| R.decipiens | SAV F-1022 | Slovakia | KY582683 | - | KY616679 | KY471572 |
| R.decolorans | FH12196 | Slovakia | KT933992 | KT933853 | KT933924 | - |
| R.delica | FH12272 | Belgium | KF432955 | KR364224 | KR364340 | - |
| R. aff. delica | BB 12.086 | Italy | - | KU237594 | KU237879 | KU237442 |
| R. cf. delica | SA07.210 | Slovakia | - | KU237600 | KU237885 | KU237449 |
| R.delicinae | BB 06.476 | Mexico | - | KU237484 | KU237770 | KU237328 |
| R.densifolia | BB 07.344 | Slovakia | - | KU237502 | KU237788 | KU237347 |
| R.dissimulans | OSA-MY-1727 | Japan | AB291731 | AB154717 | - | - |
| R.earlei | BPL245 | USA | KT933961 | KT933820 | KT933891 | - |
| R. aff. earlei | MT s.n. | Costa Rica | - | KU237598 | KU237883 | KU237446 |
| R.eccentrica | BB 07.044 | USA | KP033479 | KP033490 | KP033501 | KU237353 |
| R. cf. eccentrica | BB 07.132 | USA | KP033478 | KP033489 | KP033500 | KU237341 |
| R.edulis | BB 08.167 | Madagascar | - | KU237564 | KU237850 | KU237412 |
| R.fellea | FH12185 | Slovakia | KT933989 | KT933850 | KT933921 | - |
| R.fistulosinae sp. ined. | BB 08.105 | Madagascar | - | KU237527 | KU237813 | KU237373 |
| R.fragilis | FH12197 | France | KT933993 | KT933854 | KT933925 | - |
| R.globispora | GENT:FH-2007-BT111 | Germany | KU928144 | - | KY616671 | KY471564 |
| R.gossypina | BB 06.002 | Madagascar | - | KU237450 | KU237736 | KU237293 |
| R.griseobrunnea | PDD:81525 | New Zealand | GU222265 | - | - | - |
| R. aff. griseobrunnea | BB 09.344 | New Caledonia | - | KU237592 | KU237877 | KU237440 |
| R.hatsikiana sp. ined. | BB 08.178 | Madagascar | - | KU237557 | KU237843 | KU237405 |
| R.herrerae | BB 06.532 | Mexico | - | KU237486 | KU237772 | KU237330 |
| R.inornata sp. ined. | BB 08.194 | Madagascar | - | KU237558 | KU237844 | KU237406 |
| R.integra | FH12172 | Slovakia | KT933984 | KT933845 | KT933916 | - |
| R.laeta | SAV F-3949 | Slovakia | KY582708 | - | KY616709 | KY471600 |
| R. laevis | JR4016 | Finland | MN130091 | MN130128 | MN380529 | MN161180 |
| R.laricina | 575/08.681 | Italy | JN944008 | JN940593 | KU237846 | - |
| R.lateriticola | BB 06.031 | Madagascar | KP033476 | KP033487 | KP033498 | KU237297 |
| R.lepida | HJB9990 | Belgium | DQ422013 | DQ422013 | DQ421954 | KY471624 |
| R. cf. liberiensis | BB 06.184 | Madagascar | - | KU237474 | KU237760 | KU237318 |
| R.lilacea | BB 07.213 | Slovakia | JN944005 | KU237498 | KU237784 | KU237343 |
| R.luteotacta | FH12187 | Slovakia | KT933991 | KT933852 | KT933923 | - |
| R. marangania | MEL2293694 | Australia | EU019930 | EU019930 | ||
| R.mariae | SFC20120922-08 | South Korea | KF361778 | KF361828 | KF361728 | - |
| R.melliolens | SAV F-4201 | Slovakia | KY582719 | - | KY616712 | KY471611 |
| R.minutula | BB 08.636 | Italy | - | KU237531 | KU237817 | KU237377 |
| R.mustelina | FH12226 | Germany | KT934005 | KT933866 | KT933937 | - |
| R.nauseosa | FH12173 | Germany | KT933985 | KT933846 | KT933917 | - |
| R.nigricans | UE20.09.2004-07 | Sweden | DQ422010 | DQ422010 | - | - |
| R.nigricans | BB 07.342 | Slovakia | - | KU237495 | KU237781 | KU237339 |
| R.ochroleuca | FH12211 | Germany | KT933996 | KT933857 | KT933928 | - |
| R.odorata | BB 07.186 | Slovakia | JN944010 | KU237518 | KU237804 | KU237364 |
| R.pallidospora | JV02-218 | Sweden | DQ422032 | DQ422032 | - | - |
| R. aff. pallidospora | MPG13-6-08 | Spain | - | KU237580 | KU237866 | KU237428 |
| R.pectinatoides | BPL276 | USA | KT933975 | KT933836 | KT933907 | - |
| R.pelargonia | r-04023 | USA | JF834348 | JF834496 | JF834445 | - |
| R.persicina | UE21.09.2003-01 | Sweden | DQ422019 | DQ422019 | DQ421960 | - |
| R.polyphylla | BB 07.134 | USA | KP033486 | KP033497 | KP033508 | KU237448 |
| R.polyphylla | BB 07.023 | USA | KP033481 | KP033492 | KP033503 | KU237403 |
| R.polyphyllinae sp. ined. | BB 09.215 | New Caledonia | - | KU237590 | KU237875 | KU237438 |
| R.pseudoaurantiophylla | BB 09.219 | New Caledonia | - | KU237591 | KU237876 | KU237439 |
| R. pumicoidea | Trappe14771 | Australia | EU019931 | EU019931 | ||
| R.raoultii | FH12222 | Germany | KT934002 | KT933863 | KT933934 | - |
| R.romellii | FH12177 | Germany | KT933987 | KT933848 | KT933919 | - |
| R.sesenagula | BB 08.117 | Madagascar | - | KU237526 | KU237812 | KU237372 |
| R. sinuata | H4755 | Australia | EU019943 | |||
| R.solaris | BB 07.282 | Slovakia | JN944007 | JN940606 | KU237835 | KU237395 |
| R.subbrevipes | RITF3136 | China | MH286460 a | MH286465 a | MH911625 a | MH911617 a |
| R.subbrevipes | RITF2946 | China | MH286462 a | MH286467 a | - | MH911618 a |
| R.subbrevipes | RITF3002 | China | MH286461 a | MH286466 a | - | MH911619 a |
| R. cf. subfistulosa | BB 08.176 | Madagascar | - | KU237542 | KU237828 | KU237388 |
| R.subnigricans | RITF2657 | China | MH911602 a | MH911612 a | - | MH911620 a |
| R.subnigricans | Li160821-05 | China | MH911603 a | - | - | - |
| R.subnigricans | YM-64 | China | MH911601 a | - | - | - |
| R.vesca | BPL284 | USA | KT933978 | KT933839 | KT933910 | - |
| R.vesicatoria | BB 07.034 | USA | - | KU237599 | KU237884 | - |
| R.violeipes | SFC20121010-06 | South Korea | KF361808 | KF361858 | KF361758 | - |
| R.zvarae | FH12175 | Germany | KT933986 | KT933847 | KT933918 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.; Li, H.; Wu, S.; Chen, Q.; Yang, G.; Zhang, J.; Liang, J.; Chen, B. Morphological and Molecular Evidence for Two New Species within Russula Subgenus Brevipes from China. Diversity 2022, 14, 112. https://doi.org/10.3390/d14020112
Song J, Li H, Wu S, Chen Q, Yang G, Zhang J, Liang J, Chen B. Morphological and Molecular Evidence for Two New Species within Russula Subgenus Brevipes from China. Diversity. 2022; 14(2):112. https://doi.org/10.3390/d14020112
Chicago/Turabian StyleSong, Jie, Haijiao Li, Shijun Wu, Qianqian Chen, Guang Yang, Jinyun Zhang, Junfeng Liang, and Bin Chen. 2022. "Morphological and Molecular Evidence for Two New Species within Russula Subgenus Brevipes from China" Diversity 14, no. 2: 112. https://doi.org/10.3390/d14020112
APA StyleSong, J., Li, H., Wu, S., Chen, Q., Yang, G., Zhang, J., Liang, J., & Chen, B. (2022). Morphological and Molecular Evidence for Two New Species within Russula Subgenus Brevipes from China. Diversity, 14(2), 112. https://doi.org/10.3390/d14020112






