Morphological and Molecular Characterizations of Three Species of the Genus Synura (Synurales, Chrysophyceae) from China
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Culture
2.2. DNA Extraction, Amplification, and Sequencing
2.3. Phylogenetic Analysis
2.4. Morphological Observations
3. Results
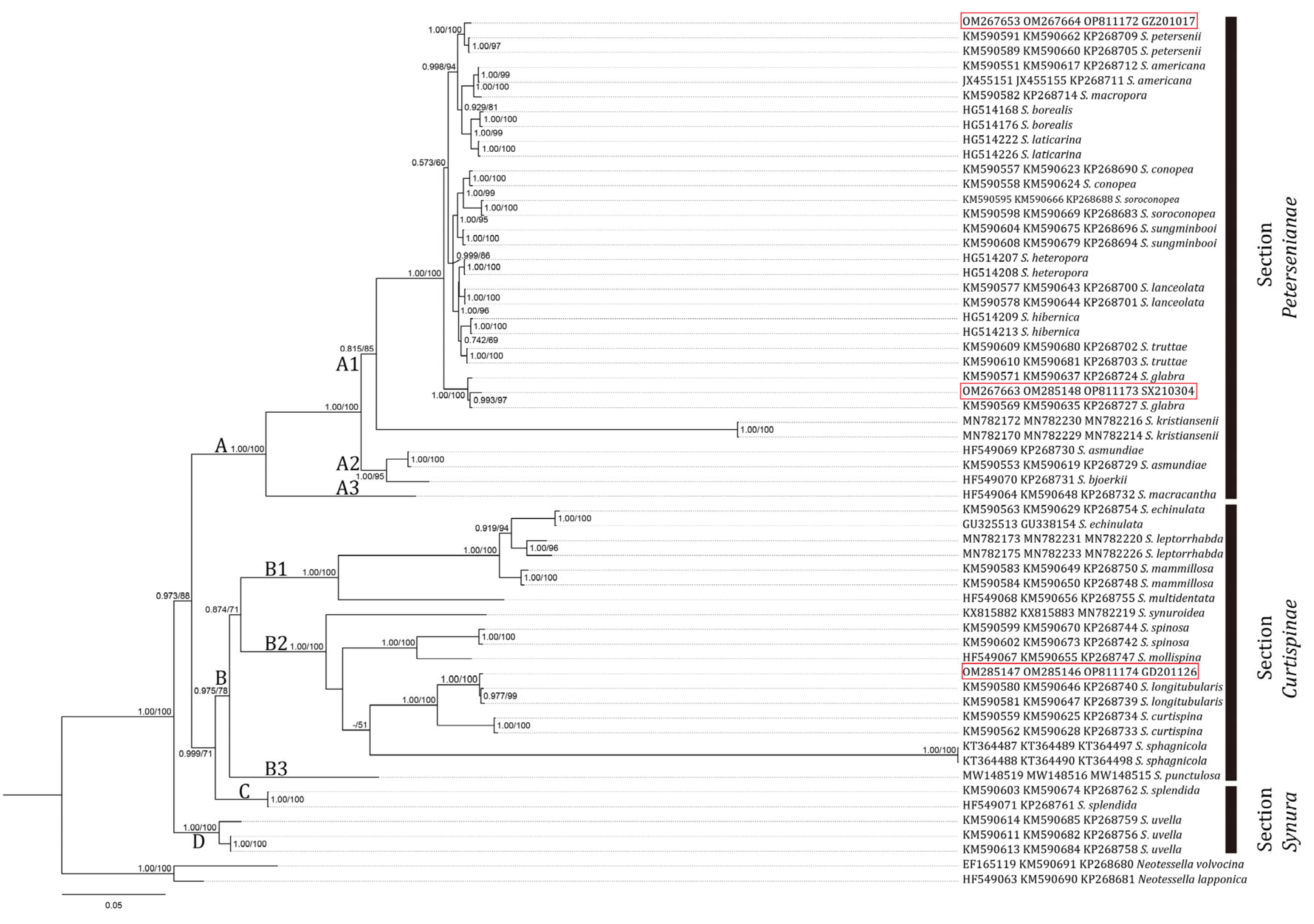
3.1. Molecular Analysis
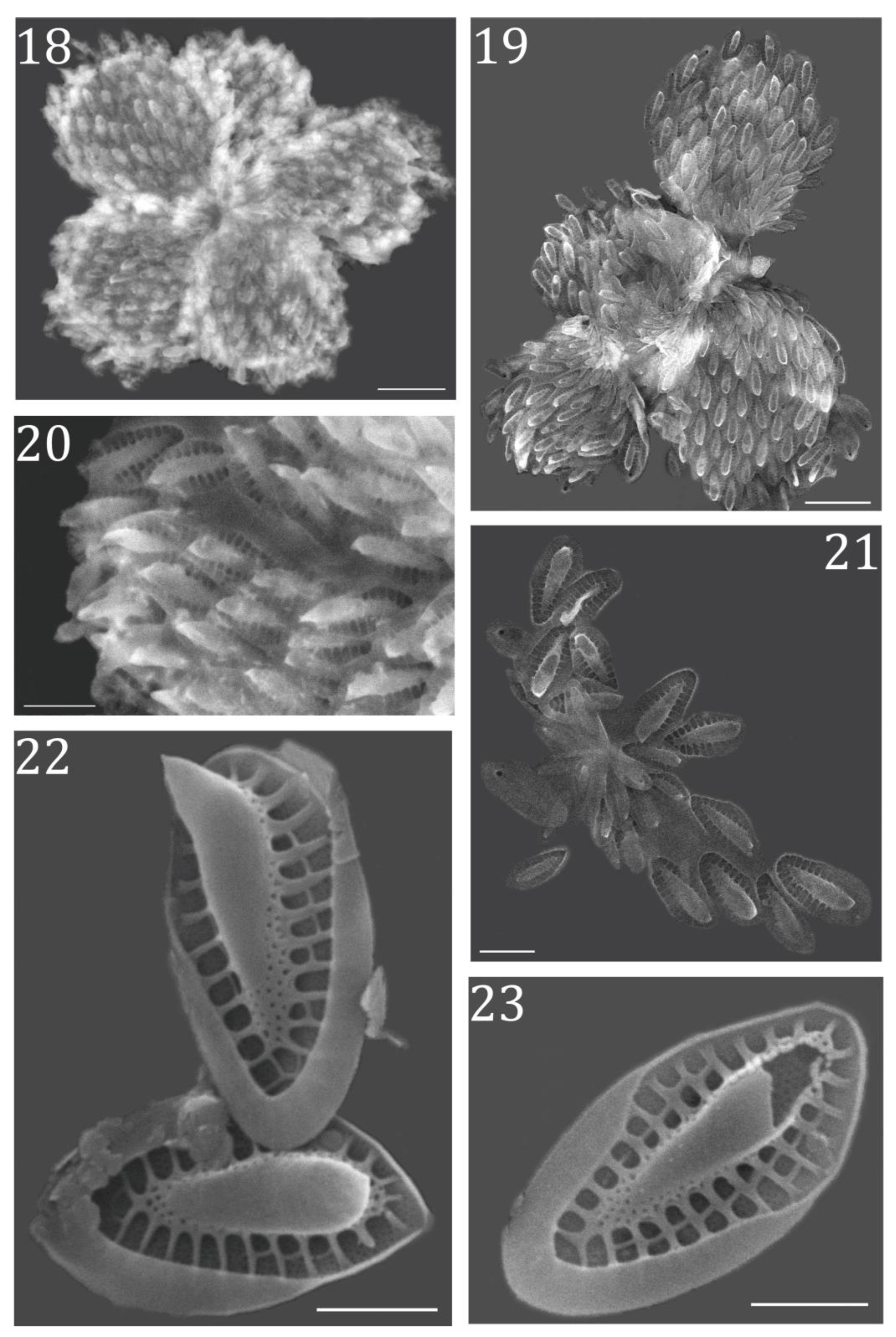
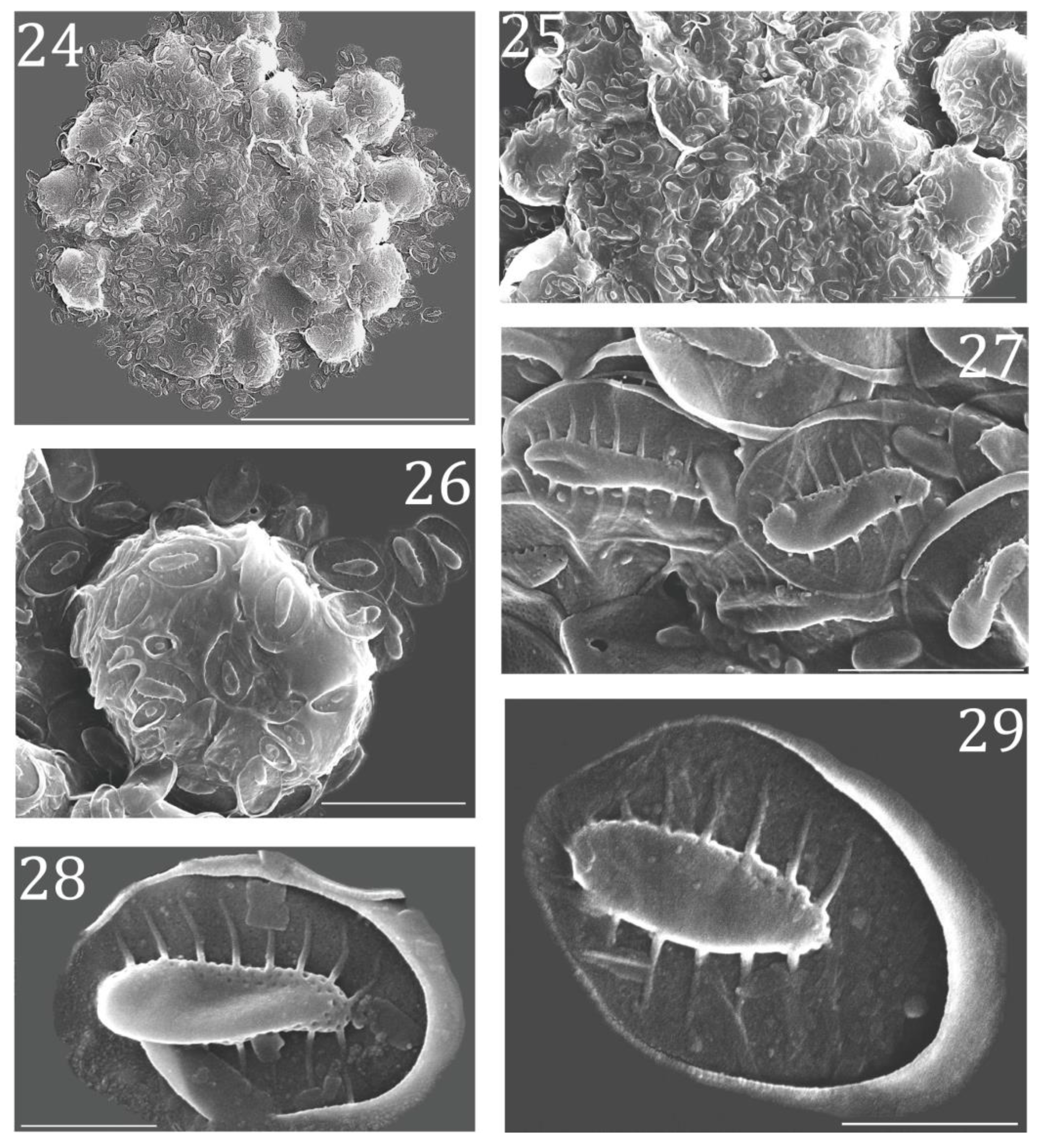
3.2. Morphological Characterization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ehrenberg, C.G. Dritter Beitrag zur Erkenntnis Grosser Organisation in er Richtung des Kleinsten Raumes. Abh. Konigl. Akad. Wiss. 1834, 1833, 145–336. [Google Scholar]
- Kristiansen, J.; Preisig, H.R. (Eds.) Encyclopedia of Chrysophyte Genera; Bibliotheca Phycologica: Berlin, Germany, 2001. [Google Scholar]
- Guiry, M.D.; Guiry, G.M. AlgaeBase. World-Wide Electronic Publication, National University of Ireland, Galway. 2021. Available online: https://www.algaebase.org/ (accessed on 5 December 2022).
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 9 October 2022).
- Wei, Y.X. Chrysophyta. In Flora Algarum Sinicarum Aquae Dulcis, Tomus ⅩⅩⅠ; Beijing Science Press: Beijing, China, 2018; pp. 127–141. [Google Scholar]
- Kristiansen, J.; Preisig, H.R. Chrysophyte and Haptophyte Algae: Pt. 2 Synurophyceae. In Süsswasserflora von Mitteleuropa; Büdel, B., Gärtner, G., Krienitz, L., Preisig, H.R., Schagerl, M., Eds.; Spektrum Akademischer Verlag: Berlin, Germany, 2007; pp. 1–252. [Google Scholar]
- Jo, B.Y.; Kim, J.I.; Škaloud, P.; Siver, P.A.; Shin, W. Multigene Phylogeny of Synura (Synurophyceae) and Descriptions of Four New Species Based on Morphological and DNA Evidence. Eur. J. Phycol. 2016, 51, 1–18. [Google Scholar] [CrossRef]
- Škaloud, P.; Škaloudová, M.; Jadrná, I.; Bestová, H.; Pusztai, M.; Kapustin, D.; Siver, P.A. Comparing Morphological and Molecular Estimates of Species Diversity in the Freshwater Genus Synura (Stramenopiles): A Model for Understanding Diversity of Eukaryotic Microorganisms. J. Phycol. 2020, 56, 574–591. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.B.; Hansen, J.B. On the Scales of Some Synura Species; Biologiske Meddelelser udgivet af Det Kongelige Danske Videnskabernes Selskab Series; Munksgaard: Copenhagen, Denmark, 1956; Volume 23, pp. 3–27. [Google Scholar]
- Balonov, I.M.; Kuzmin, G.V. Vidy Roda Synura Ehrenberg (Chrysophyta) v Vodokhranilishchakh Volzhskogo Kaskada. Bot. Zhurnal 1974, 59, 1675–1686. [Google Scholar]
- Škaloud, P.; Kristiansen, J.; Škaloudová, M. Developments in the Taxonomy of Silica-Scaled Chrysophytes—From Morphological and Ultrastructural to Molecular Approaches. Nord. J. Bot. 2013, 31, 385–402. [Google Scholar] [CrossRef]
- Asmund, B. Studies on Chrysophyceae from Some Ponds and Lakes in Alaska vi Occurrence of Synura Species. Hydrobiologia 1968, 31, 497–515. [Google Scholar] [CrossRef]
- Cronberg, G. Scaled Chrysophytes from the Tropics. Nova Hedwig. 1989, 95, 191–232. [Google Scholar]
- Petersen, J.B.; Hansen, J.B. On the Scales of Some Synura Species II; Biologiske Meddelelser udgivet af Det Kongelige Danske Videnskabernes Selskab Series; Munksgaard: Copenhagen, Denmark, 1958; Volume 23, pp. 1–13. [Google Scholar]
- Péterfi, L.S.; Momeu, L. Remarks on the Taxonomy of Some Synura Species Based on the Fine Structure of Scales. Muz. Brukenthal Stud. Comun. Stiint. Nat. 1977, 21, 15–23. [Google Scholar]
- Škaloud, P.; Škaloudová, M.; Procházková, A.; Němcová, Y. Morphological Delineation and Distribution Patterns of Four Newly Described Species within the Synura Petersenii Species Complex (Chrysophyceae, Stramenopiles). Eur. J. Phycol. 2014, 49, 213–229. [Google Scholar] [CrossRef]
- Takahashi, E. Electron Microscopical Studies of the Synuraceae (Chrysophyceae) in Japan: Taxonomy and Ecology; Tokai University Press: Tokyo, Japan, 1978. [Google Scholar]
- Siver, P.A.; Jo, B.Y.; Kim, J.I.; Shin, W.; Lott, A.M.; Wolfe, A.P. Assessing the Evolutionary History of the Class Synurophyceae (Heterokonta) Using Molecular, Morphometric, and Paleobiological Approaches. Am. J. Bot. 2015, 102, 921–941. [Google Scholar] [CrossRef]
- Jo, B.Y.; Shin, W.; Kim, H.S.; Siver, P.A.; Andersen, R.A. Phylogeny of the Genus Mallomonas (Synurophyceae) and Descriptions of Five New Species on the Basis of Morphological Evidence. Phycologia 2013, 52, 266–278. [Google Scholar] [CrossRef]
- Škaloud, P.; Kynčlová, A.; Benada, O.; Kofroňová, O.; Škaloudová, M. Toward a Revision of the Genus Synura, Section Petersenianae (Synurophyceae, Heterokontophyta): Morphological Characterization of Six Pseudo-Cryptic Species. Phycologia 2012, 51, 303–329. [Google Scholar] [CrossRef]
- Jo, B.Y.; Han, S.K. Newly Recorded Species of the Genus Synura (Synurophyceae) from Korea. J. Ecol. Environ. 2017, 41, 1. [Google Scholar] [CrossRef]
- Wee, J.L.; Fasone, L.D.; Sattler, A.; Starks, W.W.; Hurley, D.L. ITS/5.8S DNA Sequence Variation in 15 Isolates of Synura Petersenii Korshikov (Synurophyceae). Nova Hedwig. 2001, 122, 245–258. [Google Scholar]
- Kynčlová, A.; Škaloud, P.; Škaloudová, M. Unveiling Hidden Diversity in the Synura Petersenii Species Complex (Synurophyceae, Heterokontophyta). Nova Hedwig. Beih. 2010, 136, 283–298. [Google Scholar] [CrossRef]
- Siver, P.A. Synurophyte Algae. In Freshwater Algae of North America; Academic Press: Cambridge, MA, USA, 2003; pp. 523–557. ISBN 978-0-12-741550-5. [Google Scholar]
- Kristiansen, J. On the Occurrence of the Species of Synura (Chrysophyceae): With 6 Figures in the Text. SIL Proc. 1922–2010 1975, 19, 2709–2715. [Google Scholar] [CrossRef]
- Siver, P.A. The Distribution and Variation of Synura Species (Chrysophyceae) in Connecticut, USA. Nord. J. Bot. 1987, 7, 107–116. [Google Scholar] [CrossRef]
- Skvortzov, B.V. Harbin Chrysophyta, China boreali-orientalis. Bull. Herb. North-East. For. Acad. 1961, 3, 1–70. [Google Scholar]
- Kristiansen, J. Silica-Scaled Chrysophytes from China. Nord. J. Bot. 1989, 8, 539–552. [Google Scholar] [CrossRef]
- Kristiansen, J.; Tong, D. Silica-scaled chrysophytes of Wuhan, a preliminary note. J. Wuhan Bot. Res. 1988, 6, 97–100. [Google Scholar]
- Kristiansen, J.; Tong, D. Tong Studies on Silica-Scaled Chrysophytes from Wuhan, Hangzhou and Beijing, P.R. China. Nova Hedwig. 1989, 49, 183–202. [Google Scholar]
- Pang, W.T.; Wang, Y.F.; Wang, Q.X. Stomatocyst of Synura Petersenii. Acta Bot. Boreali Occident. Sin. 2012, 32, 921–923. [Google Scholar]
- Pang, W.; Wang, Q. A New Species, Synura Morusimila Sp. Nov. (Chrysophyta), from Great Xing’an Mountains, China. Phytotaxa 2013, 88, 55–60. [Google Scholar] [CrossRef]
- Wei, Y.X.; Yuan, X.P.; Kristiansen, J. Silica-Scaled Chrysophytes from Hainan, Guangdong Provinces and Hong Kong Special Administrative Region, China. Nord. J. Bot. 2014, 32, 881–896. [Google Scholar] [CrossRef]
- Wei, Y.X.; Yuan, X.P. Studies on Silica-Scaled Chrysophytes from the Daxinganling Mountains and Wudalianchi Lake Regions, China. Nova Hedwig. 2015, 101, 299–312. [Google Scholar] [CrossRef]
- Nakayama, T.; Watanabe, S.; Mitsui, K.; Uchida, H.; Inouye, I. The Phylogenetic Relationship between the Chlamydomonadales and Chlorococcales Inferred from 18SrDNA Sequence Data. Phycol. Res. 2010, 44, 47–55. [Google Scholar] [CrossRef]
- Jo, B.Y.; Shin, W.; Boo, S.M.; Han, S.K.; Siver, P.A. Studies on Ultrastructure and Three-Gene Phylogeny of the Genus Mallomonas (Synurophyceae). J. Phycol. 2011, 47, 415–425. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplificationand Direct Sequencing of Fungal Ribosomal RNA Genes for Phylo-Genetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An Integrated and Scalable Desktop Platform for Streamlined Molecular Sequence Data Management and Evolutionary Phylogenetics Studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Playfair, G.I. Freshwater Algae of the Lismore District: With an Appendix on the Algal Fungi and Schizomycetes. Proc. Linn. Soc. N. S. Wales 1915, 40, 310–362. [Google Scholar] [CrossRef]
- Playfair, G.I. New and Rare Freshwater Algae. Proc. Linn. Soc. N. S. Wales 1918, 43, 497–543. [Google Scholar]
- Playfair, G.I. Australian Freshwater Flagellates. Proc. Linn. Soc. N. S. Wales 1921, 46, 99–146. [Google Scholar] [CrossRef]
- Kristiansen, J. The Ultrastructural Bases of Chrysophyte Systematics and Phylogeny. CRC Crit. Rev. Plant Sci. 1986, 4, 149–211. [Google Scholar] [CrossRef]
- Kristiansen, J. Golden Algae: A Biology of Chrysophytes; A.R.G. Gantner Verlag: Koeningstein, Germany, 2005; pp. 1–167. [Google Scholar]
- Preisig, H.R. A modern concept of chrysophyte classification. In Chrysophyte Algae: Ecology, Phylogeny and Development; Sandgren, C.D., Smol, J.P., Kristiansen, J., Eds.; Cambridge University Press: Cambridge, UK, 1995; pp. 46–74. [Google Scholar] [CrossRef]
- Boo, S.M.; Kim, H.S.; Shin, W.; Boo, G.H.; Cho, S.M.; Jo, B.Y.; Kim, J.-H.; Kim, J.H.; Yang, E.C.; Siver, P.A.; et al. Complex Phylogeographic Patterns in the Freshwater Alga Synura Provide New Insights into Ubiquity vs. Endemism in Microbial Eukaryotes. Mol. Ecol. 2010, 19, 4328–4338. [Google Scholar] [CrossRef] [PubMed]
- Korshikov, A.A. Studies on the Chrysomonads. I. Arch. Protistenkd. 1929, 67, 253–290. [Google Scholar]
- Siver, P.A.; Lott, A.M. The Scaled Chrysophyte Flora in Freshwater Ponds and Lakes from Newfoundland, Canada, and Their Relationship to Environmental Variables. Cryptogam. Algol. 2017, 38, 325–347. [Google Scholar] [CrossRef]
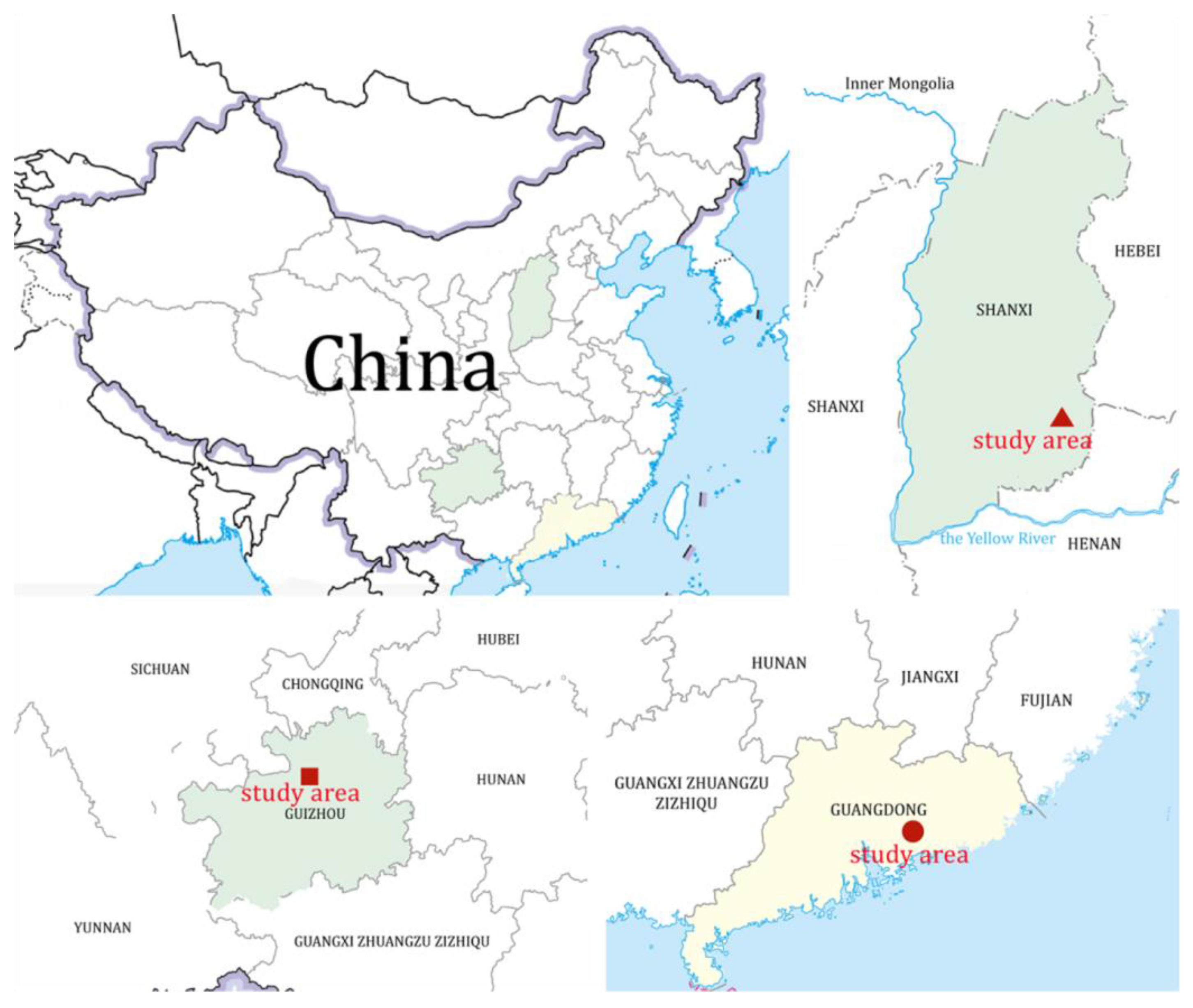


| Isolate | Locality with Longitude and Latitude | Habitat Type | Collection Date | Collector | Voucher Number |
| GD201126 | the Xianxia waterfall, Huizhou, Guangdong Province, China (23.6494° N, 113.8851° E) | a pond near a waterfall | 26 November 2020 | Nini Cui | SXU-GD201126 |
| GZ201017 | the Xinpu Wetland Park, Zunyi, Guizhou Province, China (27.7024° N, 107.0180° E) | a lake in the park | 17 October 2020 | Qi Liu | SXU-GZ201017 |
| SX210304 | the Long Korean Road, Changzhi, Shanxi Province, China (36.0619° N, 113.0049° E) | a lake near a factory | 4 May 2021 | Chen Su | SXU-SX210304 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, J.; Nan, F.; Lv, J.; Liu, Q.; Liu, X.; Xie, S.; Feng, J. Morphological and Molecular Characterizations of Three Species of the Genus Synura (Synurales, Chrysophyceae) from China. Diversity 2022, 14, 1092. https://doi.org/10.3390/d14121092
Hao J, Nan F, Lv J, Liu Q, Liu X, Xie S, Feng J. Morphological and Molecular Characterizations of Three Species of the Genus Synura (Synurales, Chrysophyceae) from China. Diversity. 2022; 14(12):1092. https://doi.org/10.3390/d14121092
Chicago/Turabian StyleHao, Junxue, Fangru Nan, Junping Lv, Qi Liu, Xudong Liu, Shulian Xie, and Jia Feng. 2022. "Morphological and Molecular Characterizations of Three Species of the Genus Synura (Synurales, Chrysophyceae) from China" Diversity 14, no. 12: 1092. https://doi.org/10.3390/d14121092
APA StyleHao, J., Nan, F., Lv, J., Liu, Q., Liu, X., Xie, S., & Feng, J. (2022). Morphological and Molecular Characterizations of Three Species of the Genus Synura (Synurales, Chrysophyceae) from China. Diversity, 14(12), 1092. https://doi.org/10.3390/d14121092








