Introducing Cyanodorina gen. nov. and Cyanodorina ovale sp. nov. (Microcystaceae, Chroococcales), a Novel Coccoid Cyanobacterium Isolated from Caohai Lake in China Based on a Polyphasic Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Sample Collection
2.2. Morphological Characterization
2.3. DNA Extraction and PCR Amplification
2.4. Phylogenetic Analysis
2.5. ITS Secondary Structure Prediction
3. Results
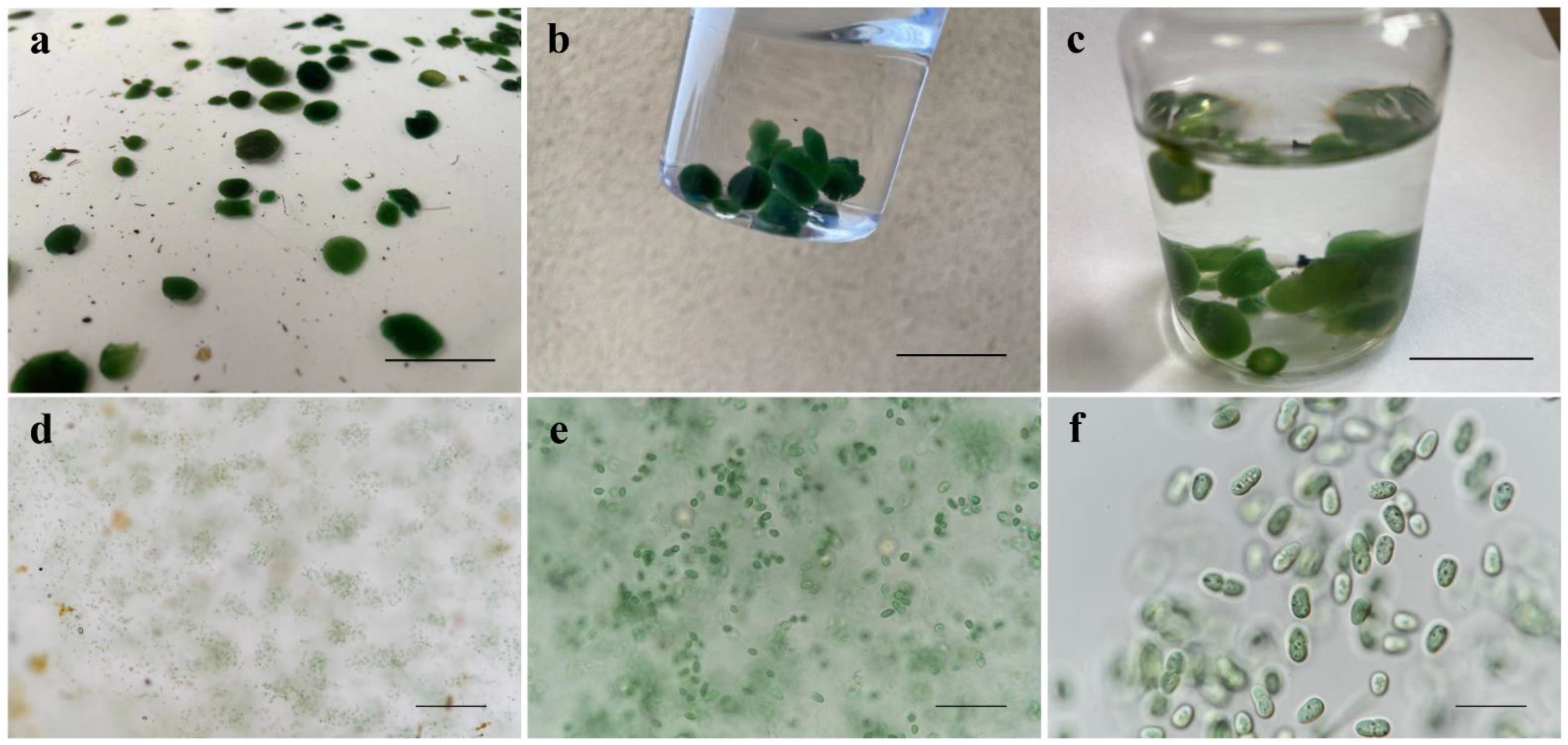
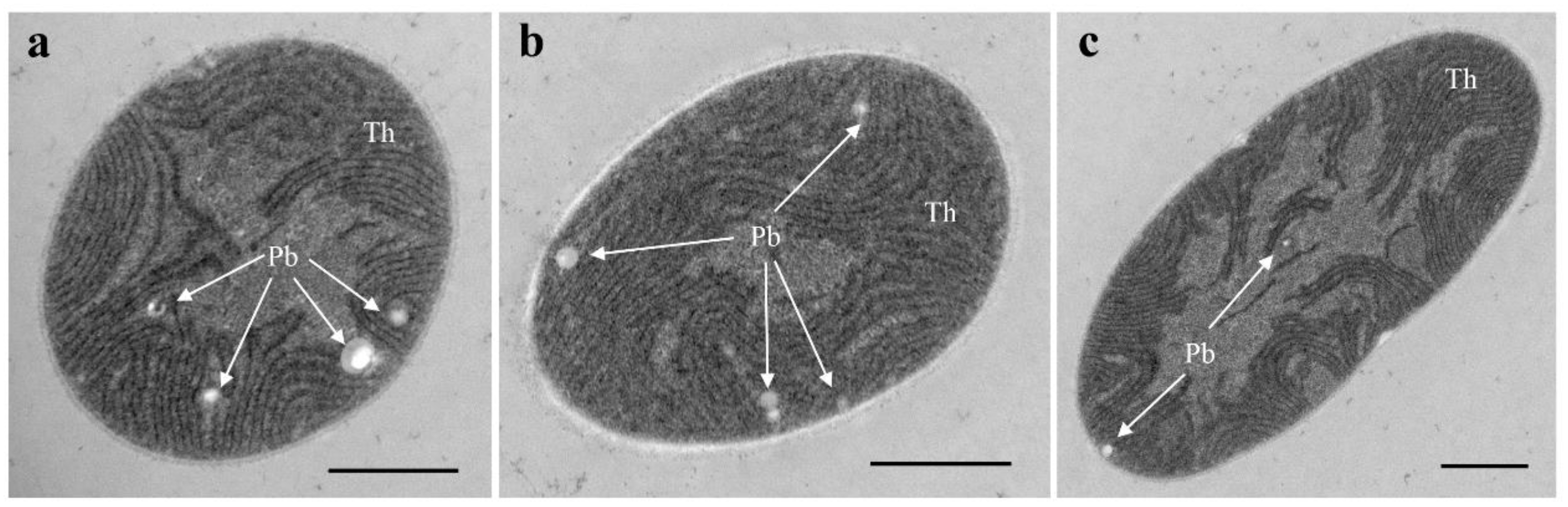
3.1. Morphological Description
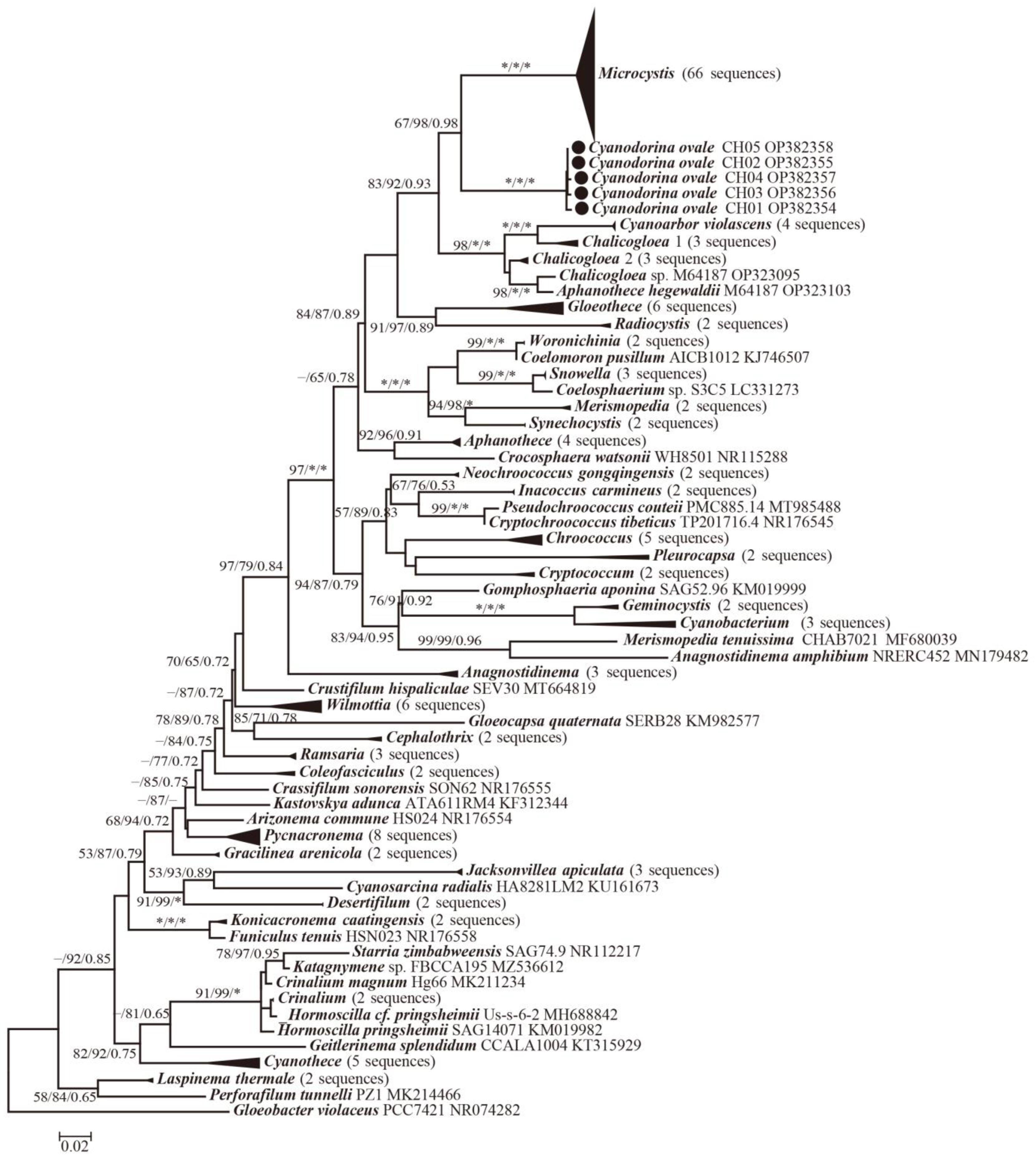
3.2. Molecular and Phylogenetic Analysis
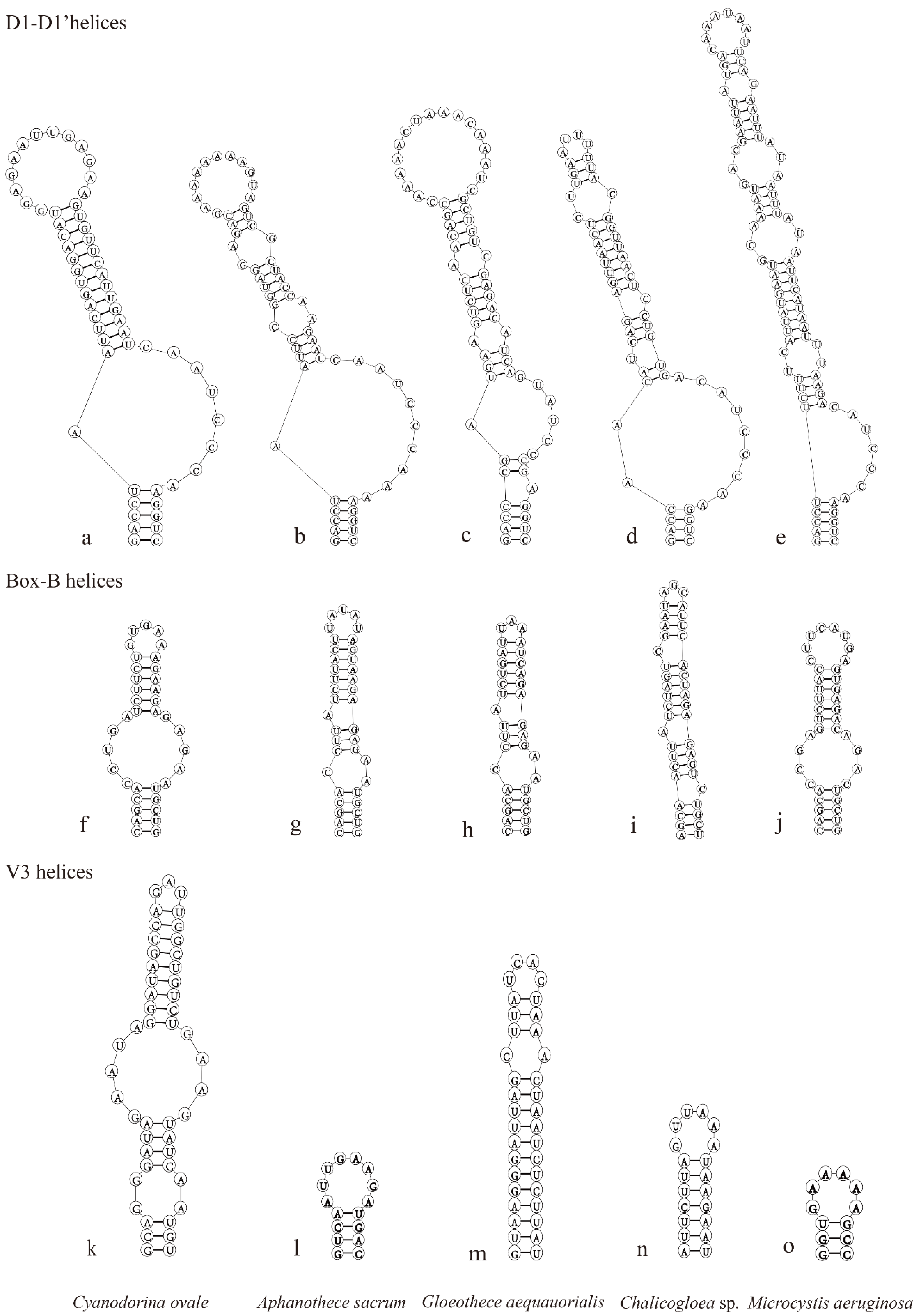
3.3. Comparison of ITS Regions between 16S and 23S rRNA Genes and Secondary Structures
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waterbury, J.B. The Cyanobacteria-Isolation, Purification and Identification. In Prokaryotes, 3rd ed.; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; Volume 4, pp. 1053–1073. [Google Scholar] [CrossRef]
- Mareš, J.; Johansen, J.R.; Hauer, T.; Zima, J., Jr.; Ventura, S.; Cuzman, O.; Tiribilli, B.; Kaštovský, J. Taxonomic resolution of the genus Cyanothece (Chroococcales, Cyanobacteria), with a treatment on Gloeothece and three new genera, Crocosphaera, Rippkaea and Zehria. J. Phycol. 2019, 55, 578–610. [Google Scholar] [CrossRef]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphyletic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Mareš, J. Multilocus and SSU rRNA gene phylogenetic analyses of available cyanobacterial genomes, and their relation to the current taxonomic system. Hydrobiologia 2018, 811, 19–34. [Google Scholar] [CrossRef]
- Harke, M.J.; Steffen, M.M.; Gobler, C.J.; Otten, T.G.; Wilhelm, S.W.; Wood, S.A.; Paerl, H.W. A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, Microcystis spp. Harmful Algae 2016, 54, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Kovačić, L.; Jezberová, J.; Komárková, J.; Kopecky., J.; Komárek, J. Ecological characteristics and polyphasic taxonomic classification of stable pigment-types of the genus Chroococcus (Cyanobacteria). Preslia 2011, 83, 145–166. [Google Scholar]
- Margheri, M.C.; Ventura, S.; Kaštovský, J.; Komárek, J. The taxonomic validation of the cyanobacterial genus Halothece. Phycologia 2008, 47, 477–486. [Google Scholar] [CrossRef]
- Komárek, J.; Kaštovský, J.; Jezberová, J. Phylogenetic and taxonomic delimitation of the cyanobacterial genus Aphanothece and description of Anathece gen. nov. Eur. J. Phycol. 2011, 46, 315–326. [Google Scholar] [CrossRef]
- Roldán, M.; Ramírez, M.; Del Campo, J.; Hernández-Mariné, M.; Komárek, J. Chalicogloea cavernicola gen. nov., sp. nov. (Chroococcales, Cyanobacteria), from low-light aerophytic environments: Combined molecular, phenotypic and ecological criteria. Int J. Syst. Evol. Microbiol. 2013, 63, 2326–2333. [Google Scholar] [CrossRef]
- Wang, Y.L.; Jia, N.N.; Geng, R.Z.; Yu, G.L.; Li, R.H. Phylogenetic insights intochroococcus-like taxa (Chroococcales, Cyanobacteria), describing Cryptochroococcus tibeticus gen. nov. sp. nov. and Limnococcus fonticola sp. nov. from Qinghai-Tibet plateau. J. Phycol. 2021, 57, 1739–1748. [Google Scholar] [CrossRef]
- Mogany, T.; Swalaha, F.M.; Allam, M.; Senzo Mtshali, P.; Ismail, A.; Kumari, S.; Bux, F. Phenotypic and genotypic characterisation of an unique indigenous hypersaline unicellular cyanobacterium, Euhalothece sp. nov. Microbiol. Res. 2018, 211, 47–56. [Google Scholar] [CrossRef]
- Gama, W.A.; Rigonato, J.; Fiore, M.F.; Sant’Anna, C.L. New insights into Chroococcus (Cyanobacteria) and two related genera: Cryptococcum gen. nov. and Inacoccus gen. nov. Eur. J. Phycol. 2019, 54, 315–325. [Google Scholar] [CrossRef]
- Rigonato, J.; Sant’Anna, C.L.; Giani, A.; Azevedo, M.T.P.; Gama, W.A.; Viana, V.F.L.; Fiore, M.F.; Werner, V.R. Sphaerocavum: A coccoid morphogenus identical to Microcystis in terms of 16S rDNA and ITS sequence phylogenies. Hydrobiologia 2018, 811, 35–48. [Google Scholar] [CrossRef]
- Strunecký, O.; Ivanova, A.P.; Mareš, J. An updated classification of cyanobacterial orders and families based on phylogenomic and polyphasic analysis1. J. Phycol. 2022; accepted. [Google Scholar]
- Boone, D.R.; Castenholz, R.W. The Archaea and the Deeply Branching and Phototrophic Bacteria. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Garrity, G., Ed.; Springer: New York, NY, USA, 2001; Volume 1, pp. 1–721. [Google Scholar]
- Komárek., J.; Anagnostidis., K. Cyanoprokaryota 1. Teil: Chroococcales. In Süsswasserflora von Mitteleuropa 19/1; Ettl, H., Gärtner, G., Heynig, H., Eds.; Gustav Fischer: Jena, Germany; Stuttgart, Germany; Lübeck, Germany; Ulm, Germany, 1998; pp. 1–548. [Google Scholar]
- Geng, R.Z.; Li, W.K.; Chao, A.M.; Guo, X.Y.; Li, H.; Yu, G.L.; Li, R.H. Establishment of a New Filamentous Cyanobacterial Genus, Microcoleusiopsis gen. nov. (Microcoleaceae, Cyanobacteria), from Benthic Mats in Open Channel, Jiangxi Province, China. Diversity 2021, 13, 548. [Google Scholar] [CrossRef]
- Clarke, J.D. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb. Protoc. 2009, 2009, pdb.prot5177. [Google Scholar] [CrossRef] [PubMed]
- Lepère, C.; Wilmotte, A.; Meyer, B. Molecular diversity of Microcystis strains (Cyanophyceae, Chroococcales) based on 16S rRNA sequences. Syst. Geogr. Plants 2000, 70, 275–283. [Google Scholar] [CrossRef]
- Edwards, U.; Rogall, T.; Blöcker, H.; Emde, M.; Böttger, E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, 232–235. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Miller, M.A.; Schwartz, T.; Pickett, B.; He, S.; Klem, E.; Scheuermann, R.H.; Passarotti, M.; Kaufman, S.; O’Leary, M.A. A RESTful API for Access to Phylogenetic Tools via the CIPRES Science Gateway. Evol. Bioinform. 2015, 11, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Boyer, S.; Johansen, J.R.; Flechtner, V.R. Phylogeny and genetic variance in terrestrial Microcoleus (Cyanophyceae) species based on sequence analysis of the 16S rRNA gene and associated 16S-23S ITS region. J. Phycol. 2002, 38, 1222–1235. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Sandrini-Neto, L.; Geraudie, P.; Santana, M.S.; Camus, L. Effects of dispersed oil exposure on biomarker responses and growth in juvenile wolfish Anarhichas denticulatus. Environ. Sci. Pollut. Res. Int. Res. 2016, 23, 21441–21450. [Google Scholar] [CrossRef]
- Comte, K.; Holland, D.P.; Walsby, A.E. Changes in cell turgor pressure related to uptake of solutes by Microcystis sp. strain8401. FEMS Microbiol. 2007, 61, 399–405. [Google Scholar] [CrossRef]
- Dvořák, P.; Poulíčková, A.; Hašler, P.; Belli, M.; Casamatta, D.A.; Papini, A. Species concepts and speciation factors in cyanobacteria, with connection to the problems of diversity and classification. Biodivers. Conserv. 2015, 24, 739–757. [Google Scholar] [CrossRef]
- Komárek, J. Several problems of the polyphasic approach in the modern cyanobacterial system. Hydrobiologia 2018, 811, 7–17. [Google Scholar] [CrossRef]
- Sciuto, K.; Rascio, N.; Andreoli, C.; Moro, I. Polyphasic characterization of ITD-01, a cyanobacterium isolated from the Ischia Thermal District (Naples, Italy). Fottea 2011, 11, 31–39. [Google Scholar] [CrossRef]
- Wang, Y.L.; Cai, F.F.; Jia, N.N.; Li, R.H. Description of a novel coccoid cyanobacterial genus and species Sinocapsa zengkensis gen. nov. sp. nov. (Sinocapsaceae, incertae sedis), with taxonomic notes on genera in Chroococcidiopsidales. Phytotaxa 2019, 409, 146–160. [Google Scholar] [CrossRef]
- Korelusová, J.; Kaštovský, J.; Komárek, J. Heterogeneity of the cyanobacterial genus Synechocystis and description of a new genus, Geminocystis. J. Phycol. 2009, 45, 928–937. [Google Scholar] [CrossRef] [PubMed]
- Castenholz, R.W.; Wilmotte, A.; Herdman, M.; Rippka, R.; Waterbury, J.B.; Iteman, I.; Hoffmann, L. Bergey’s Manual of Systematic Bacteriology; Springer Science+Business Media: New York, NY, USA, 2001. [Google Scholar]
- Nelissen, B.; Van de Peer, Y.; Wilmotte, A.; De Water, R. An early origin of plastids within the cyanobacterial divergence is suggested by evolutionary trees based on complete 16S rRNA sequences. Mol. Biol. Evol. 1995, 12, 1166–1173. [Google Scholar] [PubMed]
- Ohki, K.; Kamiya, M.; Honda, D.; Kumazawa, S.; Ho, K.K. Morphological and phylogenetic studies on unicellular dizaotrophic Cyanobacteria (Cyanophytes) isolated from the coastal waters around singapore. J. Phycol. 2008, 44, 142–151. [Google Scholar] [CrossRef]
- Komárek, J.; Cepák, V.; Kaštovský, J.; Sulek, J. What are the cyanobacterial genera Cyanothece and Cyanobacterium? Contribution to the combined molecular and phenotype taxonomic evaluation of cyanobacterial diversity. Algol. Stud./Arch. Für Hydrobiol. 2004, 113, 1–36. [Google Scholar] [CrossRef]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.-H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rpsselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Sendall, B.C.; Mcgregor, G.B. Cryptic diversity within the Scytonema complex: Characterization of the paralytic shellfish toxin producer Heterosyctonema crispum, and the establishment of the family Heteroscytonemataceae (Cyanobacteria/Nostocales). Harmful Algae 2018, 80, 158–170. [Google Scholar] [CrossRef] [PubMed]
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. Cyanodorina ovale CH01 clone1 | ||||||||||||||||||||
| 2. Microcystis aeruginosa NIES298 | 93.10 | |||||||||||||||||||
| 3. Neochroococcus gongqingensis CHAB4018 | 93.00 | 91.50 | ||||||||||||||||||
| 4. Cyanoarbor violascens C1 | 93.00 | 92.30 | 91.90 | |||||||||||||||||
| 5. Chalicogloea cavernicola CCALA975 | 92.40 | 91.30 | 91.10 | 94.80 | ||||||||||||||||
| 6. Cryptochroococcus tibeticus TP201716.4 | 92.20 | 91.20 | 94.50 | 92.00 | 91.20 | |||||||||||||||
| 7. Pseudochroococcus couteii PMC885.14 | 92.10 | 91.50 | 94.40 | 92.00 | 91.10 | 99.10 | ||||||||||||||
| 8. Gloeothece aequatorialis SAG36.87 clone7 | 91.90 | 91.90 | 92.40 | 91.80 | 91.40 | 91.10 | 91.00 | |||||||||||||
| 9. Inacoccus carmineus CCIBt3475 | 91.60 | 90.80 | 94.20 | 91.20 | 90.00 | 93.90 | 94.00 | 91.30 | ||||||||||||
| 10. Gomphosphaeria aponina SAG52.96 | 91.50 | 89.90 | 94.00 | 90.70 | 90.50 | 93.20 | 93.20 | 91.20 | 93.80 | |||||||||||
| 11. Pannus brasiliensis CCIBt3594 | 91.30 | 94.90 | 90.20 | 90.50 | 90.70 | 89.90 | 90.10 | 91.10 | 89.50 | 89.30 | ||||||||||
| 12. Aphanothece sacrum H5 | 91.30 | 90.50 | 92.60 | 91.60 | 91.60 | 92.10 | 92.60 | 91.20 | 92.90 | 93.00 | 90.80 | |||||||||
| 13. Eucapsis minor SAG 14.99 | 91.10 | 93.50 | 91.90 | 91.50 | 90.80 | 90.10 | 90.20 | 90.70 | 90.90 | 90.30 | 92.80 | 90.90 | ||||||||
| 14. Radiocystis geminata MAC1214 | 90.80 | 91.00 | 91.50 | 91.40 | 91.20 | 90.10 | 90.10 | 91.40 | 92.20 | 91.30 | 89.70 | 91.30 | 91.40 | |||||||
| 15. Chroococcus subviolaceus CCIBt3549 | 90.80 | 91.50 | 92.80 | 91.80 | 90.20 | 92.30 | 92.70 | 91.20 | 93.40 | 91.70 | 89.70 | 91.20 | 89.30 | 90.10 | ||||||
| 16. Cryptococcum komarkovae CCALA54 | 90.30 | 88.90 | 93.00 | 90.10 | 90.00 | 92.20 | 92.20 | 89.90 | 91.90 | 91.60 | 89.10 | 91.20 | 89.80 | 89.70 | 91.30 | |||||
| 17. Geitlerinema splendidum CCALA1004 | 90.00 | 87.80 | 90.30 | 88.50 | 88.80 | 90.20 | 90.50 | 88.10 | 90.40 | 89.80 | 88.50 | 90.10 | 87.60 | 88.20 | 91.20 | 89.90 | ||||
| 18. Anagnostidinema amphibium NRERC452 | 88.90 | 89.70 | 89.80 | 88.50 | 87.90 | 89.50 | 89.60 | 89.00 | 89.30 | 90.00 | 88.10 | 89.10 | 88.00 | 89.40 | 90.10 | 87.40 | 87.60 | |||
| 19. Geminocystis herdmanii PCC6308 | 88.00 | 86.60 | 88.70 | 87.00 | 87.10 | 90.00 | 89.90 | 87.00 | 89.10 | 90.20 | 86.80 | 89.10 | 86.70 | 87.30 | 87.30 | 88.40 | 87.50 | 86.70 | ||
| 20. Alborzia kermanshahica S2 | 86.90 | 85.90 | 86.50 | 89.90 | 91.50 | 85.80 | 85.80 | 86.60 | 84.90 | 85.60 | 86.00 | 86.70 | 86.90 | 86.60 | 84.70 | 85.00 | 83.40 | 82.20 | 82.50 | |
| 21. Cyanobacterium stanieri MAC3217 | 86.50 | 84.60 | 86.50 | 84.30 | 84.70 | 87.70 | 87.90 | 85.30 | 87.70 | 88.90 | 84.70 | 87.60 | 84.80 | 85.90 | 86.00 | 86.50 | 86.80 | 85.10 | 92.80 | 80.00 |
| Organisms | GenBank | ITS Total Length (nt) | D1–D1′ Helix Length (nt) | D2 Region | tRNAIle | tRNAAla | Box B Helix Length (nt) | Box A Spacer | V3 Helix Length (nt) |
|---|---|---|---|---|---|---|---|---|---|
| Cyanodorina ovale CH01 | OP382354 | 460 | 58 | CTTTCAAACTCT | + | - | 37 | GCACCTTGAAAA | 48 |
| Aphanothece sacrum FPU3 | AB116658.2 | 453 | 93 | CTTTCAAACTAG | + | - | 40 | GCACCTTGAAAA | 16 |
| Gloeothece aequauorialis SAG 36.87 | MF781064.1 | 449 | 60 | CTTTCAAACTTA | - | - | 43 | GAACCTTGAAAA | 36 |
| Chalicogloea sp. BACA0589 | OM732250.1 | 481 | 67 | CTTTCAAACTCT | + | - | 35 | GCACCTTGAAAA | 20 |
| Microcystis aeruginosa UUEX ‘B 2667 | HQ625424.1 | 404 | 63 | CTTTCAAACTAG | + | - | 39 | GAACCTTGAAAA | 12 |
| Genus | Cell Shape | Cell Size (μm) | Colony Formation | Colony Size (μm) | Sheaths | Thylakoid Arrangement | Habitat |
|---|---|---|---|---|---|---|---|
| Cyanodorina | oval | 6.54–8.92 × 4.51–5.69 | Yes | 5000–20,000 | Yes | Irregular fascicles | Freshwater, benthic |
| Microcystis | spherical or hemispherical | 1.7–8.1 | Yes | 40–3000 | Yes | Irregular fascicles | Freshwater, free floating |
| Pannus | round | 1–4 | Yes | 126.5–314.0 | Yes | Irregular fascicles | Planktonic in brackish bays and reservoirs, benthic in stagnant freshwater |
| Eucapsis | semicircle | 8–14 | Yes | <50 | Yes | - | Freshwater, benthic |
| Chalicogloea | spherical | 2.6–4.3 | Yes | <100.0 | Yes | Irregular fascicles | Salpetre Cave |
| Cyanoarbor | subspherical | 1–5 | Yes | 200.0–10,000.0 | Yes | Irregular fascicles | Salty and alkaline waters, high mts lakes, |
| Gloeothece | oval to cylindrical | 5–20 × 3–12 | Yes, rarely absent | 13.0–40.0 long, 10.0–26.0 wide | Yes | Irregular fascicles | Freshwater, terrestrial |
| Cyanothece | oval to cylindrical | 2.5–5 × 7.5–10 | No | No | Special (radial with a central network) | Freashwater, terrestrial | |
| Crocosphaera | spherical to cylindrical | 2.8–7.1 × 1.2–1.8 | No | No | Irregular fascicles | Marine | |
| Zehria | spherical to cylindrical | 2.0–5.6 × 2.0–4.0 | No | No | Irregular fascicles | Marine | |
| Rippkaea | oval to cylindrical | 4–9 × 3–5 | No | No | Irregular fascicles | Semi-terrestrial (soil of rice field) | |
| Aphanothece | oval to cylindrical | 1–12 | Yes | micro to macroscopic | Yes | Irregular fascicles | Freshwater, terrestrial |
| Cyanoaggregatum | oval or cylindrical | 2.4–3.8 × 1.4–2.0 | Yes | 92.0–340.0 long, 62.0–160.0 diameter | Yes | - | lagoon |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Li, S.; Xu, Y.; Geng, R.; Song, G.; Ma, P. Introducing Cyanodorina gen. nov. and Cyanodorina ovale sp. nov. (Microcystaceae, Chroococcales), a Novel Coccoid Cyanobacterium Isolated from Caohai Lake in China Based on a Polyphasic Approach. Diversity 2023, 15, 329. https://doi.org/10.3390/d15030329
Chen W, Li S, Xu Y, Geng R, Song G, Ma P. Introducing Cyanodorina gen. nov. and Cyanodorina ovale sp. nov. (Microcystaceae, Chroococcales), a Novel Coccoid Cyanobacterium Isolated from Caohai Lake in China Based on a Polyphasic Approach. Diversity. 2023; 15(3):329. https://doi.org/10.3390/d15030329
Chicago/Turabian StyleChen, Wei, Shuyin Li, Yuanzhao Xu, Ruozhen Geng, Gaofei Song, and Peiming Ma. 2023. "Introducing Cyanodorina gen. nov. and Cyanodorina ovale sp. nov. (Microcystaceae, Chroococcales), a Novel Coccoid Cyanobacterium Isolated from Caohai Lake in China Based on a Polyphasic Approach" Diversity 15, no. 3: 329. https://doi.org/10.3390/d15030329
APA StyleChen, W., Li, S., Xu, Y., Geng, R., Song, G., & Ma, P. (2023). Introducing Cyanodorina gen. nov. and Cyanodorina ovale sp. nov. (Microcystaceae, Chroococcales), a Novel Coccoid Cyanobacterium Isolated from Caohai Lake in China Based on a Polyphasic Approach. Diversity, 15(3), 329. https://doi.org/10.3390/d15030329






