As the Goose Flies: Migration Routes and Timing Influence Patterns of Genetic Diversity in a Circumpolar Migratory Herbivore
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Areas and Management Units
2.2. Library Preparation and Bioinformatics
2.3. Population Divergence and Nucleotide Diversity
2.4. Population Structure
3. Results
3.1. Bioinformatics
3.2. Population Divergence and Molecular Diversity
3.3. Nuclear Population Structure
4. Discussion
4.1. Historical Biogeography
4.2. Effects of Timing and Location of Annual Cycle Events and Location on Genetic Exchange
5. Future Directions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Epps, C.W.; Keyghobadi, N. Landscape genetics in a changing world: Disentangling historical and contemporary influences and inferring change. Mol. Ecol. 2015, 24, 6021–6040. [Google Scholar] [CrossRef] [PubMed]
- Ricketts, T.H. The matrix matters: Effective isolation in fragmented landscapes. Am. Nat. 2001, 158, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Hanski, I. Metapopulation dynamics. Nature 1998, 396, 41–49. [Google Scholar] [CrossRef]
- Parreira, B.; Quéméré, E.; Vanpé, C.; Carvalho, I.; Chikhi, L. Genetic consequences of social structure in the golden-crowned sifaka. Heredity 2020, 125, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Ely, C.R.; Scribner, K.T. Genetic diversity in Arctic-nesting geese: Implications for management and conservation. In Proceedings of the Trans 59th North American Wildlife and Natural Resources Conference, Anchorage, Alaska, 18–23 March 1994; Volume 59, pp. 91–110. [Google Scholar]
- Mehl, K.R.; Alisauskas, R.T.; Hobson, K.A.; Kellett, D.A. To winter east or west? Heterogeneity in winter philopatry in a central-Arctic population of king eiders. Condor 2004, 106, 241–251. [Google Scholar] [CrossRef]
- De La Cruz, S.E.W.; Takekawa, J.Y.; Wilson, M.T.; Nysewander, D.R.; Evenson, J.R.; Esler, D.; Boyd, W.S.; Ward, D.H. Spring migration routes and chronology of surf scoters (Melanitta perspicillata): A synthesis of Pacific coast studies. Can. J. Zool. 2009, 87, 1069–1086. [Google Scholar] [CrossRef]
- Ruegg, K. Genetic, morphological, and ecological characteristics of a hybrid zone that spans a migratory divide. Evolution 2008, 62, 452–466. [Google Scholar] [CrossRef] [PubMed]
- Rolshausen, G.; Segelbacher, G.; Hobson, K.A.; Schaefer, H.M. Contemporary evolution of reproductive isolation and phenotypic divergence in sympatry along a migratory Divide. Curr. Biol. 2009, 19, 2097–2101. [Google Scholar] [CrossRef] [PubMed]
- Sonsthagen, S.A.; Talbot, S.L.; Scribner, K.T.; McCracken, K.G. Multilocus phylogeography and population structure of common eiders breeding in North America and Scandinavia. J. Biogeogr. 2011, 38, 1368–1380. [Google Scholar] [CrossRef]
- Gaggiotti, O.E. Genetic threats to population persistence. Ann. Zool. Fenn. 2003, 40, 155–158. [Google Scholar]
- Agashe, D. The stablizing effect of intraspecific genetic variation on population dynamics in novel and ancestral habitats. Am. Nat. 2009, 174, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Markert, J.A.; Champlin, D.M.; Gutjahr-Gobell, R.; Grear, J.S.; Kuhn, A.; McGreevy, T.J., Jr.; Roth, A.; Bagley, M.J.; Nacci, D.E. Population genetic diversity and fitness in multiple environments. BMC Evol. Biol. 2010, 10, 205. [Google Scholar] [CrossRef]
- Berger-Tal, O.; Saltz, D. Invisible barriers: Anthropogenic impacts on inter- and intra-specific interactions as drivers of landscape-independent fragmentation. Philos. Trans. R. Soc. B 2019, 374, 20180049. [Google Scholar] [CrossRef] [PubMed]
- Convention on Biological Diversity. 2007. Available online: https://www.cbd.int/doc/meetings/cop-bureau/cop-bur-2007/cop-bur-2007-10-14-en.pdf (accessed on 15 July 2022).
- Koenig, W.D.; van Vuren, D.; Hooge, P.N. Detectability, philopatry, and the distribution of dispersal distances in vertebrates. Trends Ecol. Evol. 1996, 11, 514–517. [Google Scholar] [CrossRef]
- Lamb, J.S.; Paton, P.W.C.; Osenkowski, J.E.; Badzinski, S.S.; Berlin, A.M.; Bowman, T.; Dwyer, C.; Fara, L.J.; Gilliland, S.G.; Kenow, K.; et al. Implanted satellite transmitters affect sea duck movement patterns at short and long timescales. Condor 2020, 122, 1–16. [Google Scholar] [CrossRef]
- Bose, S.; Forrester, T.D.; Brazeal, J.L.; Sacks, B.N.; Casady, D.S.; Wittmer, H.U. Implications of fidelity and philopatry for the population structure of female back-tailed deer. Behav. Ecol. 2017, 28, 983–990. [Google Scholar] [CrossRef]
- Robertson, E.P.; Fletcher, R.J., Jr.; Cattau, C.E.; Udell, B.J.; Reichert, B.E.; Austin, J.D.; Valle, D. Isolating the roles of movement and reproduction on effective connectivity alters conservation priorities for an endangered bird. PNAS 2018, 115, 8591–8596. [Google Scholar] [CrossRef]
- Anderson, C.D.; Epperson, B.K.; Fortin, M.-J.; Holderegger, R.; James, P.M.A.; Rosenberg, M.S.; Scribner, K.T.; Spear, S. Considering spatial and temporal scale in landscape-genetic studies of gene flow. Mol. Ecol. 2010, 19, 3565–3575. [Google Scholar] [CrossRef] [PubMed]
- Vandergast, A.G.; Kus, B.E.; Preston, K.L.; Barr, K.R. Distinguishing recent dispersal from historical genetic connectivity in the coastal California gnatcatcher. Sci. Rep. 2019, 9, 1355. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. Isolation by distance under diverse systems of mating. Genetics 1946, 31, 39–59. [Google Scholar] [CrossRef]
- Avise, J.C. Molecular Markers, Natural History, and Evolution, 2nd ed.; Sinauer Associates, Inc.: Sunderland, MA, USA, 2004. [Google Scholar]
- Chabot, A.A.; Hobson, K.A.; van Wilgenburg, S.L.; McQuat, G.J.; Lougheed, S.C. Advances in linking wintering migrant birds to their breeding-ground origins using combined analyses of genetic and stable isotope markers. PLoS ONE 2012, 7, e43627. [Google Scholar] [CrossRef] [PubMed]
- Ruegg, K.C.; Anderson, E.C.; Paxton, K.L.; Apkenas, V.; Lao, S.; Siegel, R.B.; DeSante, D.F.; Moore, F.; Smith, T.B. Mapping migration in a songbird using high-resolution genetic markers. Mol. Ecol. 2014, 23, 5726–5739. [Google Scholar] [CrossRef] [PubMed]
- Luikart, G.; England, P.; Tallmon, D.; Jordan, S.; Taberlet, P. The power and promise of population genomics: From genotyping to genome typing. Nat Rev Genet 2003, 4, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Garner, B.A.; Hand, B.K.; Amish, S.J.; Bernatchez, L.; Foster, J.T.; Miller, K.M.; Morin, P.A.; Narum, S.R.; O’Brien, S.J.; Roffler, G.; et al. Genomics in conservation: Case studies and bridging the gap between data and application. Trends Ecol. Evol. 2016, 31, 81–83. [Google Scholar] [CrossRef]
- Baldassarre, G. Ducks, Geese, and Swans of North America; John Hopkins University Press: Baltimore, MD, USA, 2014. [Google Scholar]
- Ely, C.R.; Takekawa, J.Y. Geographic variation in migratory behavior of greater white-fronted geese (Anser albifrons). Auk 1996, 113, 889–901. [Google Scholar] [CrossRef][Green Version]
- Ely, C.R.; Nieman, D.J.; Alisauskas, R.T.; Schmutz, J.A.; Hines, J.E. Geographic variation in migration chronology and winter distribution of midcontinent greater white-fronted geese. J. Wildl. Manag. 2013, 77, 1182–1191. [Google Scholar] [CrossRef]
- Li, X.; Si, Y.; Ji, L.; Gong, P. Dynamic response of East Asian greater white-fronted geese to changes of environment during migration: Use of multi-temporal species distribution model. Ecol. Model. 2017, 360, 70–79. [Google Scholar] [CrossRef]
- Deng, X.; Zhao, Q.; Fang, L.; Xu, Z.; Wang, X.; He, H.; Cao, L.; Fox, A.D. Spring migration duration exceeds that of autumn migration in Far East Asian greater white-fronted geese (Anser albifrons). Avian Res. 2019, 10, 19. [Google Scholar] [CrossRef]
- Kölzsch, A.; Müskens, G.J.D.M.; Szinai, P.; Moonen, S.; Glazov, P.; Kruckenberg, H.; Wikelski, M.; Nolet, B.A. Flyway connectivity and exchange primarily driven by moult migration in geese. Mov. Ecol. 2019, 7, 3. [Google Scholar] [CrossRef]
- Ely, C.R.; Wilson, R.E.; Talbot, S.L. Genetic structure among greater white-fronted goose populations of the Pacific Flyway. Ecol. Evol. 2017, 7, 2956–2968. [Google Scholar] [CrossRef]
- Wilson, R.E.; Ely, C.R.; Talbot, S.L. Flyway structure in a circumpolar greater white-fronted goose. Ecol. Evol. 2018, 8, 8490–8507. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.D.; Madsen, J.; Boyd, H.; Kuijken, E.; Norriss, D.W.; Tombre, I.M.; Stroud, D.A. Effects of agricultural change on abundance, fitness components and distribution of two arctic-nesting goose populations. Glob. Change Biol. 2005, 11, 881–893. [Google Scholar] [CrossRef]
- Fan, Y.; Zhou, L.; Cheng, L.; Song, Y.; Xu, W. Foraging behavior of the greater white-fronted goose (Anser albifrons) wintering at Shengjin Lake: Diet shifts and habitat use. Avian Res. 2020, 11, 3. [Google Scholar] [CrossRef]
- Warren, S.M.; Fox, A.D.; Walsh, A.; O’Sullivan, P. Extended parent-offspring relationships amongst the Greenland white-fronted goose (Anser albifrons flavirostris). Auk 1993, 110, 145–148. [Google Scholar]
- Ely, C.R. Family stability in greater white-fronted geese. Auk 1993, 110, 425–435. [Google Scholar] [CrossRef]
- Weegman, M.D.; Bearhop, S.; Hilton, G.; Walsh, A.J.; Weegman, K.M.; Hodgson, D.J.; Fox, A.D. Should I stay or should I go? Fitness costs and benefits of prolonged parent-offspring and sibling associations in an Arctic-nesting goose population. Oecologia 2016, 181, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Ens, B.J.; Choudhury, C.; Black, J.M. Mate fidelity and divorce in monogamous birds. In Partnerships in Birds: The Study of Monogamy; Black, J.M., Ed.; University Press: Oxford, UK, 1996; pp. 344–401. [Google Scholar]
- Kölzsch, A.; Flack, A.; Müskens, G.J.D.M.; Kruckenberg, H.; Glazov, P.; Wikelski, M. Goose parents lead migration V. J Avian Biol. 2020, 51. [Google Scholar] [CrossRef]
- Berdahl, A.M.; Ka, A.B.; Flack, A.; Westley, P.; Codling, E.A.; Couzin, I.D.; Dell, A.I.; Biro, D. Collective animal navigation and migratory culture: From theoretical models to empirical evidence. Philosophical transactions of the Royal Society of London. Ser. B Biol. Sci. 2018, 373, 20170009. [Google Scholar] [CrossRef]
- Szczys, P.; Oswald, S.A.; Arnold, J.M. Conservation implications of long-distance migration routes: Regional metapopulation structure, asymmetrical dispersal, and population declines. Biol. Conserv. 2017, 209, 263–272. [Google Scholar] [CrossRef]
- Deng, X.; Zhao, Q.; Solovyeva, D.; Lee, H.; Bysykatova-Harmey, I.; Xu, Z.; Ushiyama, K.; Shimada, T.; Koyama, K.; Park, J.; et al. Contrasting trends in two East Asian populations of the greater white-fronted goose Anser albifrons. Wildfowl Spec. Issue 2020, 6, 181–205. [Google Scholar]
- Fox, A.D.; Weegman, M.; Bearhop, S.; Hilton, G.; Griffin, L.; Stroud, D.A.; Walsh, A.J. Climate change and contrasting plasticity in timing of passage in a two-step migration episode of an arctic-nesting avian herbivore. Curr. Zool. 2014, 60, 233–242. [Google Scholar] [CrossRef]
- Boyd, H.; Fox, A.D. Effects of climate change on the breeding success of white-fronted geese Anser albifrons flavirostris in West Greenland. Wildfowl 2008, 58, 55–70. [Google Scholar]
- Weegman, M.D.; Fox, A.D.; Hilton, G.M.; Hodgson, D.J.; Walsh, A.J.; Griffin, L.R.; Bearhop, S. Diagnosing the decline of the Greenland white-fronted goose using population and individual level techniques. Wildfowl 2017, 67, 3–18. [Google Scholar]
- Fox, A.D.; Leafloor, J.O. (Eds.) A Global Audit of the Status and Trends of Arctic and Northern Hemisphere Goose Populations (Component 2: Population accounts); Conservation of Arctic Flora and Fauna International Secretariat: Akureyri, Iceland, 2018. [Google Scholar]
- Yparraguirre, D.R.; Sanders, T.A.; Weaver, M.L.; Skalos, D.A. Abundance of Tule Geese Anser albifrons elgasi in the Pacific Flyway 2003–2019. Wildfowl 2020, 70, 30–56. [Google Scholar]
- Weegman, M.D.; Bearhop, S.; Hilton, G.; Walsh, A.J.; Fox, A.D. Conditions during adulthood affect cohort-specific reproductive success in an Arctic nesting goose population. PeerJ 2016, 4, e2044. [Google Scholar] [CrossRef]
- Stroud, D.A.; Fox, A.D.; Urquhart, C.; Francis, I.S. (Compiler) International Single Species Action Plan for the Conservation of the Greenland White-fronted Goose Anser albifrons flavirostris, 2012–2022; AEWA Technical Series No. 45; AEWA: Bonn, Germany, 2012. [Google Scholar]
- Krechmar, A.V.; Kondratyev, A.V. Waterfowl Birds of North-East Asia; NESC FEV RAS: Magadan, Russia, 2006. (In Russian) [Google Scholar]
- BirdLife International. Anser albifrons. Greater White-Fronted Goose. Available online: https://www.iucnredlist.org/species/22679881/85980652 (accessed on 16 September 2022).
- Ely, C.R.; Fox, A.D.; Alisaukas, R.T.; Andreev, A.; Bromley, R.G.; Degtyarev, A.G.; Ebbinge, B.; Gurtovaya, E.N.; Kerbes, R.; Kondratyev, A.V.; et al. Circumpolar variation in morphological characteristics of greater white-fronted geese (Anser albifrons). Bird Study 2005, 52, 104–119. [Google Scholar] [CrossRef]
- Anderson, M.G.; Rhymer, J.M.; Rohwer, F.C. Philopatry, dispersal, and the genetic structure of waterfowl populations. In Ecology and Management of Breeding Waterfowl; Batt, B.D.J., Afton, A.D., Anderson, M.G., Ankney, C.D., Johnson, D.H., Kadlec, J.A., Krapu, G.L., Eds.; University of Minnesota Press: Minneapolis, MN, USA, 1992; pp. 365–395. [Google Scholar]
- Lamb, J.S.; Paton, P.W.; Osenkowski, J.E.; Badzinksi, S.S.; Berlin, A.M.; Bowman, T.; Dwyer, C.; Fara, L.J.; Gilliland, S.G.; Kenow, K.; et al. Spatially explicit network analysis reveals multi-species annual cycle movement patterns of sea ducks. Ecol. Appl. 2019, 29, e01919. [Google Scholar] [CrossRef]
- Bolton, P.E.; West, A.J.; Cardilini, A.P.A.; Clark, J.A.; Maute, K.L.; Legge, S.; Brazill-Boast, J.; Griffith, S.C.; Rollins, L.A. Three molecular markers show no evidence of population genetic structure in the gouldian finch (Erythrura gouldiae). PLoS ONE 2016, 11, e0167723. [Google Scholar] [CrossRef]
- Sonsthagen, S.A.; Wilson, R.E.; Lavretsky, P.; Talbot, S.L. Coast to coast: High genomic connectivity in North American scoters. Ecol. Evol. 2019, 9, 7246–7261. [Google Scholar] [CrossRef]
- Supple, M.A.; Shapiro, B. Conservation of biodiversity in the genomics era. Genome Biol. 2018, 19, 131. [Google Scholar] [CrossRef]
- Bohling, J.; Small, M.; Von Bargen, J.; Louden, A.; DeHaan, P. Comparing inferences derived from microsatellite and RADseq datasets: A case study involving threatened bull trout. Conserv. Genet. 2019, 20, 329–342. [Google Scholar] [CrossRef]
- Rexer-Huber, K.; Veale, A.J.; Catry, P.; Cherel, Y.; Dutoit, L.; Foster, Y.; McEwan, J.C.; Parker, G.C.; Phillips, R.A.; Ryan, P.G.; et al. Genomics detects population structure within and between ocean basins in a circumpolar seabird: The white-chinned petrel. Mol. Ecol. 2019, 28, 4552–4572. [Google Scholar] [CrossRef] [PubMed]
- Stork, V.L.; Nason, J.; Campbell, D.R.; Fernandez, J.F. Landscape approaches to historical and contemporary gene flow in plants. Trends Ecol. Evol. 1999, 14, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.D.; Glahder, C.M.; Walsh, A.J. Spring migration routes and timing of Greenland white-fronted geese—results from satellite telemetry. OIKOS 2003, 103, 415–425. [Google Scholar] [CrossRef]
- Banks, R.C. Taxonomy of greater white-fronted geese (Aves: Anatidae). Proc. Biol. Soc. Wash. 2011, 124, 226–233. [Google Scholar] [CrossRef]
- Cunningham, S.A.; Zhao, Q.; Weegman, M.D. Increased rice flooding during winter explains the recent increase in the Pacific Flyway white-fronted goose Anser albifrons frontalis population in North America. Ibis 2021, 163, 231–246. [Google Scholar] [CrossRef]
- Alisauskas, R.T.; Fischer, J.B.; Leafloor, J.O. C5 Midcontinent Greater White-fronted Goose Anser albifrons frontalis. In A Global Audit of the Status and Trends of Arctic and Northern Hemisphere Goose Populations (Component 2: Population Accounts); Fox, A.D., Leafloor, J.O., Eds.; Conservation of Arctic Flora and Fauna International Secretariat: Akureyri, Iceland, 2018; pp. 32–34. [Google Scholar]
- Ebbinge, B.S.; Koffijberg, K.; Kruckenberg, H.; Rozenfeld, S.B.; Glazov, P.M.; Kondratyev, A. C1 Greater white-fronted goose Anser albifrons albifrons. In A Global Audit of the Status and Trends of Arctic and Northern Hemisphere Goose Populations (Component 2: Population accounts); Fox, A.D., Leafloor, J.O., Eds.; Conservation of Arctic Flora and Fauna International Secretariat: Akureyri, Iceland, 2018; pp. 23–25. [Google Scholar]
- Fox, A.D.; Francis, I.S.; Walsh, A.J.; Norriss, D.; Kelly, S. Report of the 2020/2021 International Census of Greenland White-fronted Geese. Greenland White-fronted Goose Study, Rønde Denmark and National Parks & Wildlife Service, Wexford Ireland. 2021. Available online: https://monitoring.wwt.org.uk/wp-content/uploads/2021/11/Greenland-White-fronted-Goose-Study-report-2020-21.pdf (accessed on 16 September 2022).
- Central, Mississippi, Pacific Flyway Councils. Management Plan for Midcontinent Greater White-Fronted Goose; U.S. Fish and Wildlife Service: Washington, WA, USA, 2015. [Google Scholar]
- Fujioka, M.; Lee, S.D.; Kurechi, M. Bird use of rice fields in Korea and Japan. Waterbirds 2011, 33, 8–29. [Google Scholar] [CrossRef]
- Kim, M.K.; Lee, S.; Lee, S.D. Habitat use and its implications for the conservation of the overwintering populations of bean goose Anser fabalis and greater white-fronted goose, A. albifrons in South Korea. Ornithol. Sci. 2016, 15, 141–149. [Google Scholar] [CrossRef]
- Yu, H.; Wang, X.; Cao, L.; Zhang, L.; Jia, Q.; Lee, H.; Xu, Z.; Liu, G.; Xu, W.; Hu, B.; et al. Are declining populations of wild geese in China “prisoners” of their natural habitats? Curr. Biol. 2017, 27, R376–R377. [Google Scholar] [CrossRef]
- Zhao, Q.; Cao, L.; Wang, X.; Fox, A.D. Why Chinese wintering geese hesitate to exploit farmland. Ibis 2018, 160, 703–705. [Google Scholar] [CrossRef]
- Weegman, M.D.; Walsh, A.J.; Ogilvie, M.A.; Bearhop, S.; Hilton, G.M.; Hodgson, D.J.; Fox, A.D. Adult survival and per-capita production of young explain dynamics of a long-lived goose population. Ibis 2022, 19, 2097–2101. [Google Scholar] [CrossRef]
- DaCosta, J.M.; Sorenson, M.D. Amplification biases and consistent recovery of loci in a double-digest RAD-seq protocol. PLoS ONE 2014, 9, e106713. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 14, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Malinsky, M.; Trucchi, E.; Lawson, D.J.; Falush, D. RADpainter and fineRADstructure: Population Inference from RADseq Data. Mol. Biol. Evol. 2018, 35, 1284–1290. [Google Scholar] [CrossRef]
- Petkova, D.; Novembre, J.; Stephens, M. Visualizing spatial population structure with estimated effective migration surfaces. Nat. Genet. 2016, 48, 94–100. [Google Scholar] [CrossRef]
- Novembre, J.; Stephens, M. Interpreting principal components analyses of spatial population genetic variation. Nat. Genet. 2008, 40, 646–649. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Jones, O.; Wang, J. COLONY: A program for parentage and sibship inference from multilocus genotype data. Mol. Ecol. Resour. 2010, 10, 551–555. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Alexander, D.; Lange, K. A quasi-Newton acceleration for high-dimensional optimization algorithms. Stat. Comput. 2011, 21, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Janes, J.K.; Miller, J.M.; Dupuis, J.R.; Malenfant, R.M.; Gorrell, J.C.; Cullingham, C.I.; Andrew, R.l. The K=2 conundrum. Mol. Ecol. 2017, 26, 3594–3602. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.M. pophelper: An R package and web app to analyse and visualise population structure. Mol. Ecol. Resour. 2016, 17, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G.M. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 183–195. [Google Scholar] [CrossRef]
- Ploeger, P.L. Geographical differentiation in Arctic Anatidae as a result of the isolation during the Last Glacial. Ardea 1968, 56, 1–159. [Google Scholar]
- Lanier, H.C.; Guderson, A.M.; Weksler, M.; Fedorov, V.B.; Olson, L.E. Comparative phylogeography highlights the double-edged sword of climate change faced by arctic and alpine-adapted mammals. PLoS ONE 2015, 10, E0118396. [Google Scholar] [CrossRef]
- Sonsthagen, S.A.; Wilson, R.E.; Talbot, S.L. Species-specific responses to landscape features shaped genomic structure within Alaska galliformes. J. Biogeogr. 2022, 49, 261–273. [Google Scholar] [CrossRef]
- Carrara, P.E.; Ager, T.A.; Baichtal, J.F. Possible refugia in the Alexander Archipelago of southeastern Alaska during the late Wisconsin glaciation. Can. J. Earth Sci. 2007, 44, 229–244. [Google Scholar] [CrossRef]
- Fedorov, V.B.; Trucchi, E.; Goropashnaya, A.V.; Waltari, E.; Whidden, S.E.; Stenseth, N.C. Impact of past climate warming on genomic diversity and demographic history of collared lemmings across the Eurasian Arctic. Proc. Natl. Acad. Sci. USA 2020, 117, 3026–3033. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.D. The Greenland White-Fronted Goose (Anser albifrons flavirostris): The Annual Cycle of a Migratory Herbivore on the European Continental Fringe. Ph.D. Thesis, National Environmental Research Institute, Kalø, Denmark, 2003. [Google Scholar]
- Gotfredsen, A.B. Former occurrences of geese (Genera Anser and Branta) in ancient West Greenland: Morphological and biometric approaches. Acta Zool. Crac. 2002, 45, 179–204. [Google Scholar]
- Margold, M.; Stokes, C.R.; Clark, C.D. Reconciling records of ice streaming and ice margin retreat to produce a palaeogeographic reconstruction of the deglaciation of the Laurentide Ice Sheet. Quat. Sci. Rev. 2018, 189, 1–30. [Google Scholar] [CrossRef]
- Buehler, D.M.; Baker, A.J.; Piersma, T. Reconstructing paleoflyways of late Pleistocene and early Holocene red knot Calidris canutus. Ardea 2006, 94, 485–498. [Google Scholar]
- Ketterson, E.D.; Fudickar, A.M.; Atwell, J.W.; Greives, T.J. Seasonal timing and population divergence: When to breed, when to migrate. Curr. Opin. Behav. Sci. 2015, 6, 50–58. [Google Scholar] [CrossRef]
- Hendry, A.P.; Day, T. Population structure attributable to reproductive time: Isolation by time and adaptation by time. Mol. Ecol. 2015, 14, 901–916. [Google Scholar] [CrossRef]
- Friesen, V.L.; Smith, A.L.; Gómez-Díaz, E.; Bolton, M.; Furness, R.W.; González-Solís, J.; Monteiro, L.R. Sympatric speciation by allochrony in a seabird. Proc. Natl. Acad. Sci. USA 2007, 104, 18589–18594. [Google Scholar] [CrossRef]
- Bauer, S.; Lisovski, S.; Hahn, S. Timing is crucial for consequences of migratory connectivity. Oikos 2016, 125, 605–612. [Google Scholar] [CrossRef]
- Warren, S.M.; Fox, A.; Walsh, A.; O’Sullivan, P. Age of first pairing and breeding among Greenland white-fronted geese. Condor 1992, 94, 791–793. [Google Scholar] [CrossRef]
- Ely, C.R.; Dzubin, A.X. Greater white-fronted goose (Anser albifrons). In The Birds of North America; Poole, A.A., Gill, F., Eds.; The Academy of Natural Sciences, Philadelphia, and The American Ornithologists’ Union: Washington, DC, USA, 1994. [Google Scholar]
- Van der Jeugd, H.P. Large barnacle goose males can overcome the social costs of natal dispersal. Behav. Ecol. 2001, 12, 275–282. [Google Scholar] [CrossRef]
- Szipl, G.; Depenau, M.; Kotrschal, K.; Hemetsberger, J.; Frigerio, D. Costs and benefits of social connectivity in juvenile greylag geese. Sci. Rep. 2019, 9, 12839. [Google Scholar] [CrossRef]
- Elgas, B. Breeding populations of Tule white-fronted geese in Northwestern Canada. Wilson Bull. 1970, 82, 420–426. [Google Scholar]
- Fowler, A.C.; Eadie, J.M.; Ely, C.R. Relatedness and nesting dispersion within breeding populations of greater white-fronted geese. Condor 2004, 106, 600–607. [Google Scholar] [CrossRef]
- Wilson, R.E.; Sonsthagen, S.A.; DaCosta, J.M.; Ely, C.R.; Sorenson, M.D.; Talbot, S.L. Identification of single Nucleotide Polymorphisms for Use in a Genetic Stock Identification System for Greater White-Fronted Goose (Anser albifrons) Subspecies Wintering in California: U.S. Geological Survey Open-File Report 2019–1040; U.S. Geological Survey: Reston, VA, USA, 2019; p. 18. [Google Scholar]
- Cohen, J.; Screen, J.; Furtado, J.; Barlow, M.; Whittleston, D.; Coumou, D.; Francis, J.; Dethloff, K.; Entekhabi, D.; Overland, J.; et al. Recent Arctic amplification and extreme mid-latitude weather. Nat. Geosci. 2014, 7, 627–637. [Google Scholar] [CrossRef]
- Ward, D.H.; Dau, C.P.; Tibbitts, T.L.; Sedinger, J.S.; Anderson, B.A.; Hines, J.E. Change in abundance of Pacific brant wintering in Alaska: Evidence of a climate warming effect? Arctic 2009, 62, 301–311. [Google Scholar] [CrossRef]
- Jonker, R.M.; Kraus, R.H.; Zhang, Q.; van Hooft, P.; Larsson, K.; van der Jeugd, H.P.; Kurvers, R.H.; van Wieren, S.E.; Loonen, M.J.; Crooijmans, R.P.; et al. Genetic consequences of breaking migratory traditions in barnacle geese Branta leucopsis. Mol. Ecol. 2013, 22, 5835–5847. [Google Scholar] [CrossRef] [PubMed]
- Weegman, M.D.; Alisauskas, R.T.; Kellett, D.K.; Zhao, Q.; Wilson, S.; Telenský, T. Local population collapse of Ross’s and lesser snow geese driven by failing recruitment and diminished philopatry. OIKOS 2022, 5, e09184. [Google Scholar] [CrossRef]
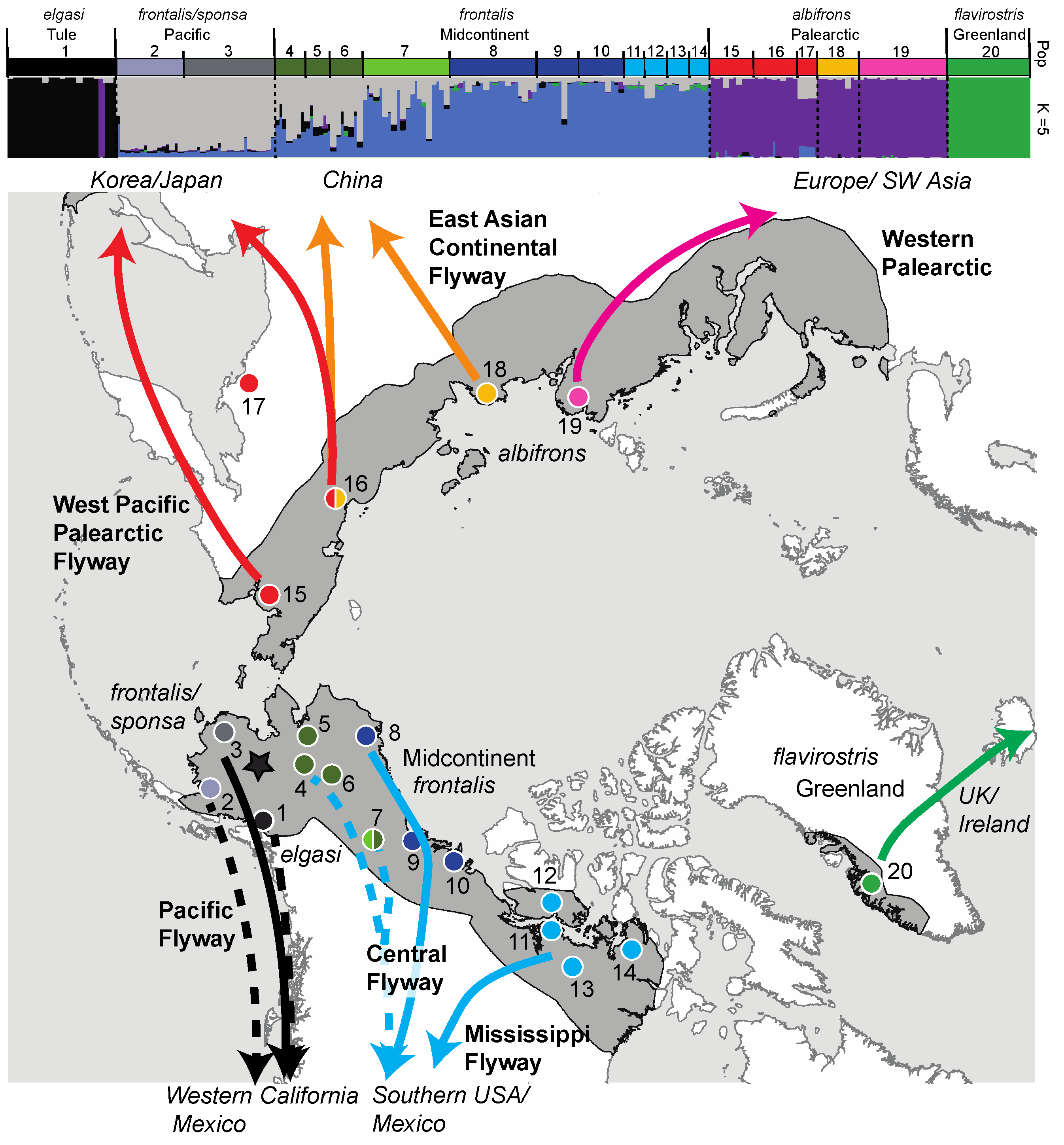
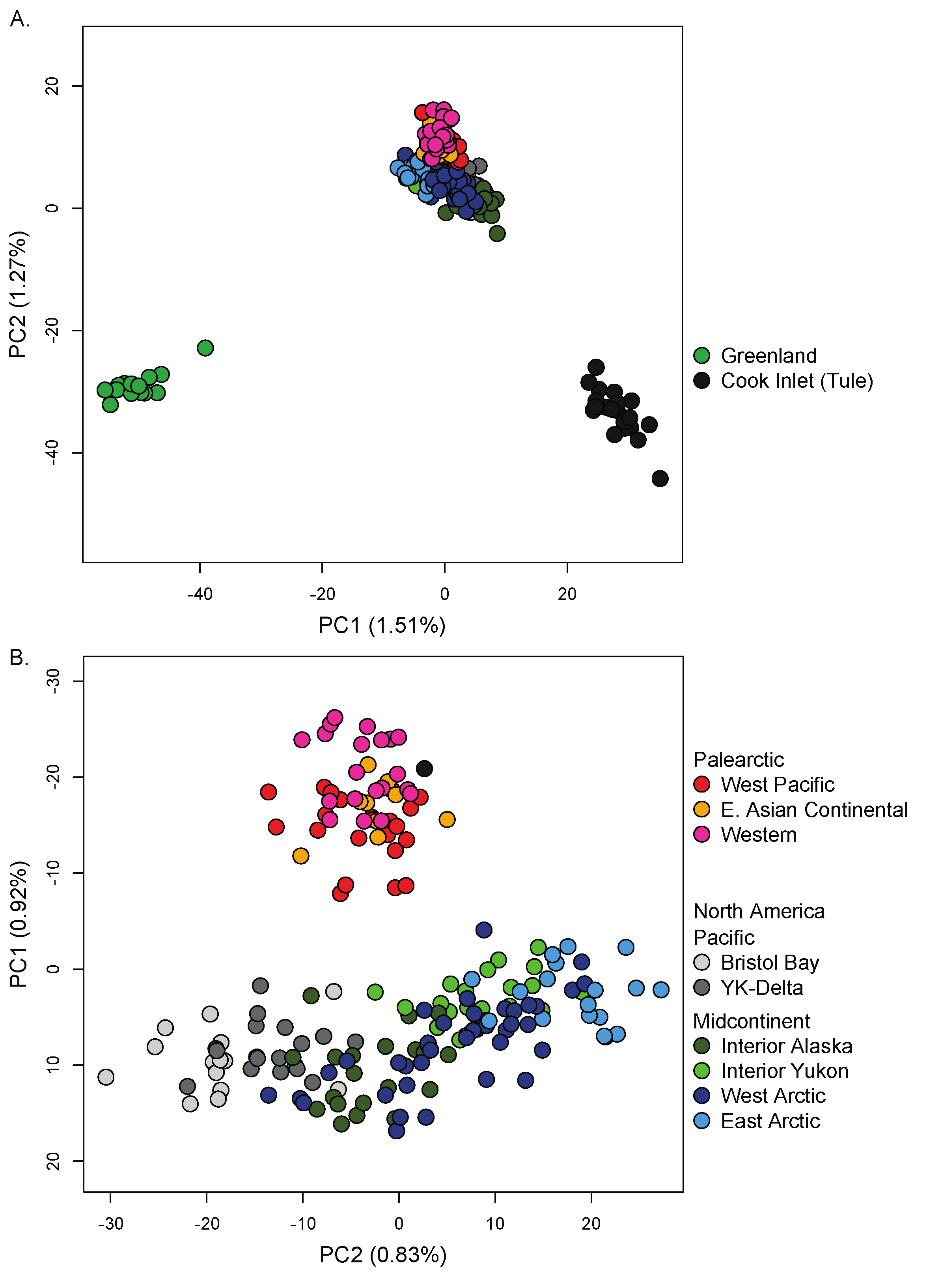
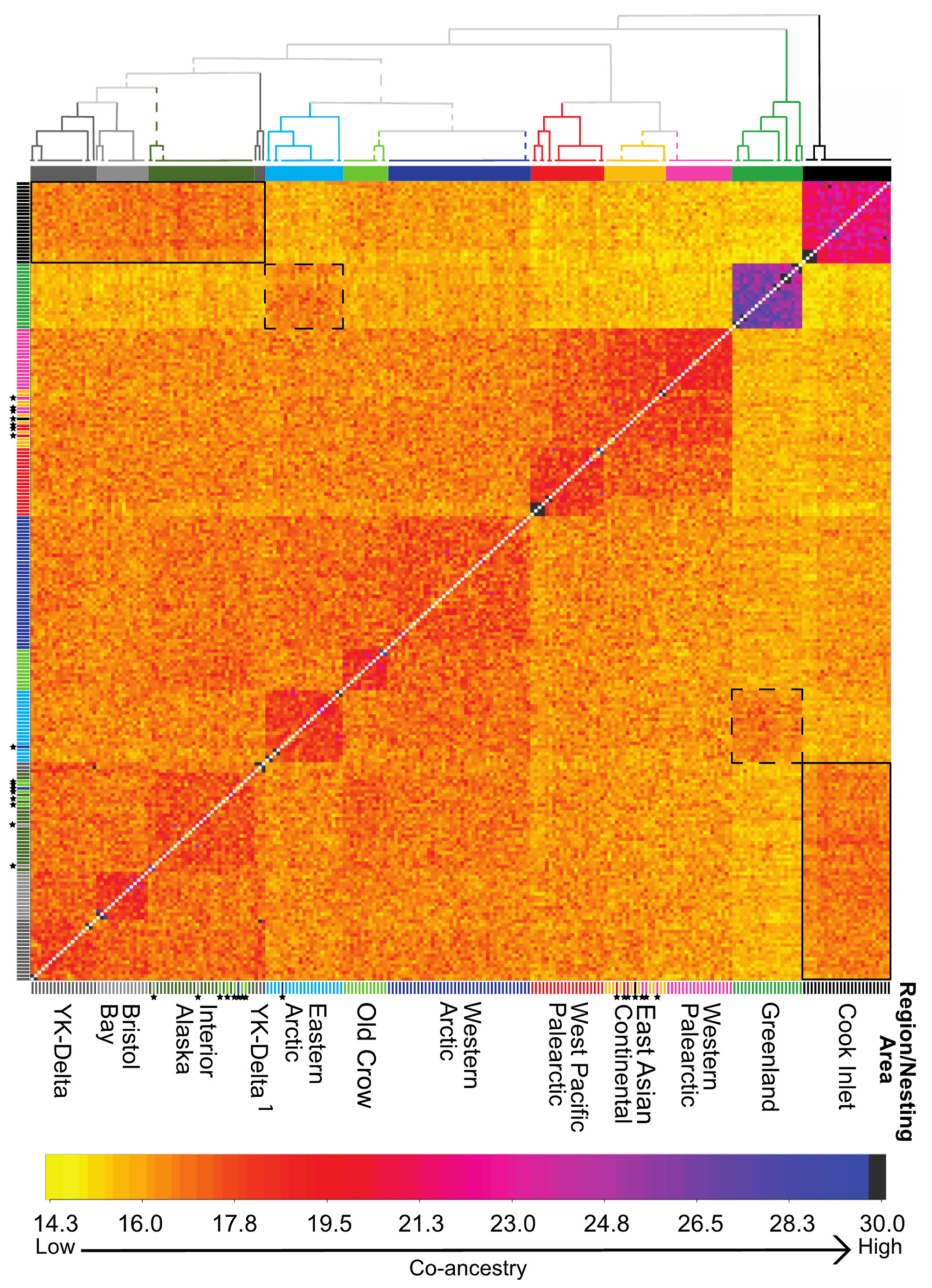
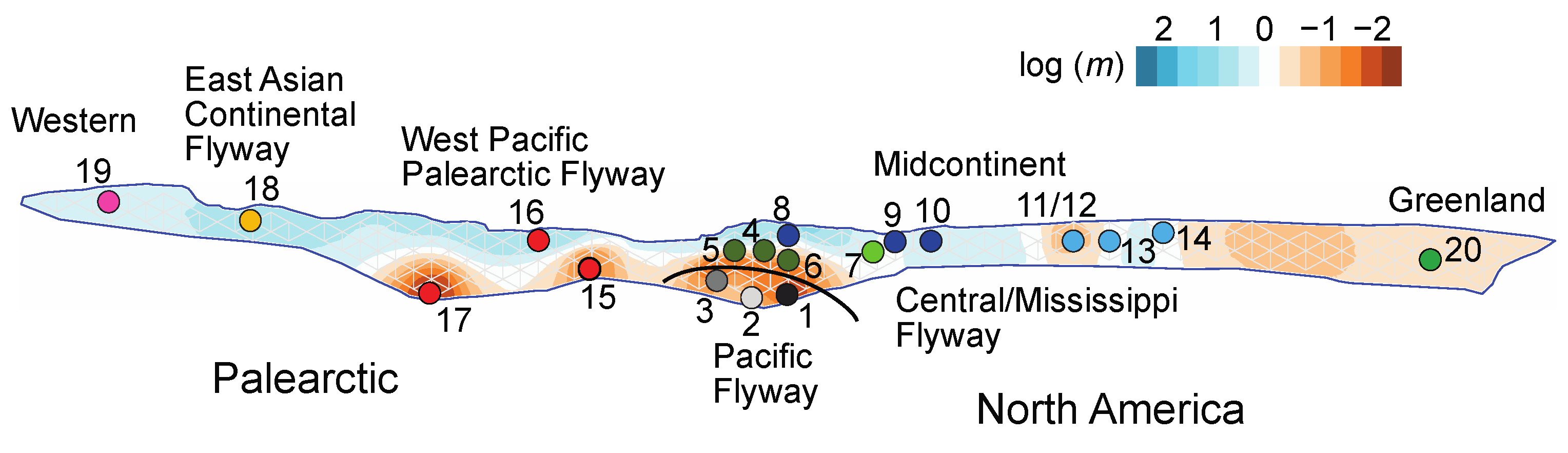
| Figure 1 Map # | Continent | Flyway | Region | Nesting Area † Taiga, * Tundra | Wintering Area | Management Unit Name and Population Size (year)source | Conservation Concerns | Subspecies | N |
|---|---|---|---|---|---|---|---|---|---|
| 1 | North America | Pacific | - | USA (Alaska): Cook Inlet † | USA: California | “Tule” ~15,000 (2019) 1 | Small stable population, poor monitoring, hunting | elgasi | 25 |
| 2 | North America | Pacific | - | USA (Alaska): Bristol Bay a,† | Western Mexico | “Pacific” 730,000 (2018) 2 | Increasing, low concern, over- abundance increasing causing conflict with “Tule” | frontalis/sponsabc | 15 |
| 3 | North America | Pacific | - | USA (Alaska): Yukon-Kuskokwim Delta * | USA: California | frontalis/sponsabc | 21 | ||
| 4 | North America | Central | Interior | USA (Alaska): Koyukuk † | USA: southern interior and Mexico | “Midcontinent” 2,000,000–3,000,000 (2016) 3 | Increasing, low concern | frontalis | 7 |
| 5 | North America | Central | Interior | USA (Alaska): Selawik † | USA: southern interior and Mexico | frontalis | 6 | ||
| 6 | North America | Central | Interior | USA (Alaska): Kanuti † | USA: southern interior and Mexico | frontalis | 7 | ||
| 7 | North America | Central | Interior | Canada (Yukon): Old Crow † | USA: southern interior and Mexico | frontalis | 20 | ||
| 8 | North America | Central | Western Arctic | USA (Alaska): North Slope * | USA: southern interior and Mexico | frontalis | 20 | ||
| 9 | North America | Central | Western Arctic | Canada (Northwest Territories): MacKenzie River Delta * | USA: southern interior and Mexico | frontalis | 10 | ||
| 10 | North America | Central | Western Arctic | Canada (Northwest Territories): Anderson River * | USA: southern interior and Mexico | frontalis | 10 | ||
| 11 | North America | Central/Mississippi | Eastern Arctic | Canada (Nunavut): Kiillinnguyaq * | USA: southern interior and Mexico | frontalis | 5 | ||
| 12 | North America | Central/Mississippi | Eastern Arctic | Canada (Nunavut): Victoria Island * | USA: southern interior and Mexico | frontalis | 5 | ||
| 13 | North America | Central/Mississippi | Eastern Arctic | Canada (Nunavut): Queen Maud Gulf * | USA: southern interior and Mexico | frontalis | 5 | ||
| 14 | North America | Central/Mississippi | Eastern Arctic | Canada (Nunavut): Rasmussen Basin * | USA: southern interior and Mexico | frontalis | 5 | ||
| 15 | Asia | West Pacific Palearctic | - | Russia: Anadyr Lowlands * | Korea and Japan | “West Pacific” 402,000–424,000 (2020) 4 | Increasing, low concern | albifrons | 10 |
| 16 | Asia | West Pacific Palearctic | - | Russia: Kolyma River Delta * | Korea, Japan, China | albifrons | 10 | ||
| 17 | Asia | West Pacific Palearctic | - | Russia: Magadan † | ? | ? | Data deficient | albifrons | 4 |
| 18 | Asia | East Asian Continental Palearctic | - | Russia: Lena River * | China | “East Asian Continental” 48,000 (2020) 4 | Decreasing thought due to wintering habitat loss | albifrons | 10 |
| 19 | Asia/ Europe | Western Palearctic | - | Russia: Kanin to Taimyr Peninsula * | Europe and SW Asia | “European” 1,400,000 5 | Over-abundance increasing causing conflict | albifrons | 20 |
| 20 | North America | Western Palearctic | Greenland | Greenland * | Ireland/United Kingdom | “Greenland” 20,200 6 | Depressed reproductive success (climate related) failing to balance natural mortality | flavirostris | 19 |
| Western Palearctic | East Asian Continental Palearctic | West Pacific Palearctic | Greenland | Midcontinent | Pacific | Cook Inlet Basin | |
|---|---|---|---|---|---|---|---|
| Western Palearctic | - | 0.041 | 0.184 | 0.103 | 0.076 | 0.160 | 0.474 |
| East Asian Continental Palearctic | 0.007 (0.251) | - | 0.248 | 0.204 | 0.084 | 0.249 | 0.599 |
| West Pacific Palearctic | 0.007 (0.351) | 0.009 (0.515) | - | 0.237 | 0.111 | 0.181 | 0.159 |
| Greenland | 0.053 (0.610) | 0.057 (0.593) | 0.057 (0.711) | - | 0.060 | 0.135 | 0.524 a |
| Midcontinent | 0.007 (0.381) | 0.009 (0.689) | 0.008 (0.214) | 0.046 (0.663) | - | 0.134 | 0.248 |
| Pacific | 0.010 (0.485) | 0.011 (0.476) | 0.010 (0.535) | 0.052 (0.574) | 0.005 (0.153) | - | 0.303 |
| Cook Inlet Basin (Tule goose) | 0.036 (0.487) | 0.038 (0.523) | 0.035 (0.539) | 0.078 (0.698) | 0.025 (0.432) | 0.027 (0.302) | - |
| Continent | Management Unit (Nesting Region/Area) | mtDNA | ddRAD-Seq |
|---|---|---|---|
| Palearctic | |||
| West Pacific Palearctic | 0.0198 (0.0104) | 0.0065 (0.0067) | |
| East Asian Continental Palearctic | 0.0047 (0.0034) a | 0.0063 (0.0066) | |
| Western Palearctic | 0.0154 (0.0084) a | 0.0066 (0.0066) | |
| Greenland | |||
| Greenland | 0.0084 (0.0050) a | 0.0058 (0.0067) | |
| North America | Midcontinent | 0.0188 (0.0098) | 0.0066 (0.0066) |
| Interior Alaska | 0.0206 (0.0107) | 0.0064 (0.0066) | |
| Interior Yukon/Old Crow | 0.0186 (0.0102) a | 0.0065 (0.0066) | |
| Western Arctic | 0.0156 (0.0084) | 0.0065 (0.0067) | |
| Eastern Arctic | 0.0143 (0.0077) | 0.0064 (0.0067) | |
| Pacific | 0.0186 (0.0099) | 0.0066 (0.0067) | |
| Bristol Bay | 0.0109 (0.0064) a | 0.0064 (0.0068) | |
| Yukon-Kuskokwim Delta | 0.0217 (0.0116) a | 0.0065 (0.0068) | |
| Cook Inlet Basin (Tule goose) | 0.0094 (0.0054) a | 0.0063 (0.0068) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilson, R.E.; Sonsthagen, S.A.; DaCosta, J.M.; Sorenson, M.D.; Fox, A.D.; Weaver, M.; Skalos, D.; Kondratyev, A.V.; Scribner, K.T.; Walsh, A.; et al. As the Goose Flies: Migration Routes and Timing Influence Patterns of Genetic Diversity in a Circumpolar Migratory Herbivore. Diversity 2022, 14, 1067. https://doi.org/10.3390/d14121067
Wilson RE, Sonsthagen SA, DaCosta JM, Sorenson MD, Fox AD, Weaver M, Skalos D, Kondratyev AV, Scribner KT, Walsh A, et al. As the Goose Flies: Migration Routes and Timing Influence Patterns of Genetic Diversity in a Circumpolar Migratory Herbivore. Diversity. 2022; 14(12):1067. https://doi.org/10.3390/d14121067
Chicago/Turabian StyleWilson, Robert E., Sarah A. Sonsthagen, Jeffrey M. DaCosta, Michael D. Sorenson, Anthony D. Fox, Melanie Weaver, Dan Skalos, Alexander V. Kondratyev, Kim T. Scribner, Alyn Walsh, and et al. 2022. "As the Goose Flies: Migration Routes and Timing Influence Patterns of Genetic Diversity in a Circumpolar Migratory Herbivore" Diversity 14, no. 12: 1067. https://doi.org/10.3390/d14121067
APA StyleWilson, R. E., Sonsthagen, S. A., DaCosta, J. M., Sorenson, M. D., Fox, A. D., Weaver, M., Skalos, D., Kondratyev, A. V., Scribner, K. T., Walsh, A., Ely, C. R., & Talbot, S. L. (2022). As the Goose Flies: Migration Routes and Timing Influence Patterns of Genetic Diversity in a Circumpolar Migratory Herbivore. Diversity, 14(12), 1067. https://doi.org/10.3390/d14121067







