Machine-Learning-Based Proteomic Predictive Modeling with Thermally-Challenged Caribbean Reef Corals
Abstract
:1. Introduction
2. Materials and Methods
2.1. Overview of Methods and Justification
2.2. The Experiment and Field Site Climatology
2.3. Sample Designations
2.4. Protein Extraction
2.5. Protein Quality Control
2.6. iTRAQ
2.7. Mass Spectrometry Data Processing
2.8. Mass Spectrometry Data Quality Control
| Quality Control Step | #Host Peptides (% of Previous Step) | #Endosymbiont Peptides (% of Previous Step) | #Host + Endosymbiont Peptides (% of Previous Step) | Host/Endosymbiont Ratio |
|---|---|---|---|---|
| Total sequenced peptides | 17,553 | 19,509 * | 37,062 (100%) | ~0.9:1 |
| Total unique peptides | 13,067 (74% *) | 13,335 (68%) | 26,412 (71%) | ~1.0:1 |
| Possessed iTRAQ tag | 3233 (25%) | 3531 (26% *) | 6764 (26%) | ~0.9:1 |
| Found in all three batches | 99 (3% *) | 68 (2%) | 167 (2.5%) | ~1.5:1 |
| Two peptides mapped to same protein | 46 (46%) | 40 (59%) | 86 (51.5%) | ~1.2:1 |
| Differentially concentrated proteins identified by response screening + stepwise discriminant analysis-derived “proteins of interest” | ||||
| 17 (40%) | 10 (25%) | 27 (31%) | ~1.7:1 | |
2.9. Data Analysis and Proteomic Predictive Modeling
3. Results and Discussion
3.1. Overview
3.2. Coral Responses to Elevated Temperatures
3.3. Proteomic Data Output and Descriptive Multivariate Analyses
3.4. Methodological Comparison
3.5. Differentially Concentrated Proteins
3.6. Stepwise Discriminant Analysis-Based Coral Biomarker Profiling
3.7. Proteomic Predictive Modeling of Coral Fragment Condition
| Comparison | df | #Proteins | Transcriptome Accession | Identity | Trend |
|---|---|---|---|---|---|
| Endosymbionts | |||||
| TIME | 1 | 2 | 1. SYMBOF_DN80090_c0_ | tyrosine decarboxylase 1-like | 5 > 31 days |
| 2. SYMBOF_DN177194_c0_g1 b | apolipoprotein B100 C terminal | 5 > 31 days | |||
| SITE | 2 | 3 | 3. SYMBOF_DN99804_c0_g1 a,b | sec34 sodium channel protein 11 | Cheeca Rocks > Little Conch |
| 4. SYMBOF_DN160797_c0_g1 b | unknown (OLQ08781.1) | The Rocks + Cheeca Rocks > Little Conch | |||
| 5. SYMBOF_DN194918_c0_g1 b | unknown | Little Conch + Cheeca Rocks > The Rocks | |||
| GENOTYPE | 5 | 1 * | SYMBOF_DN160797_c0_g1 b | unknown (see also endosymbiont #4.). | grey60 > lightyellow |
| FRAGMENT HEALTH DESIGNATION | 1 | 1 | 6. SYMBOF_DN107829_c0_g1 b | unknown (OLP80463.1) | healthy control > actively bleaching |
| Host coral | |||||
| SITE | 2 | 1 | 1. OFAVBQ_DN225385_c0_g1 c,d | sacsin | Cheeca Rocks > Little Conch + The Rocks |
| SHELF | 1 | 1 | 2. OFAVBQ_DN217378_c2_g1 | myosin 11-like | inshore > offshore |
| GENOTYPE | 5 | 1 * | OFAVBQ_DN217378_c2_g1 | myosin 11-like (see also host #2.) | grey60 + skyblue > lightyellow |
| FRAGMENT HEALTH DESIGNATION | 1 | 1 | 3. OFAVBQ_DN225503_c2_g1 | concanavalin A-like lectin/glucanase | actively bleaching > healthy control (1.5-fold) |
3.8. Proteomic Predictive Models of Colony Bleaching Susceptibility
3.9. Predicting Coral Colony Resilience with a Molecular Biology + Machine-Learning Approach
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, B.E. Coral bleaching: Causes and consequences. Coral Reefs 1997, 16, 129–138. [Google Scholar] [CrossRef]
- Grottoli, A.; Toonen, R.J.; van Woesik, R.; Vega-Thurber, R.; Warner, M.E.; McLachlan, R.; Price, J.T.; Bahr, K.D.; Baums, I.B.; Castillo, K.D.; et al. Increasing comparability among coral bleaching experiments. Ecol. Appl. 2021, 31, e02262. [Google Scholar] [CrossRef] [PubMed]
- McLachlan, R.H.; Price, J.; Solomon, S.; Grottoli, A.G. Thirty years of coral heat-stress experiments: A review of methods. Coral Reefs 2020, 39, 885–902. [Google Scholar] [CrossRef] [Green Version]
- Downs, C.A.; Mueller, E.; Phillips, S.; Fauth, J.E.; Woodley, C.M. A molecular biomarker system for assessing the health of coral (Montastrea faveolata) during heat stress. Mar. Biotechnol. 2020, 2, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, J.E.; Bartels, E.; Devlin-Durante, M.K.; Lustic, C.; Nedimyer, K.; Schopmeyer, S.; Lirman, D.; LaJeunesse, T.C.; Baums, I.B. Extensive transcriptional variation poses a challenge to thermal stress biomarker development for endangered coral. Mol. Ecol. 2018, 27, 1103–1119. [Google Scholar] [CrossRef]
- Mayfield, A.B.; Chen, C.S.; Dempsey, A.C. The molecular ecophysiology of closely related pocilloporid corals of New Caledonia. Platax 2017, 14, 1–45. [Google Scholar]
- Mayfield, A.B.; Chen, C.S.; Dempsey, A.C. Biomarker profiling in reef corals of Tonga’s Ha’apai and Vava’u Archipelagos. PLoS ONE 2017, 12, e0185857. [Google Scholar] [CrossRef] [Green Version]
- Mayfield, A.B.; Chen, C.S.; Dempsey, A.C. Identifying corals displaying aberrant behavior in Fiji’s Lau Archipelago. PLoS ONE 2017, 12, e0177267. [Google Scholar] [CrossRef] [Green Version]
- Mayfield, A.B.; Wang, L.H.; Tang, P.C.; Hsiao, Y.Y.; Fan, T.Y.; Tsai, C.L.; Chen, C.S. Assessing the impacts of experimentally elevated temperature on the biological composition and molecular chaperone gene expression of a reef coral. PLoS ONE 2011, 6, e26529. [Google Scholar] [CrossRef] [Green Version]
- Mayfield, A.B.; Aguilar, C.; Enochs, I.; Kolodziej, G.; Manzello, D.P. Shotgun proteomics of thermally challenged Caribbean reef corals. Front. Mar. Sci. 2021, 8, 660153. [Google Scholar] [CrossRef]
- Mayfield, A.B.; Wang, Y.B.; Chen, C.S.; Chen, S.H.; Lin, C.Y. Dual-compartmental transcriptomic+proteomic analysis of a marine endosymbiosis exposed to environmental change. Mol. Ecol. 2016, 25, 5944–5958. [Google Scholar] [CrossRef]
- Mayfield, A.B.; Chen, Y.J.; Lu, C.Y.; Chen, C.S. Exploring the environmental physiology of the Indo-Pacific reef coral Seriatopora hystrix using differential proteomics. Open. J. Mar. Sci. 2018, 8, 223–252. [Google Scholar] [CrossRef] [Green Version]
- Mayfield, A.B.; Chen, Y.J.; Lu, C.Y.; Chen, C.S. The proteomic response of the reef coral Pocillopora acuta to experimentally elevated temperature. PLoS ONE 2018, 13, e0192001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguilar, C.; Enochs, I.; Manzello, D.P.; Mayfield, A.B.; Kolodziej, G.; Carlton, R. Transcriptome profiling of thermotolerant corals of the Upper Florida Keys. Mol. Ecol. unpublished.
- Mayfield, A.B.; Chen, C.S. Enabling coral reef triage via molecular biotechnology and artificial intelligence. Platax 2019, 16, 23–47. [Google Scholar]
- Manzello, D.P.; Matz, M.V.; Enochs, I.C.; Valentino, L.; Carlton, R.D.; Kolodziej, G.; Serrano, X.; Towle, E.K.; Jankulak, M. Role of host genetics and heat-tolerant algal symbionts in sustaining populations of the endangered coral Orbicella faveolata in the Florida Keys with ocean warming. Glob. Change Biol. 2019, 25, 1016–1031. [Google Scholar] [CrossRef] [Green Version]
- Gintert, B.E.; Manzello, D.P.; Enochs, I.C.; Kolodziej, G.; Carlton, R.D.; Gleason, A.C.R.; Gracias, N. Marked annual coral bleaching resilience of an inshore patch reef in the Florida Keys: A nugget of hope, aberrance, or last man standing? Coral Reefs 2018, 37, 533–547. [Google Scholar] [CrossRef] [Green Version]
- Siebeck, U.; Marshall, N.; Klüter, A.; Hoegh-Guldberg, O. Monitoring coral bleaching using a colour (sp.) reference card. Coral Reefs 2006, 25, 453–460. [Google Scholar] [CrossRef]
- Mayfield, A.B. Proteomic signature of corals from thermodynamic reefs. Microorganisms 2020, 8, 1171. [Google Scholar] [CrossRef]
- Desoubeaux, G.; Chauvin, D.; Piqueras, M.C.; Bronson, E.; Bhattacharya, S.K.; Sirpenski, G.; Bailly, E.; Cray, C. Translational proteomic study to address host protein changes during aspergillosis. PLoS ONE 2018, 13, e0200843. [Google Scholar] [CrossRef]
- Musada, G.R.; Dvoriantchikova, G.; Myer, C.; Ivanov, D.; Bhattacharya, S.K.; Hackam, A.S. The effect of extrinsic Wnt/β-catenin signaling in Muller glia on retinal ganglion cell neurite growth. Dev. Neurobiol. 2020, 80, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, A.B.; Wang, Y.B.; Chen, C.S.; Chen, S.H.; Lin, C.Y. Compartment-specific transcriptomics in a reef-building coral exposed to elevated temperatures. Mol. Ecol. 2014, 23, 5816–5830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McRae, C.; Mayfield, A.B.; Fan, T.Y.; Huang, W.B.; Cote, I. Differing proteomic responses to high-temperature exposure between adult and larval reef corals. Front. Mar. Sci. 2021, 8, 716124. [Google Scholar] [CrossRef]
- Reeke, G.N.; Becker, J.W.; Cunningham, B.A.; Wang, J.L.; Yahara, I.; Edelman, G.M. Structure and function of concanavalin A. Adv. Exp. Med. Biol. 1975, 55, 13–33. [Google Scholar] [PubMed]
- Vidal-Dupiol, J.; Ladrière, O.; Meistertzheim, A.L.; Fouré, L.; Adjeroud, M.; Mitta, G. Physiological responses of the scleractinian coral Pocillopora damicornis to bacterial stress from Vibrio corallilyticus. J. Exp. Biol. 2011, 214, 1533–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gates, R.D.; Baghdasarian, G.; Muscatine, L. Temperature stress causes host cell detachment in symbiotic cnidarians: Implications for coral bleaching. Biol. Bull. 1992, 182, 324–332. [Google Scholar] [CrossRef]
- Bay, R.A.; Rose, N.H.; Logan, C.A.; Palumbi, S.R. Genomic models predict successful coral adaptation if future ocean warming rates are reduced. Sci. Adv. 2017, 3, e1701413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuller, Z.; Mocellin, V.J.L.; Morris, L.A.; Cantin, N.; Shepherd, J.; Sarre, L.; Peng, J.; Liao, Y.; Pickrell, J.; Andolfatto, P.; et al. Population genetics of the cooral (sp.). Acropora millepora: Toward genomic prediction of bleaching. Science 2020, 369, eaba4674. [Google Scholar]
- Cunning, R.; Bay, R.A.; Gillette, P.; Baker, A.C.; Traylor-Knowles, N. Comparative analysis of the Pocillopora damicornis genome highlights role of immune system in coral evolution. Sci. Rep. 2018, 8, 16134. [Google Scholar] [CrossRef] [Green Version]
- Parisi, M.G.; Parrinello, D.; Stabili, L.; Cammarata, M. Cnidarian immunity and the repertoire of defense mechanisms in anthozoans. Biology 2020, 9, 283. [Google Scholar] [CrossRef]
- Shmueli, G. To explain or to predict? Stat. Sci. 2011, 25, 289–310. [Google Scholar]
- Ramos-Silva, P.; Kaandorp, J.; Huisman, L.; Marie, B.; Zanella-Cléon, I.; Guichard, N.; Miller, D.J.; Marin, F. The skeletal proteome of the coral Acropora millepora: The evolution of calcification by co-option and domain shuffling. Mol. Biol. Evol. 2013, 30, 2099–2112. [Google Scholar] [CrossRef] [PubMed]
- Roach, T.N.F.; Dilworth, J.; Christian, M.H.; Jones, D.; Quinn, R.A.; Drury, C. Metabolomic signatures of coral bleaching history. Nat. Ecol. Evol. 2021, 5, 495–503. [Google Scholar] [CrossRef]
- Maynard, J.A.; Anthony, K.R.N.; Marshall, P.A.; Masiri, I. Major bleaching events can lead to increased thermal tolerance in corals. Mar. Biol. 2008, 155, 173–182. [Google Scholar] [CrossRef]
- Liu, G.; Heron, S.F.; Eakin, C.M.; Muller-Karger, F.E.; Vega-Rodriguez, M.; Guild, L.S.; De La Cour, J.L.; Geiger, E.F.; Skirving, W.J.; Burgess, T.F.; et al. Reef-scale thermal stress monitoring of coral ecosystems: New 5-km Global Products from NOAA Coral Reef Watch. Remote Sens. 2014, 6, 11579–11606. [Google Scholar] [CrossRef] [Green Version]
- Van Hooidonk, R.; Maynard, J.; Tamelander, J.; Gove, J.; Ahmadia, G.; Raymundo, L.; Williams, G.; Heron, S.F.; Planes, S. Local-scale projections of coral reef futures and implications of the Paris Agreement. Sci. Rep. 2016, 6, 39666. [Google Scholar] [CrossRef]
- McClanahan, T.R.; Darling, E.S.; Maina, J.M.; Muthiga, N.A.; D’agata, S.; Jupiter, S.D.; Arthur, R.; Wilson, S.K.; Mangubhai, S.; Nand, Y.; et al. Temperature patterns and mechanisms influencing coral bleaching during the 2016 El Niño. Nat. Clim. Change 2019, 9, 845–851. [Google Scholar] [CrossRef]
- Mayfield, A.B.; Bruckner, A.W.; Chen, C.H.; Chen, C.S. A survey of pocilloporids and their endosymbiotic dinoflagellate communities in the Austral and Cook Islands of the South Pacific. Platax 2015, 12, 1–17. [Google Scholar]
- Logan, C.A.; Dunne, J.P.; Eakin, C.M.; Donner, S.D. Incorporating adaptive responses into future projections of coral bleaching. Glob. Change Biol. 2014, 20, 125–139. [Google Scholar] [CrossRef]
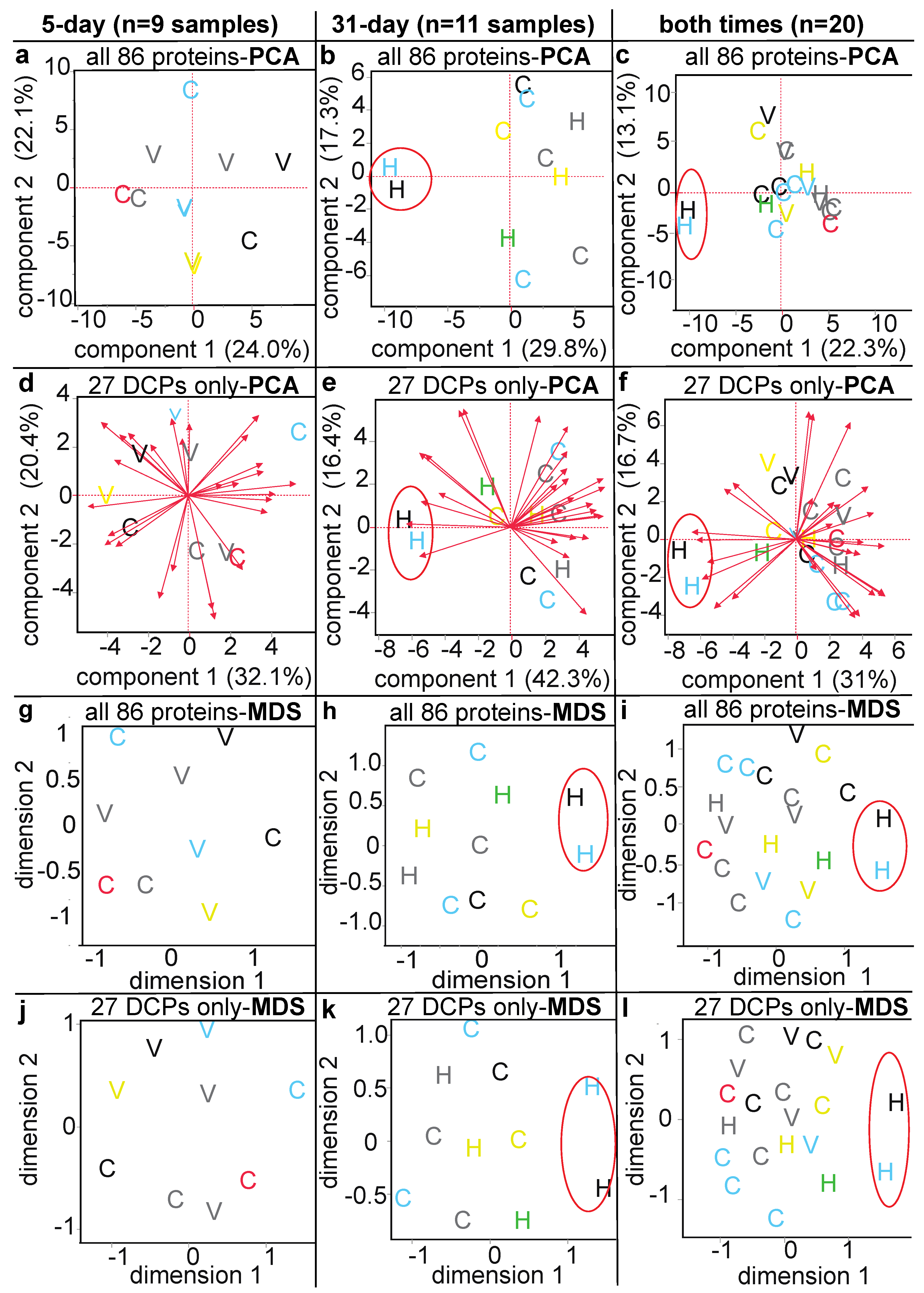
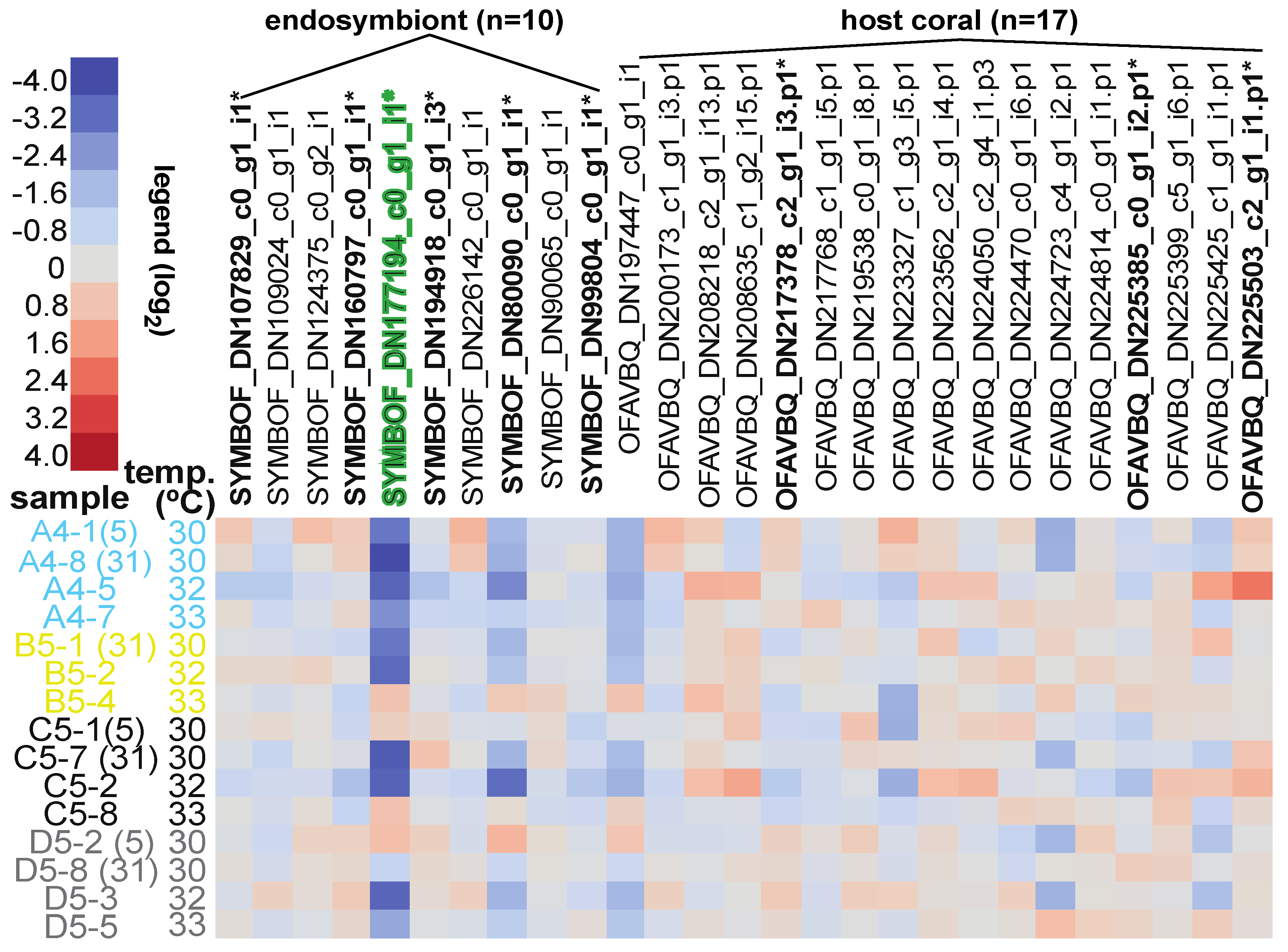
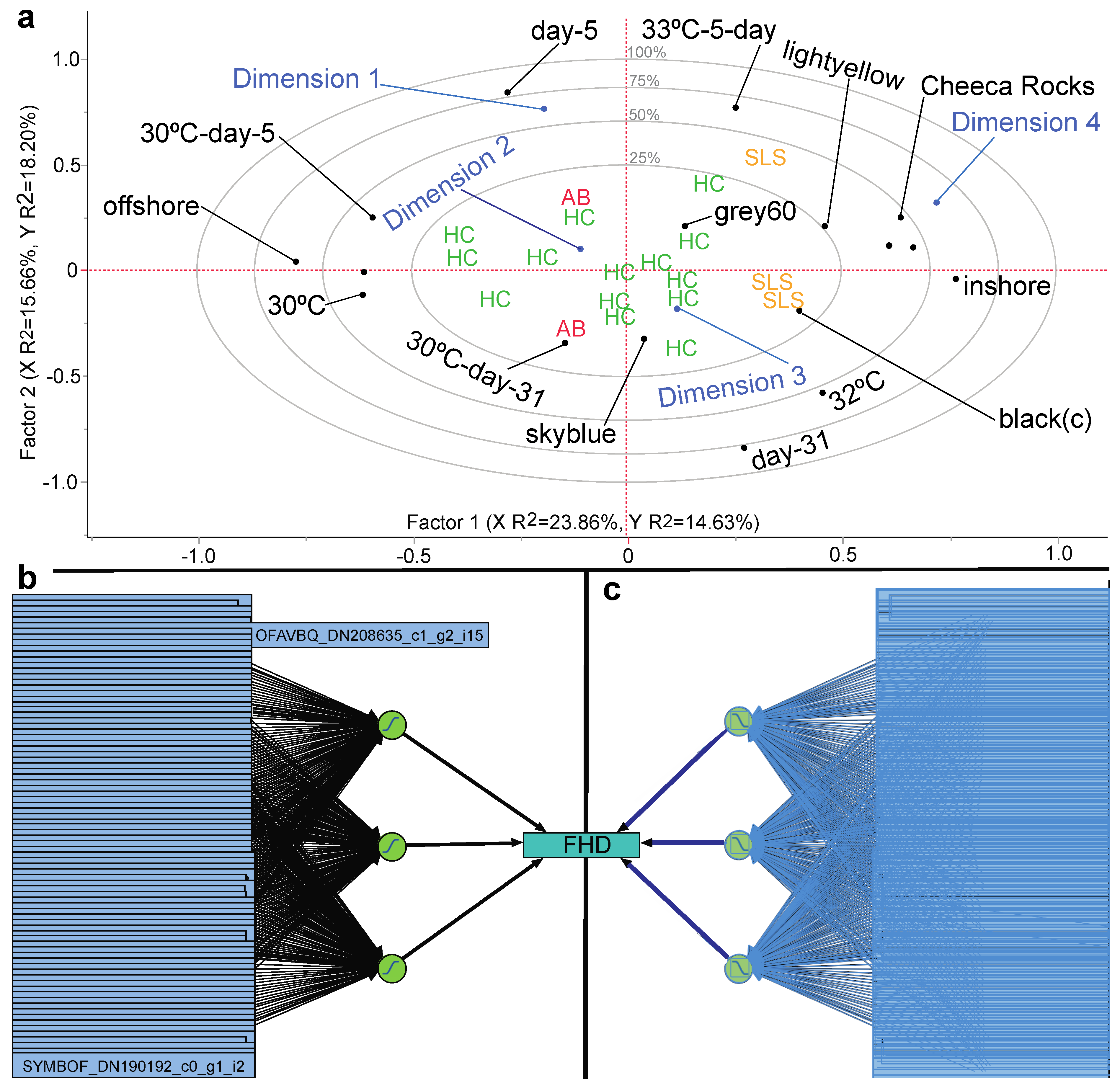
| Sample Name | Reef of Origin | Shelf | Treatment (Temp. × Time) | Genotype | Colony Code | Health Designation | Protein Loaded (µg) | iTRAQ Tag | iTRAQ Batch | |
|---|---|---|---|---|---|---|---|---|---|---|
| Colony | Fragment | |||||||||
| Normalizer | mix of all | mix of both | mix of all | mix of all | NA | NA | NA | 22 | 113 | A |
| B5-7 * | Cheeca Rocks | inshore | 30-5 | lightyellow | B5 | BLR | HC | 22 | 114 | A |
| C5-1 | Little Conch | offshore | 30-5 | black(a) | C5 | BLS | HC | 22 | 115 | A |
| B5-4 | Cheeca Rocks | inshore | 33-5 | lightyellow | B5 | BLR | SLS | 22 | 116 | A |
| A2-2 | The Rocks | inshore | 30-31 | skyblue | A2 | BLS | HC | 22 | 117 | A |
| A4-5 | The Rocks | inshore | 32-31 | skyblue | A4 | BLR | HTA | 22 | 118 | A |
| B3-1 | Cheeca Rocks | inshore | 32-31 | black(c) | B3 | BLR | HTA | 22 | 119 | A |
| C5-2 | Little Conch | offshore | 32-31 | black(a) | C5 | BLS | AB | 22 | 121 | A |
| Normalizer | mix of all | mix of both | mix of all | mix of all | NA | NA | NA | 22 | 113 | B |
| A4-1 | The Rocks | inshore | 30-5 | skyblue | A4 | BLR | HC | 22 | 114 | B |
| C2-2 | Little Conch | offshore | 30-5 | black(b) | C2 | BLS | HC | 22 | 115 | B |
| D5-2 | Cheeca Rocks | inshore | 30-5 | grey60 | D5 | BLR | HC | 22 | 116 | B |
| D6-6 | Cheeca Rocks | inshore | 33-5 | grey60 | D6 | ID | SLS | 22 | 117 | B |
| A4-8 | The Rocks | inshore | 30-31 | skyblue | A4 | BLR | HC | 22 | 118 | B |
| C5-7 | Little Conch | offshore | 30-31 | black(a) | C5 | BLS | HC | 22 | 119 | B |
| D5-3 | Cheeca Rocks | inshore | 32-31 | grey60 | D5 | BLR | HTA | 22 | 121 | B |
| Normalizer | mix of all | mix of both | mix of all | mix of all | NA | NA | NA | 22 | 113 | C |
| A4-7 | The Rocks | inshore | 33-5 | skyblue | A4 | BLR | HTA | 22 | 114 | C |
| C5-8 | Little Conch | offshore | 33-5 | black(a) | C5 | BLS | AB | 22 | 115 | C |
| D5-5 | Cheeca Rocks | inshore | 33-5 | grey60 | D5 | BLR | HTA | 22 | 116 | C |
| B5-1 | Cheeca Rocks | inshore | 30-31 | lightyellow | B5 | BLR | HC | 22 | 117 | C |
| D4-8 | Cheeca Rocks | inshore | 30-31 | grey60 | D4 | BLR | HC | 22 | 118 | C |
| D5-8 | Cheeca Rocks | inshore | 30-31 | grey60 | D5 | BLR | HC | 22 | 119 | C |
| B5-2 | Cheeca Rocks | inshore | 32-31 | lightyellow | B5 | BLR | SLS | 22 | 121 | C |
| Analytical Goal Approach | Response Variables (Y)/Predictors (X) | Acceptance Criterion (a) | Primary Finding (s) | Data Location (s) |
|---|---|---|---|---|
| Uncover multivariate treatment effects | ||||
| PERMANOVA | 86 proteins/all EP | alpha = 0.05 | Effect of fragment health designation on host proteome | Table 4 |
| Non-parametric MANOVA | MDS coordinates/ all EP | MDS stress < 0.1 and alpha = 0.05 | Effect of reef site on host and holobiont proteomes | Table 4 |
| Identify differentially concentrated proteins | ||||
| Response screening analysis | 86 proteins/all EP | FDR-p < 0.01 | 9 differentially concentrated proteins identified | Table 5 and Figure 2 |
| Stepwise discriminant analysis | 86 proteins/all EP | MPM < 15% a | 18 “proteins of interest” identified | Table 5, Figures S1 and S2 |
| Predict bleaching susceptibility | ||||
| Fragment health designation (FHD; 15/5 training/validation samples) | ||||
| Neural network | FHD/86 proteins | MPM < 10% | Only model type with high accuracy | Table 6, Tables S2 and S3 |
| Colony health designation (CHD; 15/5 training/validation samples) | ||||
| Neural network | CHD/86 proteins | MPM < 10% | More flexible than GMR models | Table 6, Tables S2 and S3 |
| GMR-lasso | CHD/86 proteins | MPM < 10% | All 86 proteins in final model | Table 6 |
| GMR-pruned forward selection | CHD/86 proteins | MPM < 10% | Three proteins in final model | Table 6 |
| Factor | df | Pseudo F | p | #Permutations | Multiple PERMANOVA Comparisons or NP-MANOVA Differences |
|---|---|---|---|---|---|
| Endosymbiont—40 proteins | |||||
| Temperature (n = 3) | 2 | 0.925 | 0.579 | 998 | |
| Temperature (n = 2) | 1 | 0.812 | 0.657 | 990 | |
| Site | 2 | 1.33 | 0.106 | 998 | |
| Shelf | 1 | 1.24 | 0.219 | 965 | |
| Genotype * | 5 | 1.037 | 0.407 | 999 | |
| Day | 1 | 1.17 | 0.300 | 994 | |
| Temperature × day | 1 | 1.095 | 0.380 | 999 | |
| Fragment health designation | 1 | 1.071 | 0.383 | 965 | |
| Host coral—46 proteins | |||||
| Temperature (n = 3) | 2 | 1.27 | 0.140 | 998 | |
| Temperature (n = 2) | 1 | 1.16 | 0.280 | 991 | |
| Site a | 2 | 1.33 | 0.128 | 998 | Significant site effect by NP-MANOVA |
| Shelf | 1 | 1.20 | 0.229 | 964 | |
| Genotype * | 5 | 1.23 | 0.153 | 998 | Black(a) ≠ grey60 ≠ skyblue ≠ lightyellow |
| Day | 1 | 1.027 | 0.381 | 993 | |
| Temperature × day | 1 | 1.092 | 0.296 | 996 | |
| Fragment health designation | 1 | 1.90 | 0.025 | 969 | Sub-lethally stressed = actively bleaching ≠ healthy control |
| Host + endosymbiont—all 86 proteins | |||||
| Temperature (n = 3) | 2 | 1.11 | 0.289 | 998 | |
| Temperature (n = 2) | 1 | 1.002 | 0.432 | 989 | |
| Site | 2 | 1.32 | 0.100 | 997 | Significant site effect by NP-MANOVA |
| Shelf | 1 | 1.18 | 0.199 | 958 | |
| Genotype * | 5 | 1.096 | 0.271 | 997 | |
| Day | 1 | 1.088 | 0.355 | 995 | |
| Temperature × day | 1 | 1.049 | 0.364 | 999 | |
| Fragment health designation | 1 | 1.47 | 0.081 | 971 | |
| Model Name | Validation | #Proteins | Type of Activation (# Nodes) | #Boosts (Learning Rate) | # Tours | Penalty Method | Training RMSE | Training MPM (%) | Validation RMSE | Validation MPM (%) | Highly Influential Protein(s) | Protein Identity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fragment health designation | ||||||||||||
| NTanH(2)-NBoost(2) | 15/5 | 86 | sigmoidal(2) | 2 (0.1) | 100 | WD | 0.02 | 0 | 0.04 | 0 | SYMBOF_DN160797_c0_g1 a SYMBOF_DN231313_c0_g1_i1 OFAVBQ_DN225382_c1_g1_i3 | integrin-linked protein kinase serine/arginine-rich splicing factor 2 unknown protein w/ HEAT repeats |
| NTanH(4)-NLinear(1)-NGaussian(1)-NBoost(2) | Kfold(5) | 86 | sigmoidal(4) linear(1) radial(1) | 2 (0.1) | 100 | WD | <0.00 | 0 | <0.00 | 0 | SYMBOF_DN177194_c0_g1 a SYMBOF_DN156077_c0_g1_i1 SYMBOF_DN102933_c0_g1_i1 | cilia & flagella-associated protein 57 tRNA (Ile)-lysidine synthase calcium-dependent protein kinase 2 |
| NTanH(2)-NLinear(4)-NGaussian(4) | Kfold(5) | 86 | sigmoidal(2) linear(4) radial(4) | 0 | 100 | absolute | <0.00 | 0 | <0.00 | 0 | OFAVBQ_DN208218_c2_g1_i13 OFAVBQ_DN220422_c1_g2_i1 OFAVBQ_DN225382_c1_g1_i3 | histone-lysine N-methyltransferase unknown unknown protein w/ HEAT repeats * |
| NTanH(3) b | hold-back(0.33) | 86 | sigmoidal(3) | 0 | 100 | WD | 0.25 | 0.08 | 0.05 | 0 | OFAVBQ_DN224050_c2_g4_i1 SYMBOF_DN156997_c0_g1_i1 SYMBOF_DN147436_c0_g1_i2 | unknown w/endonuclease domain reticulocyte-binding protein 2 homolog a unknown protein w/ PHD finger 1 domain |
| NTanH(3)-NBoost(5) | Kfold(5) | 86 | sigmoidal(3) | 5 (0.1) | 1 | squared | <0.00 | 0 | <0.00 | 0 | OFAVBQ_DN222591_c0_g1_i4 SYMBOF_DN244033_c0_g1_i1 SYMBOF_DN156077_c0_g1_i1 | PKD with egg jelly receptor c DNA helicase ETL1 unknown |
| Colony health designation | ||||||||||||
| NTanH(3)-NBoost(3) | Kfold(5) | 86 | sigmoidal(3) | 3 (0.1) | 100 | WD | 0.29 | 0.13 | 0.28 | 0 | OFAVBQ_DN217378_c2_g1 a SYMBOF_DN239782_c0_g1_i1 SYMBOF_DN99804_c0_g1 a | myosin-11-like (see Table 5) polycystin-2 sec34 sodium channel protein 11 |
| NLinear(3) | 13/6 | 86 | linear(3) | 0 | 20 | squared | 0.01 | 0 | 0.07 | 0 | SYMBOF_DN80090_c0_g1 a SYMBOF_DN177194_c0_g1 a OFAVBQ_DN217378_c2_g1 a | tyrosine decarboxylase 1-like apolipoprotein B100 C terminal myosin-11-like (see Table 5) * |
| Gen-reg with pruned forward selection | Kfold(5) | 3 | NA | NA | NA | NA | <0.00 | 0 | 0.11 | 0 | OFAVBQ_DN218976_c2_g3_i2 OFAVBQ_DN222591_c0_g1_i4 SYMBOF_DN194918_c0_g1_i3 a | unknown PKD with egg jelly receptor c unknown |
| Gen-reg-ridge regression | Kfold(5) | 86 | NA | NA | NA | NA | 0.03 | 0 | 0.40 | 0 | OFAVBQ_DN190522_c0_g2_i1 OFAVBQ_DN197447_c0_g1_i1 OFAVBQ_DN225239_c1_g1_i3 | unknown vitellogenin-2 calcineurin-binding protein cabin-1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayfield, A.B. Machine-Learning-Based Proteomic Predictive Modeling with Thermally-Challenged Caribbean Reef Corals. Diversity 2022, 14, 33. https://doi.org/10.3390/d14010033
Mayfield AB. Machine-Learning-Based Proteomic Predictive Modeling with Thermally-Challenged Caribbean Reef Corals. Diversity. 2022; 14(1):33. https://doi.org/10.3390/d14010033
Chicago/Turabian StyleMayfield, Anderson B. 2022. "Machine-Learning-Based Proteomic Predictive Modeling with Thermally-Challenged Caribbean Reef Corals" Diversity 14, no. 1: 33. https://doi.org/10.3390/d14010033
APA StyleMayfield, A. B. (2022). Machine-Learning-Based Proteomic Predictive Modeling with Thermally-Challenged Caribbean Reef Corals. Diversity, 14(1), 33. https://doi.org/10.3390/d14010033






