Abstract
Microfungi associated with woody plants on the Qinghai–Tibet Plateau (QTP) were investigated, and four collections associated with Potentilla fruticosa were obtained from Gansu and Qinghai provinces in China. Morphologically, they line well with Lentitheciaceae in having subglobose to globose ascomata with brown setae on the papilla but can be distinguished from other genera by its superficial, globose, black, thick-walled ascomata, and fusiform, hyaline (rarely pale brown), one-septate ascospores, surrounded by an entire mucilaginous sheath. The phylogenetic analyses based on a combined SSU, ITS, LSU and TEF1-α sequence data showed that four isolates formed a monophyletic clade among the genera of Lentitheciaceae, and present as a distinct lineage (sister clade to Darksidea). Therefore, we introduce a new genus Crassoascoma, with C. potentillae as the type to accommodate these taxa. Detailed description and illustration are provided, and the establishment of new taxa is justified with morphology and phylogenetic evidence.
1. Introduction
Dothideomycetes is the largest class of the phylum Ascomycota, comprising 38 orders and 210 families (including the incertae sedis) [1,2]. The morphology of this class is diverse and usually have bitunicate asci in their sexual morphs. Pleosporales is the largest order in Dothideomycetes, which includes approximately 91 families and 606 genera (including 48 genera incertae sedis) [3]. However, the numbers are constantly increasing with the establishment of new taxa [4,5,6,7].
The pleosporalean family Lentitheciaceae was introduced by Zhang et al. [8] based on multi-gene phylogeny, of which the well-supported Lentitheciaceae clade accommodated the generic type Lentithecium fluviatile, as well as L. arundinaceum, Keissleriella cladophila, Wettsteinina lacustris (CBS 618.86, morphologically could not be well-verified), two bambusicolous species Katumotoa bambusicola, Ophiosphaerella sasicola, and a possibly asexual morph species Stagonospora macropycnidia (OSC 100965/CBS 114202). With the continuous addition of new members, the family currently comprises fourteen genera: Darksidea [9], Halobyssothecium [10], Katumotoa [11], Keissleriella [12], Lentithecium [8], Murilentithecium [13], Neoophiosphaerella [14], Phragmocamarosporium [15], Pleurophoma [16], Poaceascoma [17], Pseudomurilentithecium [18], Setoseptoria [19], Tingoldiago [20] and Towyspora [21]. Members of Lentitheciaceae are characterized by lenticular to globose ascomata with brown setae or glabrous, cylindrical to clavate asci with short pedicels, and morphologically diverse ascospores, mostly narrow to broad fusiform, hyaline to brown, 1–3-septate (aseptate or muriform in some species), sometimes filiform, fasciculate, surrounded by an entire mucilaginous sheath or fusiform gelatinous sheath; the asexual morphs are stagonospora-like or dendrophoma-like [1]. Most genera have sexual morphs except for Phragmocamarosporium [15] and Towyspora [21], which only comprise coelomycetous asexual morphs. Species in Lentitheciaceae are generally saprobic on stems and twigs of herbaceous and woody plants in freshwater, terrestrial, marine environments [13,15,22,23,24,25] and endophytic Darksidea species have been isolated from semiarid habitat [9].
The Qinghai–Tibet Plateau (QTP) is the world’s largest and highest plateau with an average elevation above 4000 m. Because of its complex topography and ecological environment, the QTP is considered to be one of the biodiversity hotspots with rich biological resources. In a review of the investigation of fungi in Tibet [26], a total of 2559 species distributed in 185 families and 551 genera were counted based on published records. The Basidiomycota accounted for 66.5% of the total, followed by Ascomycota, which accounted for 29.5%, while Glomeromycota, Zygomycota and Chytridiomycota were relatively few, accounted for 2.7%, 0.8%, and 0.5%, respectively [26]. This result is probably due to previous scientific expeditions that were mainly focused on macrofungi. However, it also indicated that the research on the identification of fungi on the QTP, especially the microfungi, is limited.
This study investigated microfungi associated with decayed woody plants on the QTP. Four collections with superficial, globose, black, thick-walled ascomata and fusiform, hyaline (rarely pale brown), 1-septate ascospores surrounded by mucilaginous sheaths, were identified based on morphology and multi-gene phylogeny. A novel ascomycete Crassoascoma potentillae was recognized and illustrated, and a new genus Crassoascoma (Lentitheciaceae, Pleosporales) was introduced to accommodate the new taxon C. potentillae.
2. Materials and Methods
2.1. Collection, Isolation and Morphological Examination
Decayed branches of Potentilla fruticosa L. (Rosaceae) were collected at an altitude over 2000 m on the QTP in 2020. The samples were placed in paper envelopes and taken to the laboratory. Fungal ascomata on the host surface were examined by using a Nikon eclipse NI-ss stereoscope. Freehand sections of ascomata were made into slides mounted in water. Photomicrographs were taken by Nikon SMZ800N stereo microscope fitted with Nikon ECLIPSE Ni upright microscope with DIC objectives connected to Nikon DS-Ri2 digital camera. The measurements of ascomata, asci, and ascospores were taken by NIS-Elements Analysis D 5.21. The photo plate was processed by Adobe Photoshop CS6 software (Adobe System Inc., San Jose, CA, USA).
Single spore isolation used the same spotted pour technique as Senanayake et al. [27], and the germinated ascospores were transferred into potato dextrose agar (PDA). The colony morphology was examined after four weeks of incubation at 25 °C, and the Methuen handbook of colour was used for colour description [28].
Herbarium specimens were deposited in the herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (KUN-HKAS), Kunming, China, and Herbarium, University of Electronic Science and Technology (HUEST). The strains isolated in this study were deposited in China General Microbiological Culture Collection Center (CGMCC) and the University of Electronic Science and Technology Culture Collection (UESTCC). Taxonomic novelties were registered in MycoBank [29].
2.2. DNA Extration, PCR Amplification and Sequencing
Fungal mycelia of 28d colonies were scraped by sterile lancet and transferred into a 1.5 mL Eppendorf tube. DNA was extracted by using Ezup Column Fungi Genomic DNA Purification Kit (Sangon Biotech (Shanghai), Co., Ltd., Shanghai, China). The small subunit rDNA (SSU), internal transcribed spaces (ITS), large subunit rDNA (LSU), and translated elongated factor 1-alpha (TEF-1α) gene were amplified with universal primers NS4/NS1 [30], ITS5/ITS4, LR0R/LR5 [31], EF1-983F/EF1-2218R [32], respectively. All polymerase chain reactions (PCR) were reacted in the volume of 25 μL mixture containing 12.5 μL 2 × PCR Master Mix (Sangon Biotech (Shanghai), Co., Ltd., China), 8.5 μL ddH2O, 1 μL of each primer and 2 μL DNA template. PCR thermal cycles for four genes were performed as the following reaction conditions: initial 95 °C for 3 min, followed by 35 cycles of denaturation at 95 °C for 30 sec, elongation at 72 °C for 30 sec, and final extension at 72 °C for 10 min. The quality of PCR was tested on 1% agarose gel electrophoresis stained with ethidium bromide. PCR products were purified and sequenced with primers mentioned above by Sangon Biotech (Shanghai), Co., Ltd., China.
2.3. Sequence Alignment and Phylogenetic Analysis
The sequence chromatograms were checked by using BioEdit v.7.0.9 [33], and the forward and reverse sequences were assembled by SeqMan Pro in DNASTAR Lasergene v7.1 (DNASTAR, Inc. Madison, WI, USA). The reference sequences were retrieved from relevant publications (Table 1) and downloaded from GenBank. Sequences were aligned using MAFFT v.7 [34] and manually adjusted by BioEdit v.7.0.9 [33]. The single gene sequence datasets (SSU, ITS, LSU and TEF1-α) were concatenated with Mesquite v.3.70 [35] and the combined alignment was transformed into appropriate file formats for different analyzing programs by using a web server ALTER [36].

Table 1.
Taxa used in this study and their GenBank accession numbers. Newly generated sequences are indicated with * and the ex-type strains are in bold.
The analyses of maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI) were carried out as detailed in Dissanayake et al. [37]. The programs used in this study were PAUP v.4.0b 10 [38], raxmlGUI v. 1.3 [39] and MrBayes v3.1.2 [40,41]. Maximum likelihood analysis was performed by raxmlGUI v.1.3 [39] with GTR+I+G as the evolution model. In BI analyses, MrModeltest v. 2.3 [42] was used to select the best-fit model of nucleotide substitution for each partition. GTR+I+G is the best-fit model of ITS, LSU, TEF1-α, and HKY+I+G is the best model of SSU. Four simultaneous Markov chains were run for 1,000,000 generations and partition analysis with 100 sample frequencies, which produced 10,000 trees. The first 1000 trees were discarded as burn-in phase and the remaining 9000 trees were used for calculating posterior probabilities.
Phylograms were visualized by FigTree v.1.4.4 program [43] and edited with Adobe Illustrator CS5 v. 15.0.0 (Adobe®, San Jose, CA, USA). New sequences in the present study were deposited in GenBank (Table 1), and the final alignments were deposited in TreeBASE (www.treebase.org, accessed on 4 November 2021) with a submission number: 28957.
3. Results
3.1. Phylogenetic Analyses
The final dataset comprised of 67 taxa selected from Bambusicolaceae, Didymosphaeriaceae, Lentitheciaceae, Massarinaceae and Sulcatisporaceae in the suborder Massarineae (Pleosporales, Dothideomycetes), with Helminthosporium velutinum (MAFF 243854) and Massarina eburnean (CBS 473.64) as outgroup taxa. The combined dataset consisted of 4361 characters after alignment including gaps (SSU: 1423 bp; ITS: 731 bp; LSU: 1283 bp; TEF-1α: 924 bp). ML, MP and BI analyses were conducted and resulted in generally congruent topologies. The best scoring ML tree (Figure 1) with a final optimization likelihood value of −22,480.205214. The aligned matrix had 1375 distinct alignment patterns, and 34.57% completely undetermined characters and gaps. Estimated base frequencies were as follows: A = 0.240681, C = 0.247239, G = 0.272357, T = 0.239543; substitution rates AC = 1.235890, AG = 2.828734, AT = 1.375693, CG = 1.247991, CT = 7.529110, GT= 1.000000; gamma distribution shape parameter α = 0.468984. Maximum parsimony analyses indicated 3258 constant characters and included 317 variable characters of parsimony-uninformative and 786 characters were parsimony informative. A heuristic search yield one equally most parsimonious trees (TL = 3250, CI = 0.487, RI = 0.670, RC = 0.326, HI = 0.513).
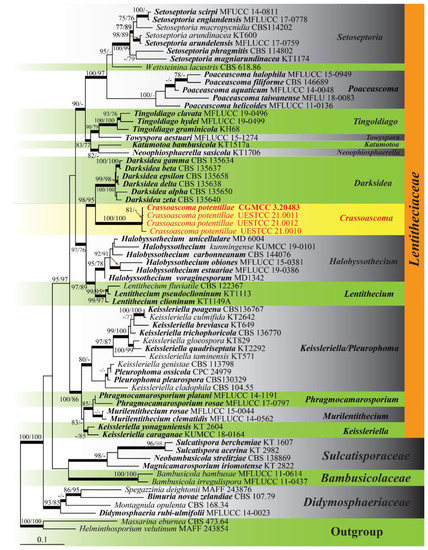
Figure 1.
RAxML analysis of Lentitheciaceae and representatives in Massarineae based on combined SSU ITS, LSU, and TEF1-α sequence data. Bootstrap support values for maximum likelihood (ML) and maximum parsimony (MP) higher than 75% were placed above the branches (ML/MP). Bayesian posterior probabilities (BYPP) greater than 0.95 were shown as bold branches. Ex-type strains were in bold and new strains generated in this study were indicated in red.
Fourteen genera are accepted in the family Lentitheciaceae and all have available sequence data [1]. However, the genus Pseudomurilentithecium [18] was excluded from our final dataset because it was unstable in the single gene phylogenetic analyses. Pseudomurilentithecium fell outside Lentitheciaceae in the LSU and TEF1-α gene trees (data not shown), but it formed an obvious long branch in this family in the multi-gene phylogeny result. The four newly generated isolates formed a well-supported monophyletic clade in Lentitheciaceae (Figure 1) and can be recognized as a new genus Crassoascoma. Moreover, the molecular data of these four isolates are identical, which supports the identification of a new species namely C. potentillae.
3.2. Taxonomy
Crassoascoma Jian K. Liu, gen. nov
MycoBank: MB 841098
Etymology: Referred to superficial thick-walled ascoma.
Saprobic on the living and decayed branches of Potentilla fruticosa L. (Rosaceae). Sexual morph: Ascomata superficial, solitary to gregarious, subglobose to globose, dark brown to black, ostiolate, with setae around the papilla. Peridium with multi-layers, comprising hyaline to brown cells of textura angularis. Hamathecium trabeculate pseudoparaphyses, filamentous and anastomosing. Asci 8-spored, clavate to subcylindrical, short pedicel. Ascospores overlapping biseriate, usually uniseriate in the lower half, fusiform, straight or slightly curved, hyaline to pale brown, 1-septate constricted at the septum and midpoint of each cell, surrounded by an entire mucilaginous sheath. Asexual morph: Undetermined.
Type species: Crassoascoma potentillae Z.P. Liu, S.N. Zhang and Jian K. Liu
Notes: The phylogenetic analyses showed that four isolates of Crassoascoma formed a monophyletic clade in Lentitheciaceae and is sister to Darksidea with high statistical support (98/95/1.00). Darksidea accommodates endophytic fungi which are characterized by 4–6-spored, clavate to ellipsoid asci and ellipsoid, aseptate ascospores [9]. Crassoascoma however, is distinct from Darksidea in having superficial, papillate, thick-walled ascomata with setae around the ostiole, clavate to subcylindrical asci, and fusiform, 1-septate ascospores surrounded by a mucilaginous sheath. Multi-gene phylogeny results also indicated a close relationship of Crassoascoma to Lentithecium and Halobyssothecium. The sexual morphs of these three genera are somewhat similar, and all have eight-spored, clavate to subcylindrical asci, fusiform ascospores with septa that constricted at median septum. However, Lentithecium contains saprobic species from aquatic habitats, usually have semi-immersed, thin-walled ascomata, and fusiform, hyaline, or yellowish-brown ascospores, with or without a sheath [8,53,54]. The superficial, black, thick-walled ascomata of Crassoascoma resemble species in Halobyssothecium [10,55], but they can be distinguished by the ascospores (hyaline to pale brown vs. versicolored; with sheath vs. lacking sheaths or appendages) and habitat (terrestrial semiarid region vs. marine).
Crassoascoma potentillae Z.P. Liu, S.N. Zhang and Jian K. Liu, sp. nov. Figure 2
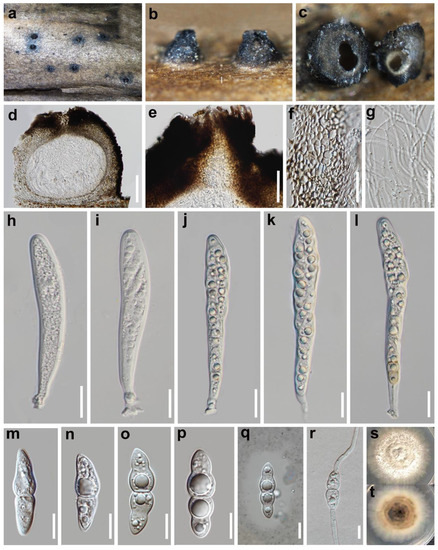
Figure 2.
Crassoascoma potentillae (HKAS 115966, holotype) (a–c) Ascomata on host surface. (d) Vertical section of ascomata. (e) Ostiole. (f) Peridium. (g) Hamathecium. (h–l) Asci, (m–p) Ascospores. (q) Ascospore stained with India ink showing the mucilaginous sheath. (r) Germinating ascospore. (s,t) Colony on PDA (4 weeks). Scale bars: d = 100 μm, e = 50μm, (f–l), r = 20 μm, (m–q) = 10 μm.
MycoBank: MB 841099
Etymology: Name “potentillae” referred to the hosts Potentilla fruticosa L. on which the fungus was collected.
Holotype: HKAS 115966
Saprobic on the epidermis of decayed wood in semiarid lands. Sexual morph: Ascomata 270–340 μm high, 270–320 μm diam. superficial, scattered, subglobose to globose, base flattened, dark brown to black, uni-loculate, rough walled, papillate. Ostiole 90–110 μm wide, 100–120 μm high, central, dark brown, with brown setae on the papilla. Peridium 28–37 μm wide, relatively thick, multi-layer, comprising pale brown to brown cells of textura angularis. Hamathecium 1.7–2.3 μm wide, trabeculate pseuoparaphyses, remotely septate, filamentous and anastomosing. Asci 139–155 × 18–20 μm (x̅ = 147 × 17 μm, n = 20), 8-spored, bitunicate, fissitunicate, clavate to subcylindrical, shortly pedicellate, apically rounded with an ocular chamber. Ascospores 31–35 × 7–8 μm (x̅ = 33 × 6.5 μm, n = 50), overlapping biseriate, usually uniseriate in the lower half, fusiform with subobtuse to acute ends, straight or slightly curved, hyaline, rarely pale brown, one-septate, the upper cell slightly bigger than the lower cell, constricted at the septum and midpoint of each cell, smooth-walled, surrounded by an entire mucilaginous sheath. Asexual morph: undetermined.
Culture characteristics: Ascospores germinated on PDA within 24 h. Colonies on PDA reaching 24 mm diam. incubated at 25 °C after 4 weeks, circular, medium dense, cottony, flat to raised, entire margins, greyish-white, thinner and velvety at the margin, pale brown to brown from the reverse.
Material examined: CHINA. Qinghai province, Huangnan autonomic prefecture, Zeku city, Maixiang district S203 road, 35°16′27″ N, 101°54′48″ E, 3315 m, on the branch of Potentilla fruticosa L. (Rosaceae), 21 July 2020, Jian-Kui Liu, HUEST 21.0015 (HKAS 115966, holotype; ex-type living culture CGMCC 3.20483 = UESTCC 21.0013); ibid., Gansu Province, Zhangye city, Minle county, 38°23′2″ N, 100°22′30″ E, 2830 m, on the branch of Potentilla fruticosa L., 2 August 2020, Jian-Kui Liu, HUEST 21.0013 (HKAS 115967, paratype); living culture UESTCC 21.0011; ibid., Gansu Province, Zhangye City, Minle County, 38°23′2″ N, 100°22′30″ E, 2830 m, on the branch of Potentilla fruticosa L., 2 August 2020, Jian-Kui Liu, HUEST 21.0014; living culture UESTCC 21.0012; ibid., Gansu Province, Jiuquan City, Sunan County, Kangle Township, 38°47′50″ N, 99°40′53″ E, 2456 m, on the branch of Potentilla fruticosa L., 2 August 2020, Jian-Kui Liu, HUEST 21.0012, living culture UESTCC 21.0010.
Notes: Four of our Crassoascoma isolates clustered together and almost identical in their ITS, TEF1-α sequence data, which indicated they can be recognized as a single species. The distinguishing characteristics of Crassoascoma potentillae are the superficial, globose, black, thick-walled ascomata with setae on the papilla, and fusiform, one-septate, hyaline (rarely pale brown) ascospores with subobtuse ends, constricted at the septum and midpoint of each cell and surrounded by a mucilaginous sheath.
4. Discussion
This study contributed to the microfungal community in semiarid lands on the Qinghai-Tibet Plateau, of which a monotypic genus Crassoascoma belongs to Lentitheciaceae (Dothideomycetes) was isolated, identified and well-described. Although the species of Lentitheciaeae are morphologically diverse, this new genus is unique in terms of the superficial, thick-walled ascomata with setae around the papilla. Along with the ascospores morphology, Crassoascoma can be clearly distinguished from other genera in Lentitheciaceae. The discovery of the interesting new genus in the special environmental habitats (e.g., high-elevation, dry, low temperate) also reflects that there may be many fungal novelties in the Qinghai-Tibet Plateau to be explored, discovered and described.
Author Contributions
Conceptualization, J.-K.L., Z.-P.L. and S.-N.Z.; methodology, S.-N.Z.; formal analysis, Z.-P.L.; resources, J.-K.L.; data curation, Z.-P.L. and R.C.; writing—original draft preparation, Z.-P.L. and S.-N.Z.; writing—review and editing, Z.-P.L.; S.-N.Z. and J.-K.L.; supervision, J.-K.L.; project administration, Q.Z. and J.-K.L.; funding acquisition, Q.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the second Tibetan Plateau Scientific Expedition and Research (STEP) Program, Grant Number 2019QZKK0503.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data are available upon request from the corresponding author. The data is not publicly available due to it its usage in the ongoing study.
Acknowledgments
Z.-P.L. thanks Na Wu for her assistance in fungal culture isolation and illustration.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; McKenzie, E.H.C.; Sarma, V.V.; Boonmee, S.; Lücking, R.; Bhat, D.J.; Liu, N.G.; et al. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 2020, 11, 1553–2107. [Google Scholar] [CrossRef]
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; McKenzie, E.H.C.; Sarma, V.V.; Lücking, R.; Boonmee, S.; Bhat, J.D.; Liu, N.G.; et al. Refined families of Dothideomycetes: Orders and families incertae sedis in Dothideomycetes. Fungal Divers. 2020, 105, 17–318. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.; Saxena, R.K.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Atienza, V.; Hawksworth, D.L.; Pérez-Ortega, S. Verrucoccum (Dothideomycetes, Dictyosporiaceae), a new genus of lichenicolous fungi on Lobaria s. lat. for the Dothidea hymeniicola species complex. Mycologia 2021, 113, 1233–1252. [Google Scholar] [CrossRef]
- Hongsanan, S.; Phookamsak, R.; Goonasekara, I.D.; Thambugala, K.M.; Hyde, K.D.; Bhat, J.D.; Suwannarach, N.; Cheewangkoon, R. Introducing a new pleosporalean family Sublophiostomataceae fam. nov. to accommodate Sublophiostoma gen. nov. Sci. Rep. 2021, 11, 9496. [Google Scholar] [CrossRef]
- Li, W.L.; Bao, D.F.; Liu, N.G.; Hyde, K.D.; Liu, J.K. Aquatisphaeria thailandica gen. et sp. nov. (Tetraplosphaeriaceae, Pleosporales) from freshwater habitat in Thailand. Phytotaxa 2021, 513, 118–128. [Google Scholar] [CrossRef]
- Pintye, A.; Knapp, D.G. Two pleosporalean root-colonizing fungi, Fuscosphaeria hungarica gen. et sp. nov. and Delitschia chaetomioides, from a semiarid grassland in Hungary. Mycol. Prog. 2021, 20, 39–50. [Google Scholar] [CrossRef]
- Zhang, Y.; Schoch, C.L.; Fournier, J.; Crous, P.W.; de Gruyter, J.; Woudenberg, J.H.C.; Hirayama, K.; Tanaka, K.; Pointing, S.B.; Spatafora, J.W.; et al. Multi-locus phylogeny of Pleosporales: A taxonomic, ecological and evolutionary re-evaluation. Stud. Mycol. 2009, 64, 85–102. [Google Scholar] [CrossRef]
- Knapp, D.G.; Kovacs, G.M.; Zajta, E.; Groenewald, J.Z.; Crous, P.W. Dark septate endophytic pleosporalean genera from semiarid areas. Persoonia 2015, 35, 87–100. [Google Scholar] [CrossRef] [Green Version]
- Dayarathne, M.C.; Wanasinghe, D.N.; Jones, E.B.G.; Chomnunti, P.; Hyde, K.D. A novel marine genus, Halobyssothecium (Lentitheciaceae) and epitypification of Halobyssothecium obiones comb. nov. Mycol. Prog. 2018, 17, 1161–1171. [Google Scholar] [CrossRef]
- Tanaka, K.; Harada, Y. Bambusicolous fungi in Japan (6): Katumotoa, a new genus of phaeosphaeriaceous ascomycetes. Mycoscience 2005, 46, 313–318. [Google Scholar] [CrossRef]
- Höhnel, F. Fragmente zur Mykologie. XXIII Mitteilung, Nr. 1154 bis 1188. Sitz. Kais. Akad. Wiss. Math.-Nat. Kl. Abt. I 1919, 128, 535–625. Available online: https://www.biodiversitylibrary.org/page/36043822 (accessed on 10 June 2021).
- Wanasinghe, D.N.; Jones, E.B.G.; Camporesi, E.; Boonmee, S.; Ariyawansa, H.A.; Wijayawardene, N.N.; Mortimer, P.E.; Xu, J.; Yang, J.B.; Hyde, K.D. An Exciting Novel Member of Lentitheciaceae in Italy from Clematis vitalba. Cryptogam. Mycol. 2014, 35, 323–337. [Google Scholar] [CrossRef]
- Tanaka, K.; Hirayama, K.; Yonezawa, H.; Sato, G.; Toriyabe, A.; Kudo, H.; Hashimoto, A.; Matsumura, M.; Harada, Y.; Kurihara, Y.; et al. Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud. Mycol. 2015, 82, 75–136. [Google Scholar] [CrossRef] [Green Version]
- Wijayawardene, N.N.; Hyde, K.D.; Bhat, D.J.; Goonasekara, I.D.; Nadeeshan, D.; Camporesi, E.; Schumacher, R.K.; Wang, Y. Additions to Brown Spored Coelomycetous Taxa in Massarinae, Pleosporales: Introducing Phragmocamarosporium gen. nov. and Suttonomyces gen. nov. Cryptogam. Mycol. 2015, 36, 213–224. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Guarro, J.; Hernández-Restrepo, M.; Sutton, D.A.; Acharya, K.; Barber, P.A.; Boekhout, T.; Dimitrov, R.A.; Dueñas, M.; et al. Fungal Planet description sheets: 320–370. Persoonia 2015, 34, 167–266. [Google Scholar] [CrossRef]
- Phookamsak, R.; Manamgoda, D.S.; Li, W.J.; Dai, D.Q.; Singtripop, C.; Hyde, K.D. Poaceascoma helicoides gen et sp. nov., a new genus with scolecospores in Lentitheciaceae. Cryptogam. Mycol. 2015, 36, 225–236. [Google Scholar] [CrossRef]
- Hyde, K.D.; Dong, Y.; Phookamsak, R.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Liu, N.G.; Abeywickrama, P.D.; Mapook, A.; Wei, D.; et al. Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020, 100, 5–277. [Google Scholar] [CrossRef] [Green Version]
- Quaedvlieg, W.; Verkley, G.J.; Shin, H.D.; Barreto, R.W.; Alfenas, A.C.; Swart, W.J.; Groenewald, J.Z.; Crous, P.W. Sizing up Septoria. Stud. Mycol. 2013, 75, 307–390. [Google Scholar] [CrossRef] [Green Version]
- Hirayama, K.; Tanaka, K.; Raja, H.A.; Miller, A.N.; Shearer, C.A. A molecular phylogenetic assessment of Massarina ingoldiana sensu lato. Mycologia 2010, 102, 729–746. [Google Scholar] [CrossRef] [Green Version]
- Li, G.J.; Hyde, K.D.; Zhao, R.L.; Hongsanan, S.; Abdel-Aziz, F.A.; Abdel-Wahab, M.A.; Alvarado, P.; Alves-Silva, G.; Ammirati, J.F.; Ariyawansa, H.A.; et al. Fungal diversity notes 253–366: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 78, 1–237. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; Jeewon, R.; Maharachchikumbura, S.S.N.; Liu, J.K.; Bhat, D.J.; Jones, E.B.G.; McKenzie, E.H.C.; Camporesi, E.; Bulgakov, T.S.; et al. Fungal diversity notes 491–602: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2017, 83, 1–261. [Google Scholar] [CrossRef]
- Hyde, K.D.; Chaiwan, N.; Norphanphoun, C.; Boonmee, S.; Camporesi, E.; Chethana, K.W.T.; Dayarathne, M.C.; de Silva, N.I.; Dissanayake, A.J.; Ekanayaka, A.H.; et al. Mycosphere notes 169–224. Mycosphere 2018, 9, 271–430. [Google Scholar] [CrossRef]
- Jiang, H.B.; Phookamsak, R.; Doilom, M.; Mortimer, P.E.; Xu, J.C.; Lumyong, S.; Hyde, K.D.; Karunarathna, S.C. Taxonomic and phylogenetic characterizations of Keissleriella bambusicola sp. nov. (Lentitheciaceae, Pleosporales) from Yunnan, China. Phytotaxa 2019, 423, 129–144. [Google Scholar] [CrossRef]
- Xu, L.; Bao, D.F.; Luo, Z.L.; Su, X.J.; Shen, H.W.; Su, H.Y. Lignicolous freshwater ascomycota from Thailand: Phylogenetic and morphological characterisation of two new freshwater fungi: Tingoldiago hydei sp. nov. and T. clavata sp. nov. from Eastern Thailand. MycoKeys 2020, 65, 119–138. [Google Scholar] [CrossRef] [Green Version]
- Pubu, C.R.; Wang, M.; Liu, X.Y. A review of investigation of fungi in Tibet. Mycosystema 2016, 35, 1025–1047. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour; Eyre Methuen: London, UK, 1978; pp. 1–252. [Google Scholar]
- Crous, P.W.; Gams, W.; Stalpers, J.A.; Robert, V.; Stegehuis, G. MycoBank: An online initiative to launch mycology into the 21st century. Stud. Mycol. 2004, 50, 19–22. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [Green Version]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Mesquite Project. Available online: https://www.mesquiteproject.org/ (accessed on 25 August 2021).
- Glez-Peña, D.; Gómez-Blanco, D.; Reboiro-Jato, M.; Fdez-Riverola, F.; Posada, D. ALTER: Program-oriented format conversion of DNA and protein alignments. Nucleic Acids Res. 2010, 38, W14–W18. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Bhunjun, C.S.; Maharachchikumbura, S.S.N.; Liu, J.K. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 2020, 11, 2652–2676. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Paesimony (*and Other Methods); Sinauer Associates: Sunderland, UK, 2002. [Google Scholar]
- Silvestro, D.; Michalak, I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2011, 12, 335–337. [Google Scholar] [CrossRef]
- Rannala, B.; Yang, Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996, 43, 304–311. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Nylander, J.A.A. MrModeltest, Version 2. Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004.
- FigTree 1.4.4. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 25 August 2021).
- Dai, D.Q.; Bhat, D.J.; Liu, J.K.; Chukeatirote, E.; Zhao, R.L.; Hyde, K.D. Bambusicola, a new genus from bamboo with asexual and sexual morphs. Cryptogam. Mycol. 2012, 33, 363–379. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Hardy, G.E.S.J.; Gené, J.; Guarro, J.; Baseia, I.G.; García, D.; Gusmão, L.F.P.; Souza-Motta, C.M.; et al. Fungal planet description sheets: 716–784. Persoonia 2018, 40, 240–393. [Google Scholar] [CrossRef]
- Devadatha, B.; Calabon, M.S.; Abeywickrama, P.D.; Hyde, K.D.; Jones, E.B.G. Molecular data reveals a new holomorphic marine fungus, Halobyssothecium estuariae, and the asexual morph of Keissleriella phragmiticola. Mycology 2020, 11, 167–183. [Google Scholar] [CrossRef] [Green Version]
- Dong, W.; Wang, B.; Hyde, K.D.; McKenzie, E.H.C.; Raja, H.A.; Tanaka, K.; Abdel-Wahab, M.A.; Abdel-Aziz, F.A.; Doilom, M.; Phookamsak, R.; et al. Freshwater Dothideomycetes. Fungal Divers. 2020, 105, 319–575. [Google Scholar] [CrossRef]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.A.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Phukhamsakda, C.; Hyde, K.D.; Jeewon, R.; Lee, H.B.; Jones, E.B.G.; Tibpromma, S.; Tennakoon, D.S.; Dissanayake, A.J.; Jayasiri, S.C.; et al. Fungal diversity notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018, 89, 1–236. [Google Scholar] [CrossRef]
- Luo, Z.L.; Bahkali, A.H.; Liu, X.Y.; Phookamsak, R.; Zhao, Y.C.; Zhou, D.Q.; Su, H.Y.; Hyde, K.D. Poaceascoma aquaticum sp. nov. (Lentitheciaceae), a new species from submerged bamboo in freshwater. Phytotaxa 2016, 253, 71–80. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Chooi, Y.H.; Gilchrist, C.L.M.; Lacey, E.; Pitt, J.I.; Roets, F.; Swart, W.J.; Cano-Lira, J.F.; Valenzuela-Lopez, N.; et al. Fungal planet description sheets: 1042–1111. Persoonia 2020, 44, 301–459. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Norphanphoun, C.; Abreu, V.P.; Bazzicalupo, A.; Chethana, K.W.T.; Clericuzio, M.; Dayarathne, M.C.; Dissanayake, A.J.; Ekanayaka, A.H.; He, M.Q.; et al. Fungal diversity notes 603–708: Taxonomic and phylogenetic notes on genera and species. Fungal Divers. 2017, 87, 1–235. [Google Scholar] [CrossRef]
- Zhang, Y.; Crous, P.W.; Schoch, C.L.; Hyde, K.D. Pleosporales. Fungal Divers. 2012, 53, 1–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyde, K.D.; Jones, E.B.G.; Liu, J.K.; Ariyawansa, H.; Boehm, E.; Boonmee, S.; Braun, U.; Chomnunti, P.; Crous, P.W.; Dai, D.Q.; et al. Families of Dothideomycetes. Fungal Divers. 2013, 63, 1–313. [Google Scholar] [CrossRef]
- Calabon, M.S.; Jones, E.B.G.; Hyde, K.D.; Boonmee, S.; Tibell, S.; Tibell, L.; Pang, K.L.; Phookamsak, R. Phylogenetic assessment and taxonomic revision of Halobyssothecium and Lentithecium (Lentitheciaceae, Pleosporales). Mycol. Prog. 2021, 20, 701–720. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).