Abstract
Different polar environments (lakes and glaciers), also in Antarctica, encapsulate brine pools characterized by a unique combination of extreme conditions, mainly in terms of high salinity and low temperature. Since 2014, we have been focusing our attention on the microbiology of brine pockets from three lakes in the Northern Victoria Land (NVL), lying in the Tarn Flat (TF) and Boulder Clay (BC) areas. The microbial communities have been analyzed for community structure by next generation sequencing, extracellular enzyme activities, metabolic potentials, and microbial abundances. In this study, we aim at reconsidering all available data to analyze the influence exerted by environmental parameters on the community composition and activities. Additionally, the prediction of metabolic functions was attempted by the phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt2) tool, highlighting that prokaryotic communities were presumably involved in methane metabolism, aromatic compound biodegradation, and organic compound (proteins, polysaccharides, and phosphates) decomposition. The analyzed cryoenvironments were different in terms of prokaryotic diversity, abundance, and retrieved metabolic pathways. By the analysis of DNA sequences, common operational taxonomic units ranged from 2.2% to 22.0%. The bacterial community was dominated by Bacteroidetes. In both BC and TF brines, sequences of the most thermally tolerant and methanogenic Archaea were detected, some of them related to hyperthermophiles.
1. Introduction
In continental Antarctica, lakes are often characterized by the presence of icing blisters, which develop annually on their surface. Differently from their Arctic counterparts, Antarctic icing blisters mostly derive from the generation of hydrostatic pressures by the progressive freezing of high salt-content water beneath the lake-ice cover during winter. In fact, even if the lake ice cover never melts, heating by direct insolation or enhanced thaw and seepage at the permafrost table probably allows free water, in the form of liquid and saline brine lenses, to accumulate beneath the lake-ice cover in warmer periods [1,2]. Studying briny ecosystems is multifaceted, and assumes special relevance for geological aspects, and also for unravelling the functional potential and roles in biogeochemical cycling of the psychrophilic lifeforms. Microorganisms inhabit extreme environments on Earth [3,4,5] that are also intriguingly similar to other worlds within our Solar system [6]. Current knowledge about the drivers of prokaryotic diversity, distribution, and metabolism in Antarctic briny systems is patchy, and therefore deserves to be deepened. The TF and BC areas in the NVL host several small perennially ice-covered lakes [7], and some of them conceal brine lenses [8,9,10]. Recently, brines from three Tarn Flat and the Boulder Clay lakes have been analyzed for the prokaryotic component by culture-dependent and -independent approaches [4,5,11,12]. Overall, the prokaryotic community included methanogens, strictly anaerobes, halophiles, and (hyper)thermophiles [4,12]. Diversification in terms of abundance, metabolic potentials, and enzymatic activities of the prokaryotic assemblages among the examined brines has been highlighted [4,5]. In the Tarn Flat brines, previous analyses have found the presence of proteolytic activity, as well as a comparatively lower alkaline phosphatase activity than in Boulder Clay. Enzymes that could degrade polysaccharides were also detected, whose hydrolytic activity rates were quantitatively different between the studied samples [4]. Conversely, in Boulder Clay, the microbial community was mostly active in the decomposition of organic phosphates and lower proteolytic and glycolytic activity rates were recorded. Moreover, decreasing patterns of aminopeptidase and phosphatase activities were observed with increasing depth of the collection site [5]. These peculiar features were ascribed to a lot of factors including brines’ historical origin, depth horizon, and time of segregation.
The prediction of microbial functions from 16S rRNA gene sequencing data has been proposed as a valuable alternative to the shotgun metagenomics approach. As an additional in-depth analysis, in this study, we used the phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt2), as a bioinformatics tool applied to 16S rRNA gene data, to infer the functional profile of prokaryotic communities in Antarctic lakes’ brines. Recently, this approach provided useful insights into predictive metagenomics of microbial communities in the thalassohaline brine of Lake Tuz, [13]. To the best of our knowledge, predictions on the metabolic functions of prokaryotes in cryosystems, such as Antarctic briny systems, have never been performed. This approach may be particularly suitable in extreme environments to rapidly survey microbial metabolic mechanisms and their possible relationships with well-known environmental constrains, allowing one to explore unique adaptation strategies, as well as to give insights into the biotechnological potentialities of microbes inhabiting unusual habitats.
In this study, we aimed at coupling previous information on the microbiological and physicochemical features of five distinct briny systems of the Northern Victoria Land and an attempt to predict functional profiles of microbial communities. All available data were collected, comparatively analyzed, and further reprocessed to answer the following questions: (i) Which environmental parameters exert a main influence on the prokaryotic communities in terms of composition, structure, and activities? (ii) Does brine separation overwhelm environmental conditions in shaping prokaryotic diversity? (iii) To what extent do the prokaryotic assemblages show differences among the brines? (iv) How does the metabolic potential of prokaryotes vary among the brines? This research provides further insights into the diversity, biotechnological potential, and ecological role of prokaryotes in these under-investigated and peculiar Antarctic cryosystems.
2. Materials and Methods
2.1. Brine Sample Collection
Five brine samples were collected, in 2014, from three perennially frozen lakes lying in the Tarn Flat (TF) and Boulder Clay (BC) areas of the NVL, Antarctica (coordinates 75°4′ S, 162°30′ E and 74°44′ S, 164°01′ E, respectively). Samples TF4 and TF5 were taken at two different depths (3.78–3.98 and 4.10–4.94 m depth, respectively) of the same borehole drilled in the TF lake. These brine pockets were separated by a thick ice layer of 12 cm [10]. Samples BC1 and BC2 were collected from two different sampling points (at 2.5 and 0.9 m depth, respectively) at Lake 16 in BC, whilst sample BC3 (depth 2.0 m) was collected from the adjacent Lake L-2 [14]. Sampling was carried out using presterilized polycarbonate bottles, sterilized peristaltic pump, and tubing.
2.2. Available Dataset
The microbiological and physicochemical results previously obtained on BC and TF lake brines, and used in this study, are summarized in Supplementary Material Table S1. Briefly, trace elements (Table S1) were analyzed by inductively coupled plasma sector field mass spectrometry (ICP-SFMS) [3,11,14]. Anions (Table S1) were analyzed by ion chromatography [3,14]. Total organic and inorganic carbon contents were determined using a Shimadzu 5050 A TOC analyser [3,14] (Table S1). The salinities and pHs of the brines were measured using refractive index and a potentiometric method, respectively [10,14] (Table S1). The bacterial viable counts (BVC) were determined by spread plating on agar plates [11]. Prokaryotic cell abundances (PA), respiration (CTC), viability (L/D), biomass (PB) (Table S1), and morphometric features (Table S1) were estimated by epiflurescence microscopy, as reported in Papale et al. [4] and Azzaro et al. [5]. Microbial enzymatic activities on proteinaceous and glucidic organic matter as well as on organic phosphates (i.e., AP, beta-GLU, and LAP) were fluorometrically estimated [4,5] (Table S1). Physiological profiles (PP) were determined by the Biolog EcoPlate™ microplate assay and spectrophotometrically measured [4,5] (Table S1). The prokaryotic (Bacteria and Archaea) community diversity and composition (Table S1) were evaluated by Ion Torrent sequencing [4,12]. The total OTU number, retrieved in each sample, was used to estimate alpha diversity by considering the Shannon, evenness, and Chao1 indices.
2.3. Predictive Functional Profiling
PICRUSt2 (phylogenetic investigation of communities by reconstruction of unobserved states) was selected among the tools for the functional prediction of the prokaryotic communities. To date, PICRUSt2 translates 16S RNA sequences into the most accurate prediction method [15] and acts as a predictor of functional metagenomic content based on the frequency of detected 16S rRNA gene sequences corresponding to genomes in regularly updated, functionally annotated genome databases [16]. It is in fact based on a new algorithm that uses short reads and recent positioning tools that insert sequences into an existing phylogenetic tree. Briefly, 16S RNA sequences with the Phred quality score less than 20 per base, and more than four consecutive low-quality base calls, were filtered by Trimmomatic (SLIDINGWINDOW 4:20). Then, denoising was carried out with DADA2 with the denoise-single command (using --p-trim-left 20 --p-trunc-len 0 --i-demultiplexed-seqs). Sequences were taxonomically classified in QIIME2 (version 2019.4) by Silva reference files (Silva release 132 full-length sequences and taxonomy references) using classify-consensus-blast.
Metagenomes from 16S data were predicted with the PICRUSt2 tool (version 2.3.0). The Hidden state prediction method with the mp (maximum parsimony) approach was used. To specify how distantly a sequence needs to be placed in the reference phylogeny, before it is excluded, the command --p-max-nsti (cut-off 2) was applied. The accuracy of metagenome predictions was tested trough the nearest sequenced taxon index (NSTI). The accuracy prediction is related to the presence of closely representative bacterial genomes. The lower values reveal a closer mean relationship. The data obtained by PICRUSt2 were examined by Kyoto Encyclopaedia of Genes and Genomes (KEGG) [17].
2.4. Statistical Analyses
Not-metric multidimensional scaling (nMDS) was performed by using Bray–Curtis similarity matrices calculated on transformed data of the relative abundances at phylum level (Primer 7, Plymouth Marine Laboratory, Roborough, UK). A Venn diagram was constructed to evidence the taxonomic sharing level between brines by using a web-based tool for the analysis of dataset of genera distribution [18].
After normalization, the entire dataset (consisting of previously obtained data plus metabolic prediction results) was then processed by calculating similarity and performing the cluster analysis. The obtained matrix was used to perform the principal component analysis (PCA) by Primer 7 software, using the origin of each Lake as a factor and overlaying vectors of the most affecting factors. Additional nMDS was performed using the Bray–Curtis similarity matrices calculated on transformed data of all biological data and the BEST Spearman rank correlation with physicochemical data (Primer 7). To achieve the main objectives, first, brines samples were compared for their main microbiological and physicochemical data (previously individually reported for BC and TF) (Table S1), and predictive functional attributes (determined in this study). In a second step, the complete dataset was statistically analyzed.
3. Results
3.1. Main Features of Analyzed Antarctic Brines: A Comparison
3.1.1. Chemical Data
From a physicochemical point of view, the brines from BC were richer in some elements (e.g., Ca, Mg, and K) than TF samples (Table S1). Specifically, samples from BC1 and BC3 showed high concentrations of Ca (362.9 and 148.1 mg L−1, respectively), K (70.6 and 18.7 mg L−1, respectively), and Mg (167.4 and 21.8 mg L−1, respectively). Conversely, BC2 had a composition that was similar to TF brines, with values ranging from 0.3 to 2.8 mg L−1. Sulfate ions were detected at a concentration of 5–10 mg L−1 in TF and BC2 brines, while in BC1 and BC3 samples they accounted for more than 1000 mg L−1. The remaining elements did not differ among all samples (Table S1). Salinity and TOC were higher in TF than in BC brines (Table S1).
3.1.2. Microbial Abundance
Prokaryotic cell abundance (PA) was in the order of 108–109 cells L−1 (Table S1). The highest values were observed in TF5 (8.1 × 109 cells L−1), followed by BC1 (6.1 × 109 cells L−1) and TF4 (5.0 × 109 cells L−1). Differently, BC2 and BC3 showed one order of magnitude lower abundance, i.e., 0.38 and 0.41 × 109 cells L−1, respectively. Cell volume (VOL) exhibited remarkable differences among the investigated brines, with the smallest and largest cell sizes in TF4 (mean value 0.04 µm3) and BC1 (0.242 µm3), respectively. In BC2, VOL was on average 0.176 µm3 while in BC3 and TF5 it was 0.139 and 0.105 µm3, respectively. In addition, the prokaryotic biomass (PB) strongly differed among the brine samples, reaching surprisingly high values of 567 and 242 µg C L−1 in BC1 and TF5, respectively.
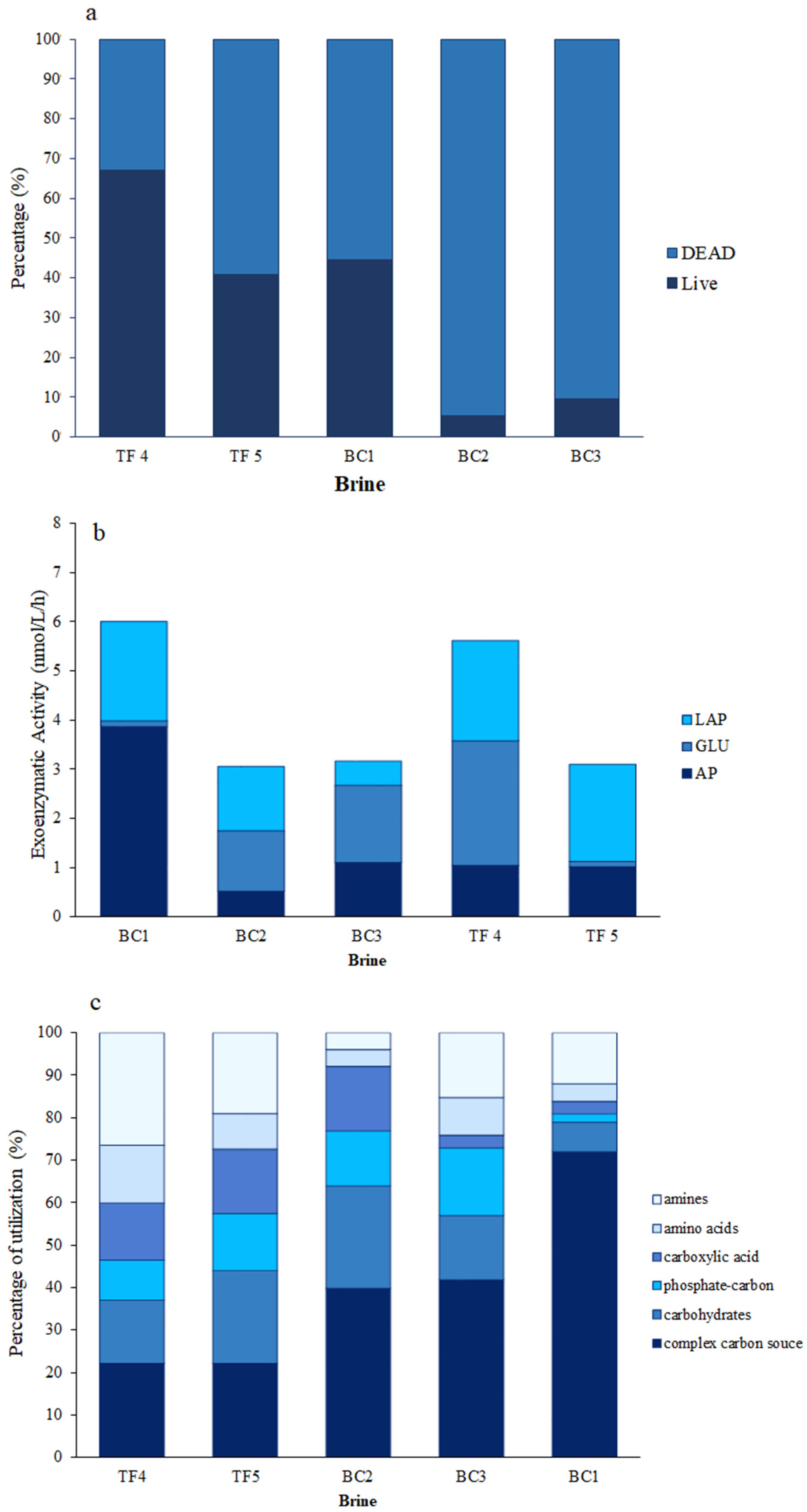
The highest percentage of live cells, accounting for 67%, was observed in TF4, while the lowest was observed in BC2 and BC3 (Figure 1a). Moreover, the percentages of live cells in BC1 and TF5 were comparable. Respiring cells (CTC) were higher in TF brines (18 and 30% of the total cells in TF4 and TF5, respectively) than in BC brines (range 1–8.5) (Table S1).
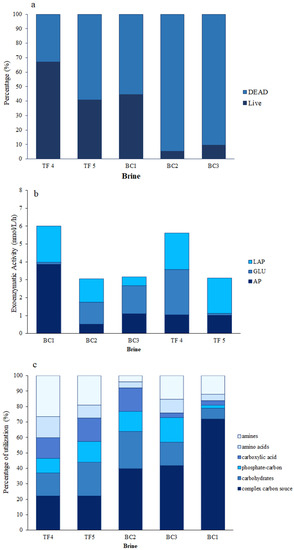
Figure 1.
Microbial abundance and activities: (a) Viable (live/dead) cells in brine samples; (b) enzymatic activities measured in brine samples; (c) percentages of carbon source utilization obtained by Biolog Ecoplate.
Concerning morphotype composition (Table S1), the prokaryotic cells were grouped into six classes: cocci, coccobacilli, vibrios, rods, curved rods, and filamentous forms (i.e., cells exceeding 4 µm in length). In this case, each morphotype also contributed differently to the composition of the microbial assemblage among the brines. Overall, the most abundant cell groups were represented by rods (range 27–74% of the total cells), coccobacilli (range 26–50%, except for BC1), and cocci (range 23–44%, except for BC3) (Table S1). Vibrios and curved rods were almost lacking or negligible. Finally, the filamentous forms with a relatively high percentage (27% of the total cells) were observed in BC1 only.
3.1.3. Microbial Activities
AP activity showed very low rates in BC2 (0.6 nmol L−1 h−1), higher in BC1 (3.9 nmol L−1 h−1), and reciprocally similar values in BC3, TF4, and TF5 (1.1, 1.1, and 1 nmol L−1 h−1, respectively) (Table S1). Differently, β-GLU activity was at low levels in BC1 and TF5 (0.1 and 0.1 nmol L−1 h−1, respectively), while it showed high values in BC2, BC3, and TF4 brines (1.2, 1.6, and 2.5 nmol L−1 h−1, respectively). Finally, LAP activity was measured with minimum value in BC3 (0.5 nmol L−1 h−1); higher rates were found in the remaining samples (ranging from 1.3 nmol L−1 h−1 in BC2 to 2.0 nmol L−1 h−1 in TF4) (Figure 1b).
Overall, the percentages of carbon source utilization obtained by Biolog Ecoplate showed that the complex carbon sources were well utilized polymers in each brine, and mainly in BC brines (Table S1). In particular, in BC3, they accounted for the 72% of the total utilized sources. Differently, amines were better used in TF than in BC. Carbohydrates were well utilized in BC1 and TF5 brines and to a lesser extent in BC2 and TF4 brines. BC1, TF5, and TF4 brines expressed discrete utilization patterns for carboxylic acids, while BC2, BC1, and TF5 brines expressed discrete utilization patterns for phosphate carbon sources. Finally, aminoacids were scantly utilized everywhere, except in TF4 (Figure 1c).
3.1.4. Prokaryotic Community Composition
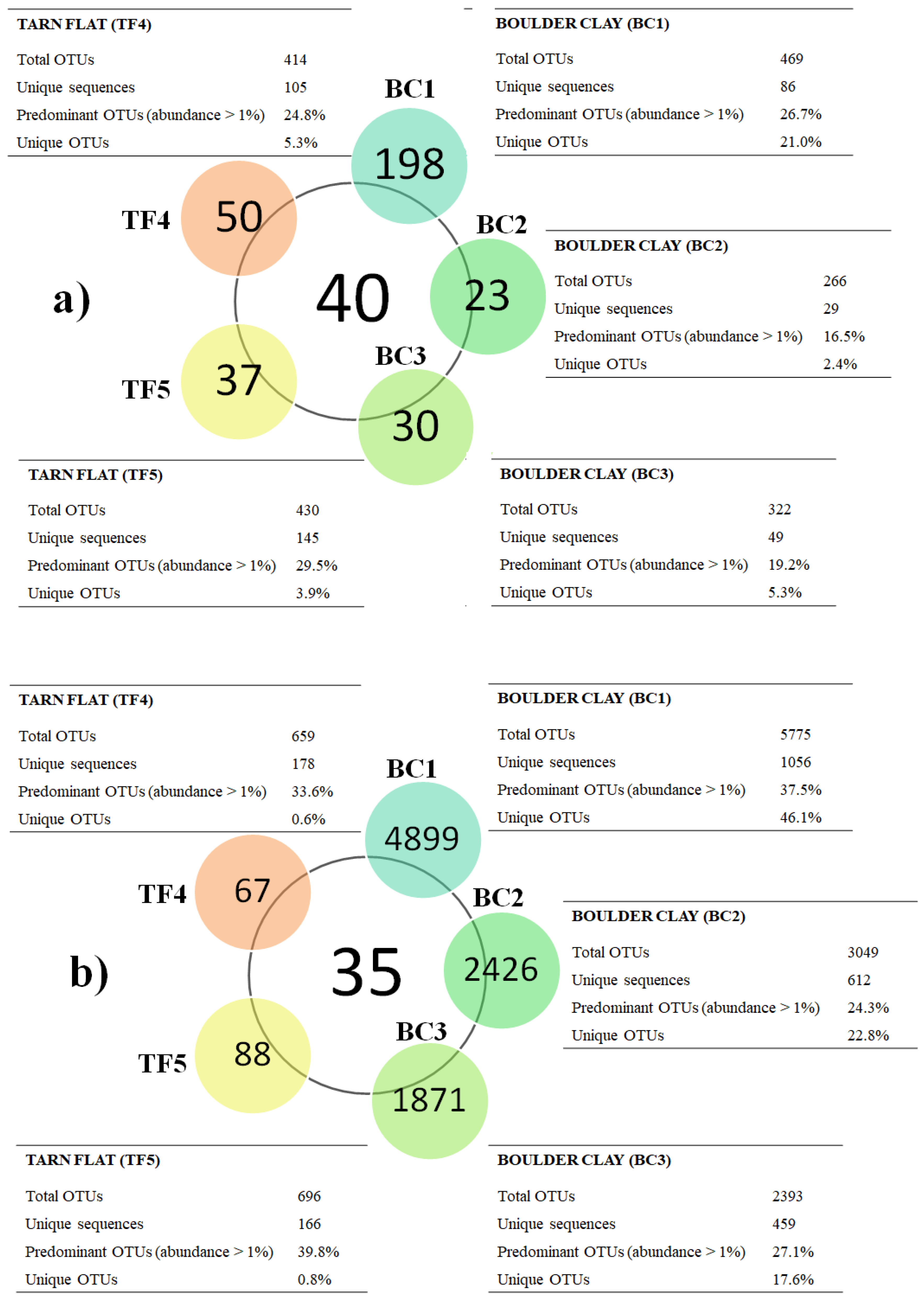
Main data on total sequence reads, quality trimming, OTU information, and diversity indices obtained for brine samples are reported in Supplementary Material Table S2 [4,12]. Overall, a comparable number of bacterial OTUs were generated in BC and TF samples using the Silva database (Figure 2a). Conversely, Archaeal reads were numerically higher in BC than TF brines (Figure 2b). In analyzed brines, the calculated alpha diversity, in terms of Shannon and Chao1 indices, showed a positive trend in TF samples. In particular, all the Archaea samples had higher values than bacteria. In terms of OTU composition, all brines shared 40 bacterial OTUs, while 19 OTUs were shared only between TF brines (Figure 2a and Figure S1a). Similarly, all brines shared 35 archaeal OTUs, with 19 OTUs that were shared among BC brines and 59 between TF brines (Figure 2b and Figure S1b).
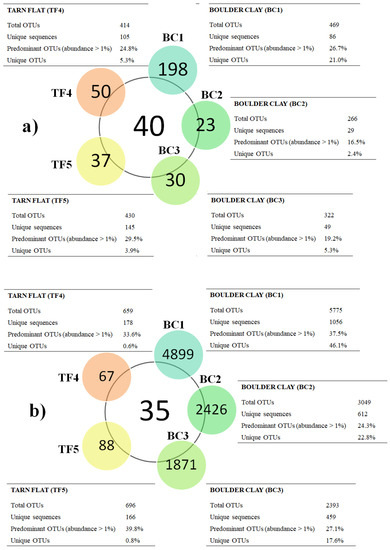
Figure 2.
Diagram comparing bacterial (a) and archaeal (b) communities. Central circles show the number of OTUs shared among brines, while the lateral-colored circles report unique OTUs per brine. The number of total OTUs and unique sequences, as well as the percentages of the predominant and unique OTUs, for each brine, are shown in the tables, which are located close to the sample names.
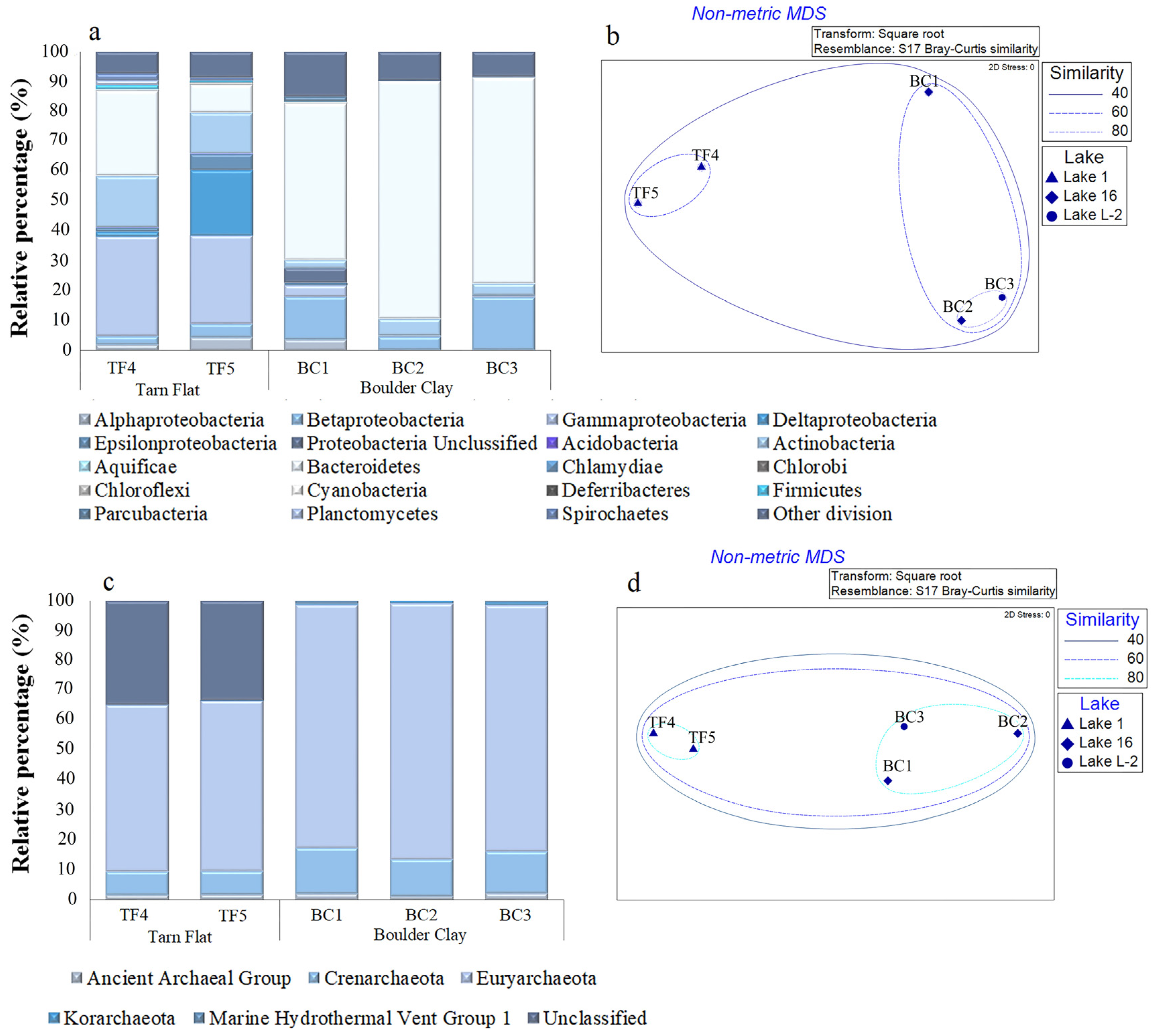
With respect to the bacterial community composition (Figure 3a,b and Table S1), TF and BC brines mainly differed for the predominant phyla (Gammaproteobacteria and Bacteroidetes, respectively), a higher abundance of Actinobacteria in TF than in BC, and the almost exclusive occurrence of Delta- and Epsilonproteobacteria sequences in TF brines. Alphaproteobacterial abundances were comparable in TF5 and BC1 samples. Betaproteobacteria were particularly abundant in BC1 and BC3 brine samples, whereas in BC2, they showed lower relative percentages, but similar to TF brines (Figure 3a and Table S1). As shown in Figure 3b, brines BC2 and BC3 grouped together in a cluster with 80% of similarity, mainly due to the highest and similar Bacteroidetes abundance. Together with BC1, they formed a bigger cluster with a similarity of 60%, while brines from TF clustered separately in a 60% similarity group.
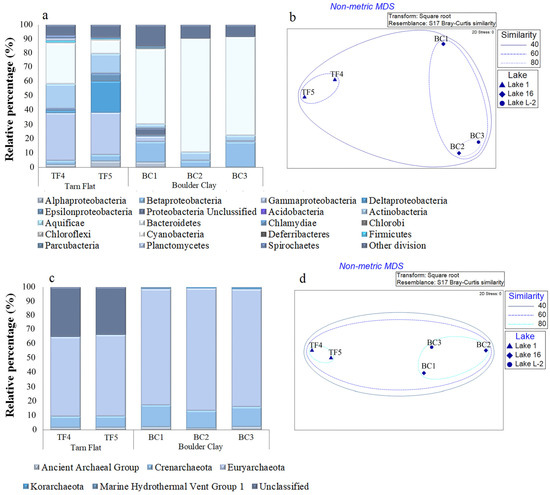
Figure 3.
Bacterial (a) and archaeal (c) community composition at phylum level in TF and BC brines. Nonmetric multidimensional scaling analysis (nMDS) computed on transformed and clustered abundance data for bacterial (b) and archaeal (d) taxonomic groups at the phylum level.
The archaeal community composition (Figure 3c,d) was relatively similar in TF and BC brines. They harbored almost the same taxonomic groups, even if differences in abundance values occurred (Figure 3c and Table S1). In all the brine samples, the archaeal community was mainly represented by Euryarchaeota, with higher abundance in BC than TF brine samples. Crenarchaeota displayed a concentration almost twice in BC than that in TF brines. Two other archaeal groups, i.e., the Ancient Archaeal Group and Korarchaeota, were considerably less abundant in all brines, with the highest percentages (ca. 2%) in BC3 (Figure 3c and Table S1). According to the distribution of archaeal phyla, BC and TF brines clustered in two distinct groups (with a similarity of 80% each). A bigger cluster grouped TF and BC brines with a similarity of 60% (Figure 3d).
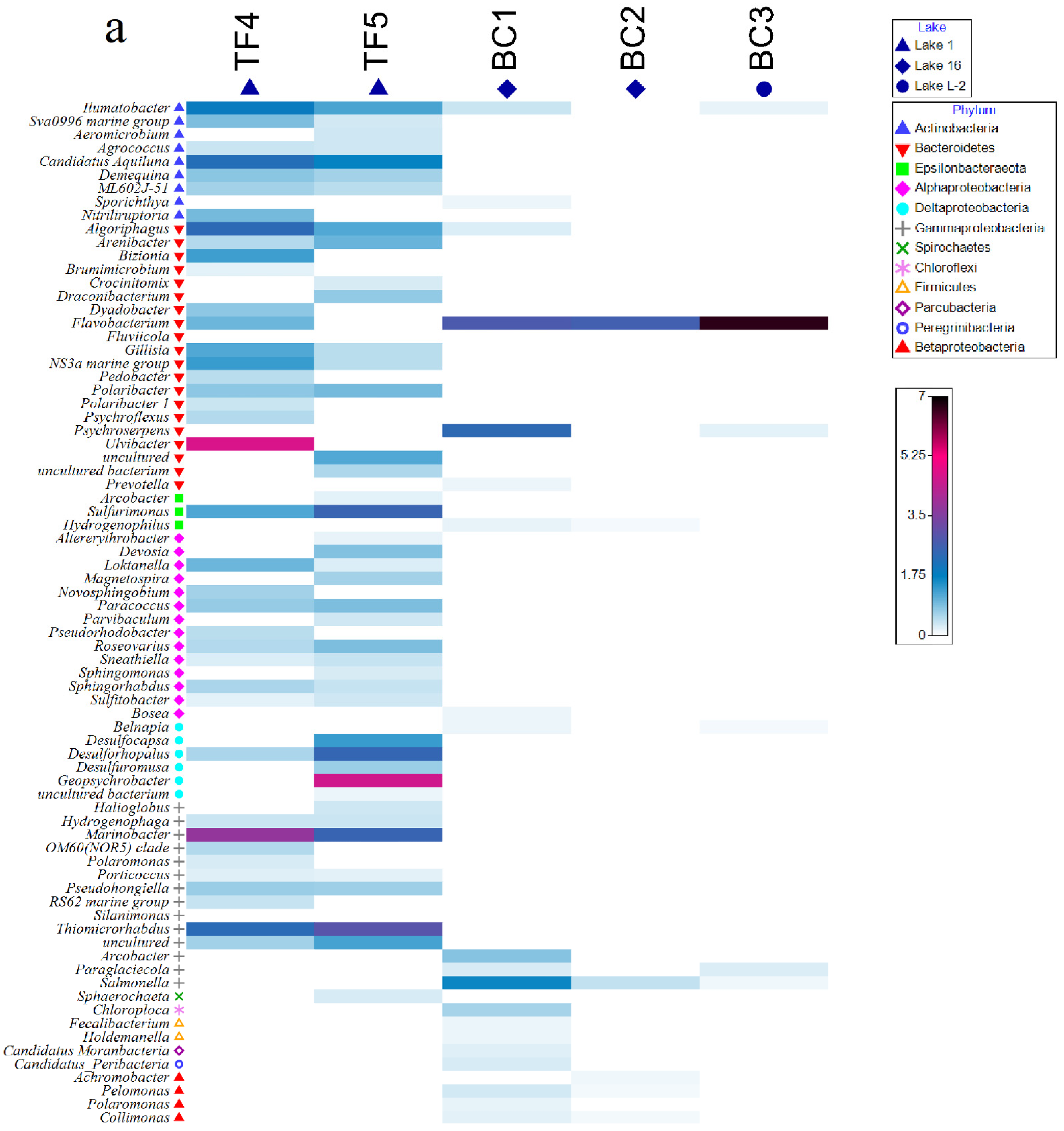
At the genus level, TF and BC brines strongly differed (Figure 4 and Supplementary Material Figure S2). With respect to bacteria, Flavobacterium, Psychroserpens, and Salmonella were the relatively most abundant genera in BC brines (relative abundances ranging from ca. 2 to 44% of the total community), BC1 and BC3 shared with TF4 sequences related to Flavobacterium, whereas Illumatobacter was a common genus among BC1, BC3, and TF brines. The highest genus-sharing level was observed among BC3 and both TF brines (i.e., Psychroserpens, Prevotella, Bosea, Bradyrhizobium, Belnapia, Arcobacter, Paraglaciecola with TF4; Hydrogenophilus, Pelomonas and Collimonas with TF5; Flavobacterium and Salmonella were common genera to BC3 and both TF brines) (Figure 4a and Supplementary Material Figure S2a).
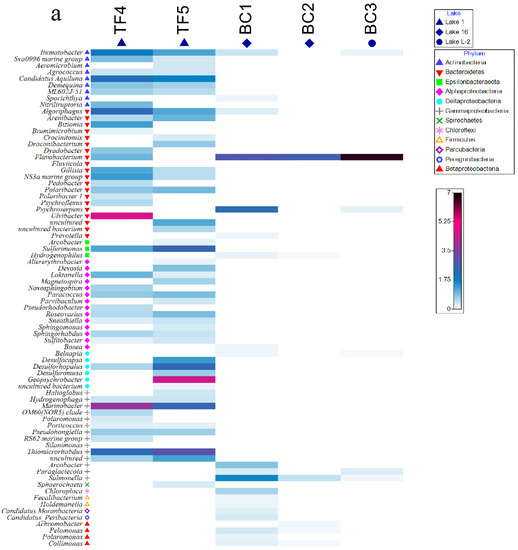
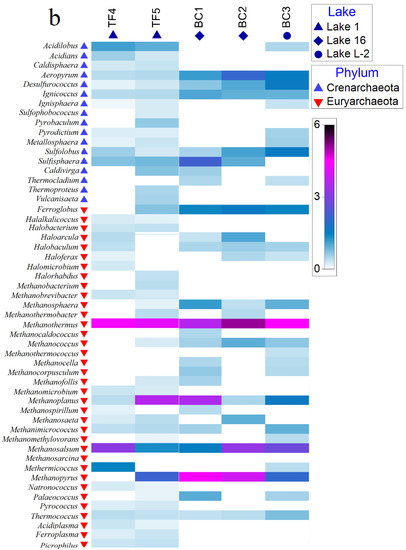
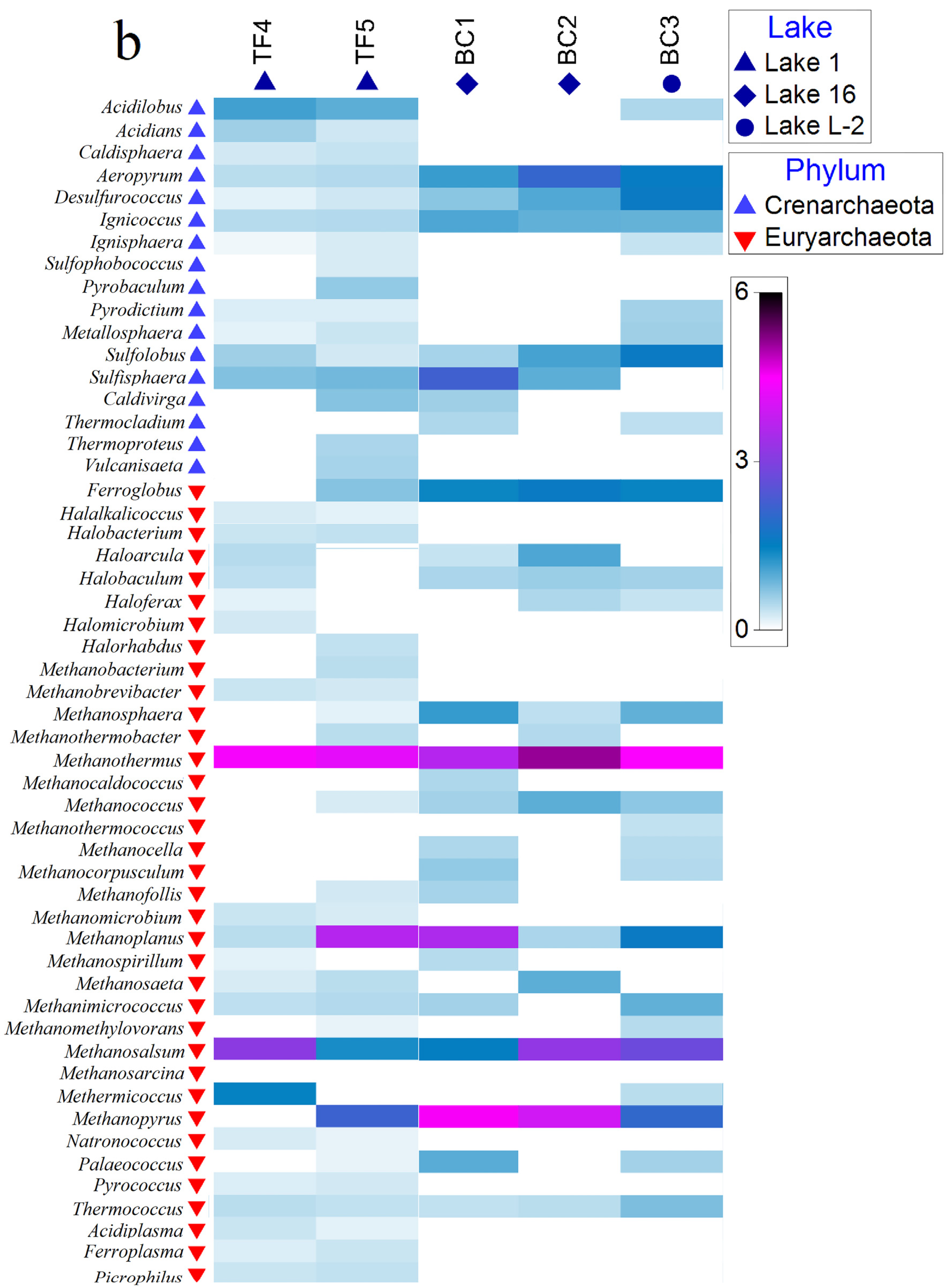
Figure 4.
Shade matrix plots showing (a) the bacterial community structure at the genus level and (b) the archaeal community structure at the genus level. Shading intensity within the matrix indicates the square root transformed relative abundance of each genus.
Archaeal genera were distributed quite homogeneously among TF and BC brines. In particular, the genera Aeropyrum, Desulfurococcus, Ignicoccus, Sulfolobus, Methanothermus, Methanoplanus, Methanosalsum, and Thermococcus were common to all brines. Among them, Methanothermus was the most abundant genus in all brines (relative abundances ranging from ca. 13 to 25% of the total community for BC brines and from 17 to 19% of the total community for TF brines), whereas Aeropyrum (together with Ferroglobus and Methanopyrus) was more abundant in BC (Figure 4b and Supplementary Material Figure S2b).
3.1.5. Overall Comparison among Brine Samples with Respect to Previous Data
The PCA computed on the dataset showed the spatial separation of brine samples (Supplementary Material Figure S3). The two main components explained the 91.1% of the total variance, with PC1 and PC2 accounting for the 61.8 and 29.3% of the variance, respectively. The first component was mainly expressed by the Ca concentration and PB values (negative correlation), while the second component was mainly expressed by the S, TOC, and PB values (positive correlation). BC1 was distinct from other groups. A bigger cluster was composed of all brines (with the exception of BC1) and included two subclusters (i.e., BC2 plus BC3 and TF4 plus TF5).
3.2. Predicted Functional Genes
3.2.1. General Aspects
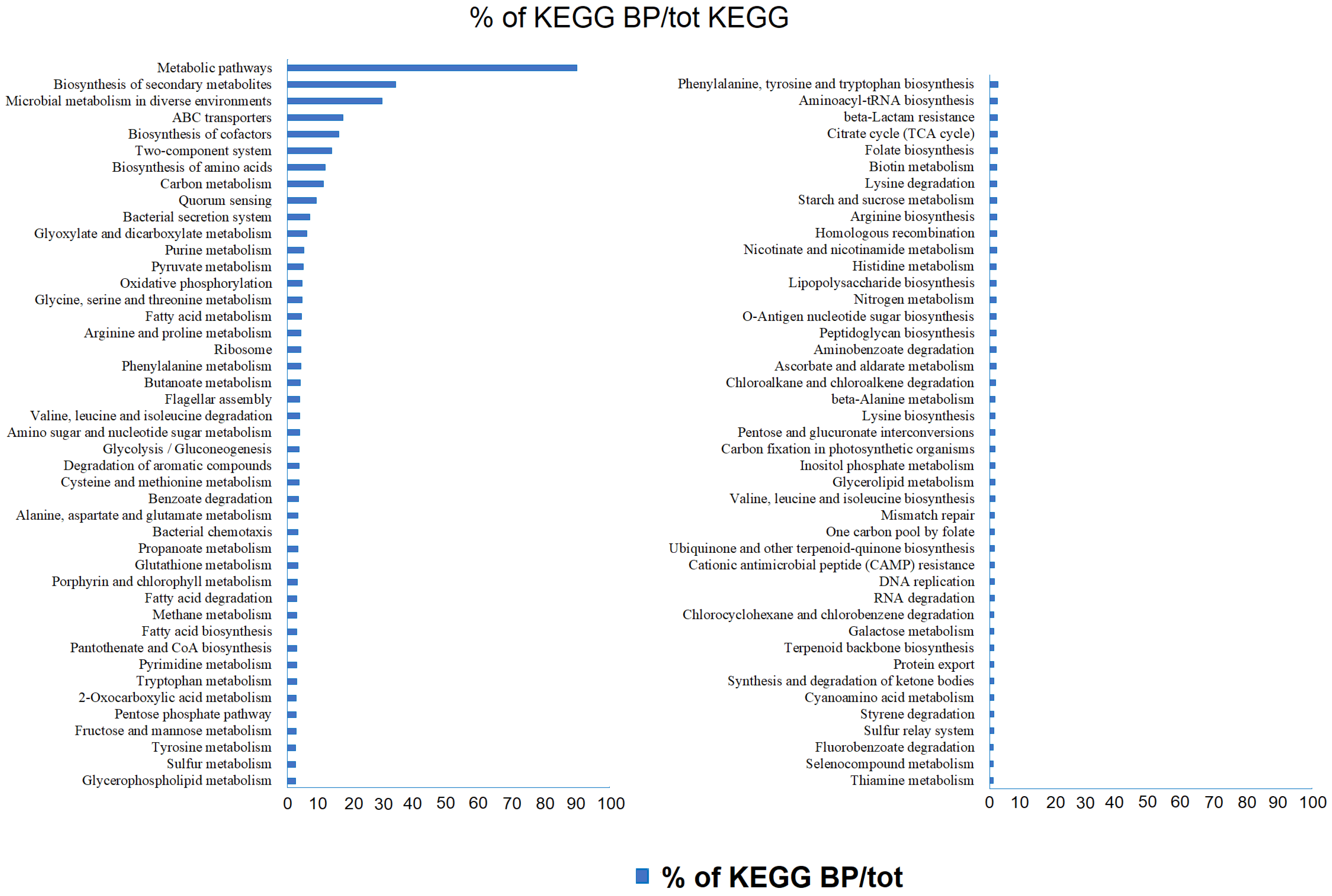
The KEGG pathway database accounted for a total of 1333 predicted KEGG orthologs (Kos, i.e., sets of homologous sequences) in the brine samples. Among the total KOs associated with metabolic processes, 642 (48.16%) had an abundance >0.1% in at least one sample. Overall, the predicted gene sequences annotated based on KEGG pathways resulted in 130 different biological processes, mostly related to functions such as “metabolic pathways” (range ≈ 18–19% of the total bacterial pathways in each sample), followed by “biosynthesis of secondary metabolites” (range ≈ 8–9% of the total bacterial pathways in each sample), “microbial metabolism in diverse environments” (≈5% of the total bacterial pathways in each sample), “biosynthesis of amino acids” (≈4% of the total bacterial pathways in each sample), “biosynthesis of cofactors” (≈4% of the total bacterial pathways in each sample), “ABC transporters” (range ≈ 1–2% of the total bacterial pathways in each sample), and “two components system” (range ≈ 1–2% of the total bacterial pathways in each sample).
The results for predicted biological processes in which unique KEGG showed relative abundance >0.01% for the whole briny prokaryotic community are reported in Figure 5.
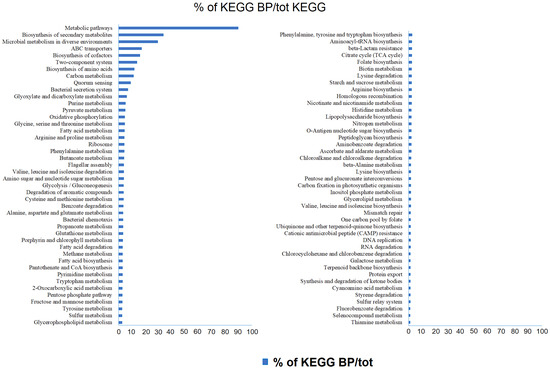
Figure 5.
Percentages of unique KEGGs on the total retrieved KEGGs related to predicted biological processes (abundance >0.01%).
Less represented pathways were related to degradation processes (≈1.5%), methane metabolism (0.6%), sulfur metabolism (≈0.5%), and quorum sensing (≈1.5%). The percentages of retrieved KOs, i.e., related to a single pathway, were calculated.
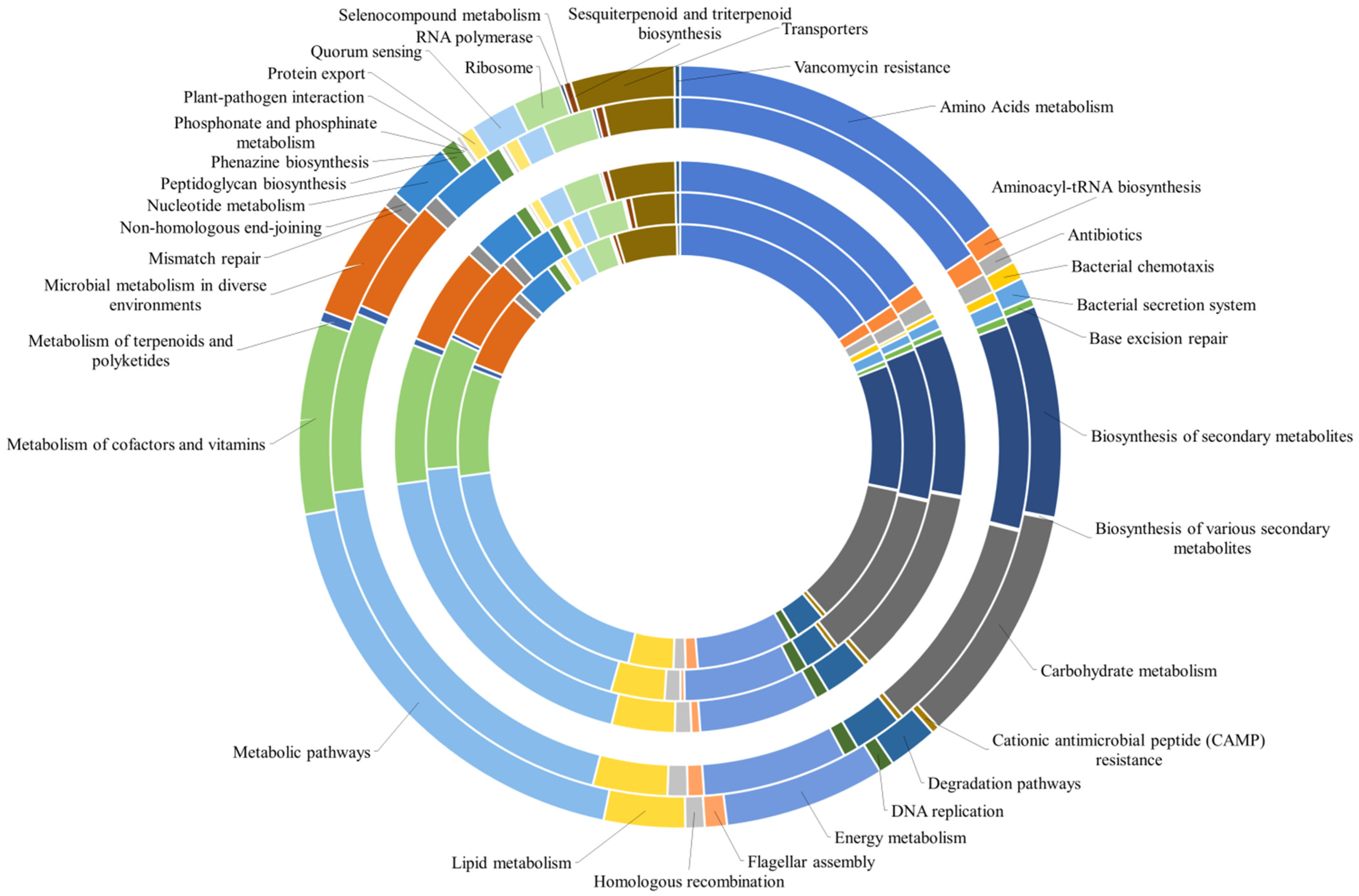
The occurrence of generic metabolic pathways, which included more specific processes, did not allow one to fully appreciate all the predicted functions within the analyzed communities. Therefore, all data were reclassified by grouping together pathways correlated to the same specific biological function (as reported in Supplementary Material Table S3). The relative abundance of predicted genes identified by the PICRUSt analysis within the prokaryotic communities is reported in Figure 6. The biological processes related to metabolic pathways were responsible for the higher number of KOs (approximately 19% in almost all samples) than the other detected biological processes. The second most represented category included all processes related to the “metabolism of amino acids”, with a global value of 15% of predicted genes in all brines, followed by the “carbohydrate metabolism” (10% of predicted genes in all samples). “Biosynthesis of secondary metabolites”, “metabolism of cofactors and vitamins”, “energy metabolism” and “microbial metabolism in different environments” accounted for about 9, 7, 6, and 5% of total predicted genes, respectively. The most represented or interesting metabolic pathways are detailed below.
Figure 6.
Relative abundance of predicted genes of the most abundant pathways identified in the bacterial and archaeal populations by the PICRUSt analysis. The pathways are presented according to KEGGs. Brines are in the order BC1 (as the innermost sample)→BC2→BC3→TF4→TF5 (as the outermost sample).
3.2.2. Metabolism of Amino Acids
The generic pathway “biosynthesis of aminoacids” was retrieved at similar percentages (between 3.8% and 4.1% of the total predicted genes) in all brines. Overall, the most prominent predicted pathways were glycine, serine, and threonine metabolism > cysteine and methionine metabolism > phenylalanine, tyrosine, and tryptophan biosynthesis > alanine, aspartate, glutamate, arginine, lysine, valine, leucine, and isoleucine degradation. The molecular process identified with KEGG16370 (pfkB, 6-phosphofructokinase 2) was more abundant in BC than in TF brines, whereas the process L-serine/L-threonine ammonia lyase (SDS, KEGG17989) was particularly represented in BC1, absent in BC2 and BC3, and less abundant in TF brines (Figure S4a).
3.2.3. Metabolism of Carbohydrates
The carbohydrate metabolism was mainly mediated by predicted pathways such as pyruvate metabolism, glyoxylate and dicarboxylate metabolism, and glycolysis/gluconeogenesis, followed by the 2-oxocarboxylic acid metabolism, citrate cycle (TCA cycle), butanoate, and propanoate metabolism (equally represented in all brine samples). Some predicted molecular processes were better represented in TF5, as in the case of glycolate oxidase iron-sulfur subunit (glcF, KEGG11473), glycolate oxidase (glcD, KEGG00104), and formate dehydrogenase major subunit (fdoG, KEGG00123) (Figure S4b).
3.2.4. Metabolism of Cofactors and Vitamins
The biosynthesis of cofactors was mainly supported by the activity of a number of enzymes, as follows: 3-oxoacyl-[acyl-carrier protein] reductase (fabG, KEGG00059) and 3-oxoacyl-[acyl-carrier-protein] synthase II (fabF, KEGG09458), both involved in the synthesis of fatty acids; aldehyde dehydrogenase (NAD+) (ALDH, KEGG00128); dihydrolipoamide dehydrogenase (DLD, KEGG00382); branched-chain amino acid aminotransferase (KEGG00826); dihydroorotase (URA4, KEGG01465); oxygen-independent coproporphyrinogen III oxidase (hemN, KEGG02495). The molecular processes were similarly represented in all samples. In addition to the biosynthesis of cofactors, other well represented predicted biological processes included folate biosynthesis, nicotinate, and nicotinamide metabolism, one carbon pool by folate and porphyrin and chlorophyll metabolism, with percentages ranging from 0.4% to 0.8% on the total of KOs retrieved in all samples (Figure S4c).
3.2.5. Energy Metabolism
The energy metabolism category was mainly represented by a high number of KOs involved in the “carbon metabolism” and “oxidative phosphorylation” (3 and 1% of the retrieved KOs, respectively) (Figure S4d). In addition, KEGGs also involved in the “methane and sulfur metabolism” were detected. Molecular functions related to methane metabolism were found in all samples. Molecular functions related to sulfur metabolism were detected in all samples. Among them, the most represented were serine O-acetyltransferase (cysE, KEGG00640), thiosulfate/3-mercaptopyruvate sulfurtransferase (TST, KEGG01011), and cysteine synthase (cysK, KEGG01738) involved in the cysteine and sulfur metabolism (in the case of the enzyme form cysK use thiosulfate instead of sulfide, to produce cysteine). Moreover, the molecular process sulfide:quinone oxidoreductase (sqr, KEGG17218), involved in the sulfide oxidation pathways responsible for the sulfide-dependent reduction of quinones, was detected in all samples.
3.2.6. Transporters
The category of transporters was represented by three predicted pathways such as ABC transporters, two-components system, and, to a lesser extent, by the phosphotransferase system (PTS) (Figure S4e). The pathway of ABC transporters, responsible for the transport of substrates across the cell membrane, was similarly represented in all brine samples, with predominant for molecular processes involved in the amino acids and phospholipid transport. In detail, two KEGGs related to amino acid transport via ATP-binding protein (livG, branched-chain amino acid transport system ATP-binding protein, KEGG01995 and livF, branched-chain amino acid transport system, ATP-binding protein, KEGG01996), two KEGGs related to amino acid transport via permease protein (livH, branched-chain amino acid transport system permease protein, KEGG01997 and livM, branched-chain amino acid transport system permease protein, KEGG01998), and one KEGG related to amino acid transport via substrate-binding protein (livK, branched-chain amino acid transport system substrate-binding protein, KEGG01999) were found in all samples. In total, three KEGGs related to phospholipid transport were detected in the brine samples, namely phospholipid/cholesterol/gamma-HCH transport system ATP-binding protein (mlaF, KEGG02065), phospholipid/cholesterol/gamma-HCH transport system permease protein (mlaE, KEGG02066), and phospholipid/cholesterol/gamma-HCH transport system substrate-binding protein (mlaD, KEGG02067).
Some molecular processes were more represented in BC1 brine samples (ccoN, cytochrome c oxidase cbb3-type subunit I, KEGG00404; ccoO, cytochrome c oxidase cbb3-type subunit II, KEGG00405; dctB, two-component system, NtrC family, C4-dicarboxylate transport sensor histidine kinase DctB, KEGG10125; dctD, two-components system, NtrC family, C4-dicarboxylate transport response regulator DctD, KEGG10126), while cytochrome c (CYC, KEGG08738) was highly represented in BC samples as compared with in TF brines.
3.2.7. Degradation Pathways
The predictive analysis also showed the presence of several pathways involved in different degradation processes. The most abundant calculated on the total of all KOs retrieved for degradation pathways were the benzoate degradation and RNA degradation pathways. The most represented molecular processes detected within the RNA degradation are linked to the action of two helicases involved in the maintenance of the genome and ribosome assembly, namely ATP-dependent DNA helicase RecQ (recQ, KEGG03654) and ATP-dependent RNA helicase RhlE (rhlE, KEGG11927). Finally, although to a lower extent, a series of genes involved in the degradation of aliphatic hydrocarbons (chloroalkanes), terpenes (i.e., limonene and pinene), aromatic compounds (i.e., chlorobenzene, styrene, toluene, xylene, and naphthalene) were predicted with similar percentages in all samples (Figure S4f).
3.2.8. Biosynthesis of Secondary Metabolites
The most represented molecular functions associated with the “biosynthesis of secondary metabolites” pathway were the 3-oxoacyl-[acyl-carrier protein] reductase (KEGG00059); aldehyde dehydrogenase (NAD+) (ALDH, KEGG00128); the glyceraldehyde 3-phosphate dehydrogenase (GAPDH, KEGG00134); acetyl-CoA C-acetyltransferase (ACAT, KEGG00626); and acyl carrier protein (acpP, KEGG02078). Some of them were common to other predicted metabolic pathways, and therefore involved in carbohydrate and amino acid metabolism. The molecular function beta-glucosidase (bglX, KEGG05349) was mostly represented in the BC2 brine sample.
3.2.9. Microbial Metabolism in Diverse Environments
The “microbial metabolism in diverse environments” pathway involves all processes related to different kinds of metabolism, i.e., carbohydrate metabolism, energy metabolism, amino acid metabolism, and xenobiotic degradation. In the brine samples, the most represented molecular functions were related to carbohydrate metabolism, such as aldehyde dehydrogenase (NAD+) (ALDH, KEGG00128); GAPDH, glyceraldehyde 3-phosphate dehydrogenase (KEGG00134); succinate-semialdehyde dehydrogenase/glutarate-semialdehyde dehydrogenase (gabD, KEGG00135); dihydrolipoamide dehydrogenase (DLD, KEGG00382); acetyl-CoA C-acetyltransferase (ACAT, KEGG00626); 4-hydroxy-tetrahydrodipicolinate synthase (dapA, KEGG01714); and glutamine synthetase (glnA, KEGG1915).
3.3. Statistical Analyses on the Entire Dataset
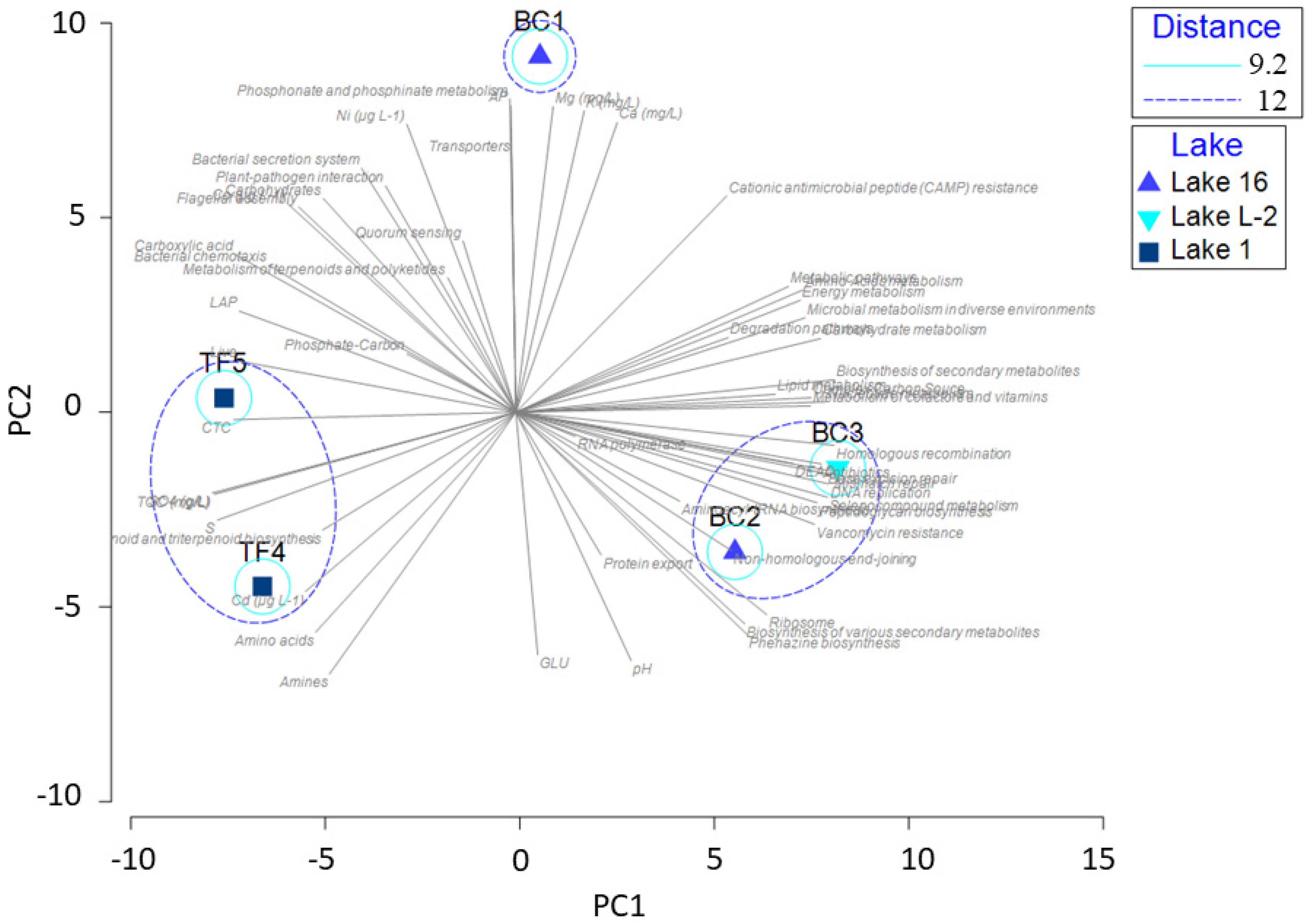
The PCA, shown in Figure 7, was computed on the entire dataset, including results from enzymatic activities, physicochemical parameters, microbiological data, archaeal and bacterial community composition, and predictive analysis of metabolic pathways. The following spatial clustering of brines was obtained: a group including TF4 and TF5 brines; a group including BC2 and BC3 brines; BC1, with a strong positive correlation with some chemical elements (i.e., Ca, Mg, Ni, and K), and some specific substrates (i.e., carbohydrates and carboxylic acids). The two principal components explained 77.9% of the total variance, with PC1 accounting for 48.8% of the variance and the PC2 accounting for 29.1% of the variance. The overlapping of the vectors related to the predicted metabolic pathways showed a higher positive correlation among the BC brines.
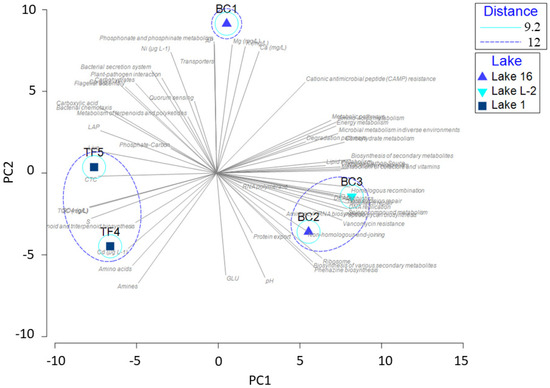
Figure 7.
Principal component analysis computed on the entire dataset, including results from enzymatic activities, physicochemical parameters, microbiological data, archaeal and bacterial community composition, and predictive analysis of the metabolic pathways of brine samples.
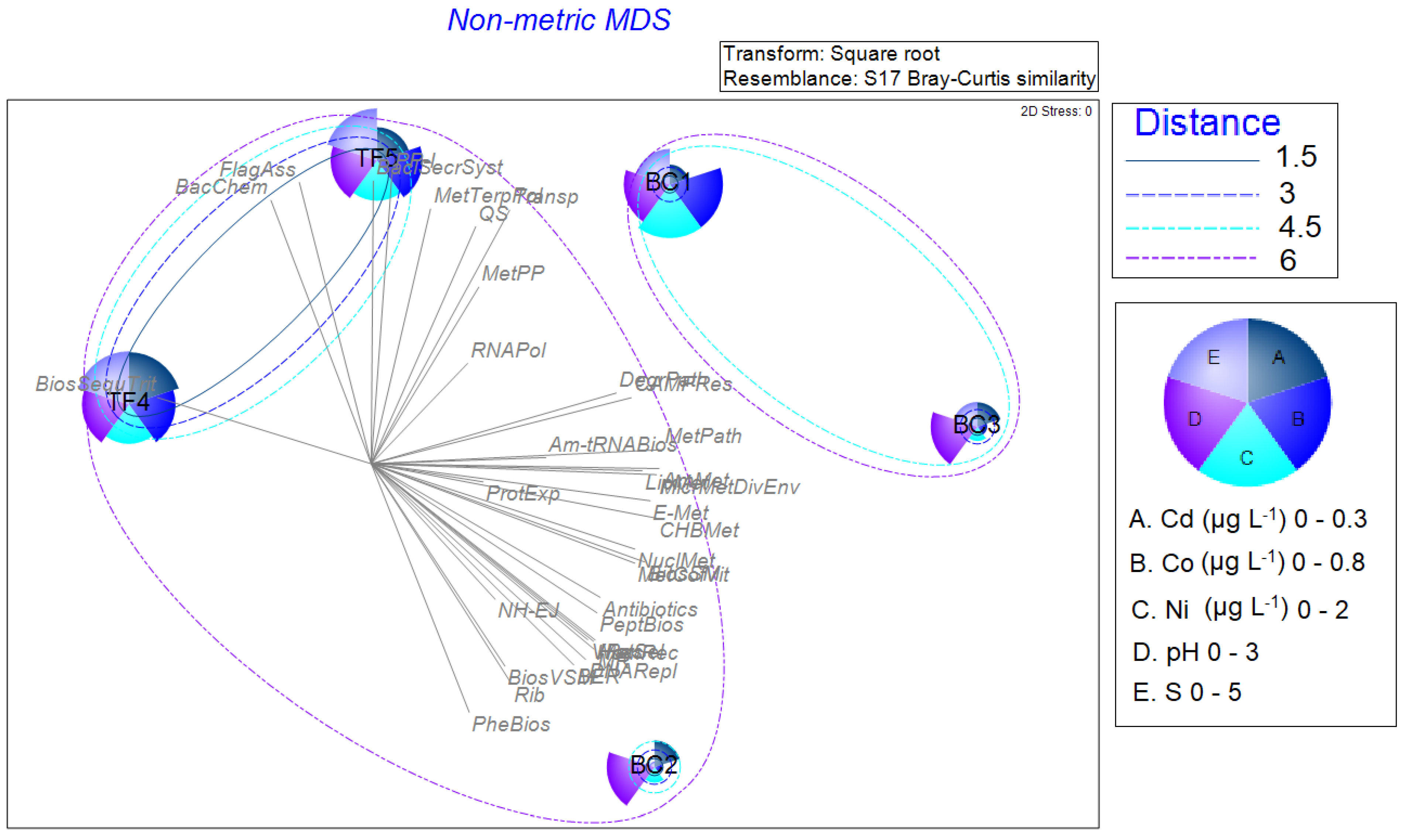
The nMDS, shown in Figure 8, was performed by using all normalized biological data (enzymatic activities, microbiological data, archaeal and bacterial community composition, and predictive analysis), by superimposing the cluster analysis performed on physicochemical results. It evidenced a slight difference in brine sample separation, with BC2 that was included in a bigger cluster together with TF4 and TF5 brines, and BC1 and BC3 samples grouping together in another cluster. The bubble plots show the parameters having the most influence (Co, Cd, Ni, pH, and S) on the multidimensional scaling, as determined by the BEST Spearman rank correlation test.
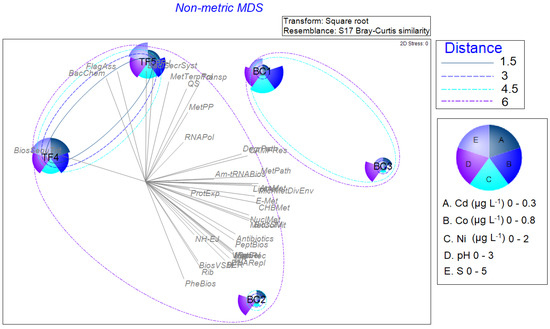
Figure 8.
Non-metric multidimensional scaling analysis computed on normalized biological data (enzymatic activities, microbiological data, archaeal and bacterial community composition, and predictive analysis), by superimposing the cluster analysis performed on the physicochemical results. The bubble plots show the parameters having the most influence on the multidimensional scaling, as determined by the BEST Spearman rank correlation test.
4. Discussion
This study was devised with the intention of contributing to the poor existing knowledge on the microbiology of Antarctic briny systems. Complex datasets, which were previously obtained for three perennially ice-covered Antarctic lakes, were merged and compared. Then, available information was integrated with the analysis of predictive functional metabolic profiles of prokaryotes inhabiting such underexplored cryoenvironments. Physicochemically, three subsystems were distinguished: (1) TF4 plus TF3, (2) BC2, and (3) BC1 plus BC3. This latter was more positively affected by the concentration of certain chemicals (i.e., Ca, Mg, K, and Ni), while TF brines were more affected by salinity and TOC than BC. The contextual analysis of physicochemical and biological data confirms the separation between the two systems (BC and TF), with slight differences. Remarkable differences were observed among the analyzed brines in relation to their microbiological parameters, as described below.
4.1. Microbial Abundance, Biomass, and Morphometric Traits
Microbial abundance was comparable in both TF brines and BC1, where it was higher than BC2 and BC3. The observed discrepancy could depend on several interacting factors such as the possible competition among organisms inhabiting the same environment (i.e., grazers) or the availability of trophic resources. For instance, the specific occurrence of filamentous forms in BC1 and the high PA in TF5 contributed to modulate the microbial biomass in these brines. In terms of VOL, diverse community structures were observed, with larger sizes in BC than in TF brines. These features were presumably related to stress, starvation, or dormant cells in extremely cold and salty environments such as permafrost below the active layer [19,20].
In terms of morphotypes, cocci and rods were differently relevant in TF and BC brines, respectively. Specifically, these aforementioned shapes are connected with inputs of organic substances less or more difficult to be decomposed [21]. In fact, prokaryotic cells modify their size and shape to meet different environmental conditions [22,23]. In this context, studies of the genomic diversity could help to understand the intriguing properties of these cryo ecosystems. For instance, Lo Giudice et al. [12] observed in BC1 brine the massive presence of Flavobacterium spp., a bacterial strain with a mean size of 0.4 µm3.
By comparing the brines for their prokaryotic viability, the viable and respiring prokaryotic communities were more abundant in TF4, TF5, and BC1 samples than in BC2 and BC3, where high proportions of dead cells were present. These clear differences among the brines make us assume that the more limited the resources, the more complex the community is. Overall, the respiring cells were always lower than the LIVE ones by L/D, with the exception of BC3 in whih their percentages were reciprocally similar. This disagreement between L/D and CTC results could be due to the different approaches used to recognize the living cells based on the cell membrane integrity (L/D) and the active respiring capacity (CTC), respectively. Cellular respiratory activity and cell membrane integrity are among the most suitable parameters for evaluating the viability of cells in a microbial assemblage [24]. According to Posch et al. [25], the low percentages of CTC+ cells could be attributed to alive but dormant cells or with respiration rates below the detection limit of the method.
4.2. Microbial Activities
The occurrence of viable and respiring prokaryotic cells implies the capability to metabolize in situ thermogenic or biogenic sources, as reported by Murray et al. [8]. In fact, the organic carbon resources contained in the brines could enable microbial populations to survive for millennia, as suggested in Lake Vida. In our study, the microbial assemblages of each brine were found to be able to potentially metabolize the carbon sources. The main characteristic was the good utilization of the complex carbon sources in all the brines, probably owing to the easy degradability and antifreeze characteristics of these compounds [26]. In cryosystems, they influenced the adaptability of the prokaryotic community to freezing conditions, avoiding the intracellular ice crystal growth, and the consequent cell damage [27,28]. Carbohydrates were also efficiently used in almost all brines, and mostly in BC1 and TF5. They are functional biomolecules that allow energy storage, particularly important to survive in extreme environments [29]. Differently, the capability to utilize amines, mainly in TF4, suggested the occurrence of nitrogen compounds, nitrifying bacteria, and nitrogen fixation [30]. Finally, the discrete utilization of carboxylic acids, and among them the pyruvic acid (particularly in BC1), reinforced the results on the occurrence of methanogens, as it was determined by molecular analyses (see below).
Substrate utilization is strictly dependent on the production of enzymes. Under extremely cold conditions, such as those in icy brines, enzymes work at freezing temperatures [31] or under saturating salt conditions [32]. Nevertheless, their activity under both salinity and temperature extremes has been the subject of limited investigations [33]. Bacteria isolated from cold environments have been shown to produce several extracellular enzymes, such as leucine aminopeptidases or proteases [34,35], amylase [36,37], and esterase [38]. The peculiar characteristics of sea brines such as low temperature and high salt increase the viscosity of such matrices, limiting the diffusion processes [39], as well as reducing water activity within them, which is critical for enzyme activities [33]. The LAP, AP, and ß-GLU activity rates detected in our studies [4,5] suggested that the microbial communities inhabiting the brines of TF and BC lakes had the ability to produce a relatively wide spectrum of enzymes involved in the degradation of organic polymers.
Extracellular enzyme activity is the first step for the decomposition of high molecular weight organic matter into monomers or small molecules that microbial communities can uptake [40]. The low molecular weight compounds released by the active hydrolytic process could be beneficial to the brine communities and also to the surrounding communities, enabling them to exploit the available organic matter. Within sea-ice brines, enhanced enzyme production may help balance this resource need, where characteristic extremes of temperature and salinity already act to reduce growth rates [41]. Bacterial use of extracellular enzymes to degrade high molecular weight material under extreme conditions, such as in the brines, also involves chemotaxis, halotaxis, and chemohalotaxis that bacteria may adopt to position themselves in response to environmental gradients encountered within the sea-ice brine network; the study of organic matter decomposition in this context can give insights into the potential bacterial activity in an extraterrestrial ice layer [41]. Active proteolytic activity suggests that brine pockets are enriched in proteinaceous material, such as observed in sea-ice brines [42].
4.3. Prokaryotic Community Compositions
The analyzed brine systems hosted peculiar communities of microorganisms, sulfate-reducing bacteria, and methanogenic Archaea, as has been previously discussed [4,12]. However, diversity analyses on the bacterial and archaeal communities highlighted a clear separation between the Tarn Flat and Boulder Clay brine systems, thus, reflecting the above discussed data on microbial abundances and activities. For Bacteria, main differences relied on the predominance of Bacteroidetes in BC brines, and the exclusive occurrence of Delta- and Epsilonproteobacteria in TF brines (particularly in TF5). More markedly, BC and TF brines differed at the genus level. The highest genus-sharing (mainly dependent on Bacteroidetes affiliates) was observed among BC3 and both TF brines. The separation of the BC3 brines from both BC1 and BC2 samples (deriving from an adjacent lake) suggests the occurrence of lake-specific bacterial communities. For Archaea, even if the same taxonomic groups were detected, they occurred at a different extent, with Euryarchaeota that were more abundant in BC than in TF brines.
4.4. Prediction of the Metabolic Profiles
To the best of our knowledge, the prediction of metabolic profiles has never been reported for microbial communities inhabiting brines of perennially ice-covered Antarctic lakes. The PICRUSt software compares the identified 16S rRNA gene sequences to those of known genome-sequenced species, thereby, estimating the possible gene contents of the uncultured microbial communities [43]. Although this tool has been designated and validated for humane microbiomes [44], its accuracy to detect the phylogenetic proximity of the reference genomes to the environmental strains has been evaluated. The authors assessed the prediction efficiency of the bioinformatics tool to compare potential functions of samples driving from a wider variety of habitats [16]. According to the authors, the method is valid for application on 16S datasets providing valuable accuracy measure, for example, the nearest sequenced taxon index (NSTI, in our study it ranged between 0.08 and 0.1).
In our study, a very high rate of putative biological processes derived from the predictive analysis, revealing a microbial metabolism mainly based on the processes of synthesis and biodegradation of organic molecules. BC and TF brines did not show a sharp separation in terms of predicted metabolic profile, suggesting that, despite the differences encountered at both the taxonomic and environmental levels, the communities globally envisaged similar metabolic capacities. However, some small point variations delineate the possible presence of specific molecular processes.
Among the best represented metabolic pathways, the biosynthesis of amino acids was identified as one of the most predicted in the microbial communities. As part of processes related to amino acid metabolism, some pathways were strongly predicted in BC brines, such as those for 6-phosphofructokinase 2 and L-serine/L-threonine ammonia-lyase. This latter protein is involved in the first step of the sub-pathway synthesizing 2-oxobutanoate from L-threonine, and in the L-isoleucine biosynthesis (also involved in the amino acid biosynthesis).
The molecular function beta-glucosidase (bglX, KEGG05349), detected within the metabolic pathway of carbohydrate metabolism, was mostly represented in the BC2 brine. This process is responsible for the hydrolysis of the glycosidic bonds in various glycosides and oligosaccharides with the release of glucose [44]. Differently, other molecular processes were better represented in TF brines, glycolate oxidase iron-sulfur subunit (involved in the oxidation of glycolate to glyoxylate, which generally requires using glycolate as a sole carbon source) and the formate dehydrogenase major subunit. This enzyme is also involved in the energy metabolism pathway and belongs to a set of enzymes able to catalyse the formate oxidation by using a second substrate as electron acceptor, for example, NAD+. Generally, this kind of dehydrogenases are involved in the metabolic processes of methylotrophic microorganisms and are crucial in the catabolism of C1 compounds, such as methanol [45].
The most prominent energy-generating metabolic pathways were predicted to be carbon metabolism, methane metabolism, and oxidative phosphorylation, indicating that ATP was generated by electron transfer to terminal acceptor, i.e., oxygen, nitrate, or sulfate. In particular, the prediction of genes encoding enzymes involved in methane metabolism confirmed the considerations by Stibal et al. [46], who highlighted the occurrence of methanogens in polar subglacial systems. The results reinforce our observation on microbial activities and are in line with the abundance of sequences related to Methanopyrales in TF5 brine [4] and strictly anaerobic methanogens in the BC active community [12], respectively.
Most of the studies related to the N-cycling functional markers in Antarctica have focused on the core genes involved in N-fixation, nitrification, and denitrification processes [47]. Nitrate is, here, predicted to be reduced through two dissimilatory membrane-bound respiratory (Nar) nitrate reductases, which generate a transmembrane proton motive force allowing ATP synthesis. Membrane-bound nitrate reductases are generally associated with denitrification and anaerobic nitrate respiration processes. Ammonia is then probably used as raw material for L-glutamate synthesis, as suggested by the predicted glnA gene, which catalyses the ATP-dependent biosynthesis of glutamine from glutamate and ammonia.
Sulfur metabolism was predicted as a common pathway for energy in all brines. This seemed to be mainly based on assimilatory sulfate reduction, a process entirely occurring in plants and microorganisms, necessary for the formation of sulfur-containing amino acids. The processes are mediated through sulfite reduction by reductases encoded from genes of cysJIH operon, almost equally predicted in all brine samples. A crucial step in the assimilatory sulfate reduction metabolic pathway is the formation of 3′-phosphoadenosine-5′-phosphosulfate [48]. The molecular process is catalysed by the enzyme phosphoadenosine phosphosulfate reductase, detected in this study through the prediction of the encoding cysH gene. The occurrence of such molecular processes mediating sulfur metabolism is also supported by the prediction of the gene cysK, encoding for the cysteine synthase, and by the occurrence of genes encoding for enzymes involved in sulfate transportation, such as cysP, cysU, cysW, and cysA. Interestingly, the sulfide quinone reductase (SQR) activity, here, detected at similar extent in all samples, was found to be widely distributed among prokaryotes, and the protein sequence comparison leads to the conclusion that SQR is a phylogenetically very old enzyme that was acquired early in evolution [49]. These findings are in line with previous evidence of the presence of sulfur cycling microorganisms in Antarctic subglacial environments [50,51,52,53,54,55], able to support sulfur transformations providing energy for growth.
Overall, the predictive analysis alone did not separate the briny microbial communities at a metabolic potential level. The number of samples was exiguous, due to the low accessibility and the difficulty to collect from such remote and peculiar ecosystems. However, in this study, we try to use some statistical elaboration to highlight possible distinguishing features among brines, which should be considered cautiously. TF and BC brines appeared distinguished, probably by differences intrinsic to individual systems. Interestingly, the spatial distribution obtained by analysing the entire dataset reflected that obtained from the elaboration of the sole physicochemical data (Figure 7 and Supplementary Material Figure S3) but suggested that biological parameters tend to separate more markedly BC2 and BC3 brines from TF4 and TF5 that, instead, grouped all together in a bigger cluster in the physicochemical level analysis. The BEST Spearman rank correlation test detected five parameters, namely Cd, Co, Ni concentrations, and pH and salinity values, as those that most affected the brine systems. Except for pH, all these parameters affected mostly TF4, TF5, and BC1 brines.
5. Conclusions
The brine systems differed in geochemical conditions and phylogenetic diversity of the hosted microorganisms, but they shared a similar metabolic potential even at conditions commonly considered prohibitive for life. The environmental parameters, mainly salinity and metal concentrations, affected these habitats by shaping the taxonomic composition of the microbial communities. The observation of microbial communities that can grow and survive in icy systems on Earth is an important tool for expanding our awareness of life subsistence and persistence under challenging conditions. The collective ability of brine microbial communities to catabolize several carbon sources, and to play a pivotal role in nutrient cycling, strongly suggests a nutritional versatility which could provide a fitness advantage. Bioinformatics tools have given a new edge to the research approach. Exploratory analyses can be very useful to focus more in-depth studies appropriately and are of greater value especially for remote sites that are difficult to access. Here, the predictive analysis gained more insights into the ecological role of microbial communities in perennially ice-covered Antarctic lakes and prospected environmental parameters shaping their structure.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/d13070323/s1, Figure S1: Venn diagram for (a) Bacteria and (b) Archaea, Figure S2: Venn diagram showing the sharing level of bacterial (a) and archaeal (b) genera between BC and TF brine samples, Figure S3: Principal component analysis computed on the previous dataset including physicochemical data related to BC and TF brine samples, Figure S4: Circos diagrams showing the connection among brine samples and metabolic pathways. The ribbon size represents the cell value corresponding to a brine/metabolic pathway segment pair, while the stacked bars show ribbon contribution for each segment, Table S1: Microbiological and physicochemical data previously reported for brines, Table S2: The sequence reads, good quality reads, observed number of OTUs, and Shannon diversity, Evenness, and Chao 1 indices per sample of the bacterial and archaeal 16S rRNA gene datasets. Data are from Papale et al. [4] and Lo Giudice et al. [12], Table S3: Relative abundance (% on the total predicted pathways) of predicted metabolic pathways identified in prokaryotic populations produced by the PICRUSt analysis. The icons are referred to percentages within the same subgroup (in bold).
Author Contributions
Conceptualization, A.L.G.; methodology, all authors; software, C.R. and M.P.; formal analysis, M.P., C.R., G.C. and G.M.; investigation, all authors; data curation, all authors; writing—original draft preparation, A.L.G., C.R. and M.P.; writing—review and editing, all authors; supervision, A.L.G.; funding acquisition, M.A. and M.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Programma Nazionale di Ricerche in Antartide (PNRA): PNRA16_00194—A1 “Climate Change and Permafrost Ecosystems in Continental Antarctica” and PNRA18_00186-E “Interactions between permafrost and ecosystems in Continental Antarctica”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data on bacterial abundances and activities are available upon request. Ion Torrent sequence data obtained from this study have been registered as NCBI Bioproject PRJNA435985.
Conflicts of Interest
The authors declare no conflict of interest.
References
- French, H.M.; Guglielmin, M. Frozen ground phenomena in the vicinity of Terra Nova Bay, Northern Victoria Land, Antarctica: A preliminary report. Geogr. Ann. 2000, 82, 513–526. [Google Scholar] [CrossRef]
- Guglielmin, M.; Lewkowicz, A.G.; French, H.M.; Strini, A. Lake-ice blisters, Terra Nova Bay area, Northern Victoria Land, Antarctica. Geogr. Ann. Ser. A Phys. Geogr. 1999, 91, 99–111. [Google Scholar] [CrossRef]
- Borruso, L.; Sannino, C.; Selbmann, L.; Battistel, D.; Zucconi, L.; Azzaro, M.; Turchetti, B.; Buzzini, P.; Guglielmin, M. A thin ice layer segregates two distinct fungal communities in Antarctic brines from Tarn Flat (Northern Victoria Land). Sci. Rep. 2018, 8, 6582. [Google Scholar] [CrossRef]
- Papale, M.; Lo Giudice, A.; Conte, A.; Rizzo, C.; Rappazzo, C.; Maimone, G.; Caruso, G.; La Ferla, R.; Azzaro, M.; Gugliandolo, C.; et al. Microbial assemblages in pressurized Antarctic brine pockets (Tarn Flat, Northern Victoria Land): A hotspot of biodiversity and activity. Microorganisms 2019, 7, 333. [Google Scholar] [CrossRef]
- Azzaro, M.; Maimone, G.; La Ferla, R.; Cosenza, A.; Rappazzo, A.C.; Caruso, G.; Paranhos, R.; Cabral, A.S.; Forte, E.; Guglielmin, M. The prokaryotic community in an extreme Antarctic environment: The brines of Boulder Clay lakes (Northern Victoria Land). Hydrobiologia 2021, 848, 1837–1857. [Google Scholar] [CrossRef]
- Preston, L.; Dartnell, L. Planetary habitability: Lessons learned from terrestrial analogues. Int. J. Astrobiol. 2014, 13, 81–98. [Google Scholar] [CrossRef]
- Porcino, N.; Cosenza, A.; Azzaro, M. A review on the geochemistry of lakes in Victoria Land (Antarctica). Chemosphere 2020, 251, 126229. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.E.; Kenig, F.; Fritsen, C.H.; McKay, C.P.; Cawley, K.M.; Edwards, R.; Kuhn, E.; McKnight, D.M.; Ostrom, N.E.; Peng, V.; et al. Microbial life at −13 °C in the brine of an ice-sealed Antarctic lake. Proc. Natl. Acad. Sci. USA 2012, 109, 20626–20631. [Google Scholar] [CrossRef] [PubMed]
- Dugan, H.A.; Doran, P.T.; Wagner, B.; Kenig, F.; Fritsen, C.H.; Arcone, S.A.; Kuhn, E.; Ostrom, N.E.; Warnock, J.P.; Murray, A.E. Stratigraphy of Lake Vida, Antarctica: Hydrologic implications of 27 m of ice. Cryosphere 2015, 9, 439–450. [Google Scholar] [CrossRef]
- Forte, E.; Dalle Fratte, M.; Azzaro, M.; Guglielmin, M. Pressurized brines in continental Antarctica as a possible analogue of Mars. Sci. Rep. 2016, 6, 33–58. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, C.; Conte, A.; Azzaro, M.; Papale, M.; Rappazzo, A.C.; Battistel, D.; Roman, M.; Lo Giudice, A.; Guglielmin, M. Cultivable bacterial communities in brines from perennially ice-covered and pristine Antarctic lakes: Ecological and biotechnological implications. Microorganisms 2020, 8, 819. [Google Scholar] [CrossRef]
- Lo Giudice, A.; Conte, A.; Papale, M.; Rizzo, C.; Azzaro, M.; Guglielmin, M. Prokaryotic diversity and metabolically active communities in brines from two perennially ice-covered Antarctic lakes. Astrobiology 2021, 21, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Oyewusi, H.A.; Wahab, R.A.; Edbeib, M.F.; Mohamad, M.A.N.; Hamid, A.A.A.; Kaya, Y.; Huyop, F. Functional profiling of bacterial communities in Lake Tuz using 16S rRNA gene sequences. Biotechnol. Biotechnol. Equip. 2021, 35, 1–10. [Google Scholar] [CrossRef]
- Sannino, C.; Borruso, L.; Mezzasoma, A.; Battistel, D.; Zucconi, L.; Selbmann, L.; Azzaro, M.; Onofri, S.; Turchetti, B.; Buzzini, P.; et al. Intra- and inter-cores fungal diversity suggests interconnection of different habitats in an Antarctic frozen lake (Boulder Clay, Northern Victoria Land). Environ. Microbiol. 2020, 22, 3463–3477. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.N.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An improved and extensible approach for metagenome inference. bioRxiv 2019, 672295. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, 353–361. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- La Ferla, R.; Azzaro, M.; Michaud, L.; Caruso, G.; Lo Giudice, A.; Paranhos, R.; Cabral, A.S.; Conte, A.; Cosenza, A.; Maimone, G.; et al. Prokaryotic abundance and activity in permafrost of the Northern Victoria Land and Upper Victoria Valley (Antarctica): A study case. Microb. Ecol. 2017, 74, 402–415. [Google Scholar] [CrossRef]
- Papale, M.; Conte, A.; Mikkonen, A.; Michaud, L.; La Ferla, R.; Azzaro, M.; Caruso, G.; Paranhos, R.; Cabral, A.S.; Maimone, G.; et al. Prokaryotic assemblages within permafrost active layer at Edmonson Point (Northern Victoria Land, Antarctica). Soil Biol. Biochem. 2018, 123, 165–179. [Google Scholar] [CrossRef]
- Kalcheva, H.; Beshkova, M.; Pehlivanov, L.; Kalcev, R. Bacterioplankton dynamics and the influence of environmental factors on it in the Srebarna Lake. In Proceedings of the The Third International Scientific Conference BALWOIS, Ohrid, Republic of Macedonia, 27–31 May 2008. [Google Scholar]
- Young, K.D. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 2006, 70, 660–703. [Google Scholar] [CrossRef]
- Gentile, G.; Maimone, G.; La Ferla, R.; Azzaro, M.; Catalfamo, M.; Genovese, M.; Santisi, S.; Maldani, M.; Macrì, A.; Cappello, S. Phenotypic variations of Oleispira antarctica RB8T in different growth conditions. Curr. Microbiol. 2020, 77, 3414–3421. [Google Scholar] [CrossRef]
- Rezaeinejad, S.; Ivanov, V. Heterogeneity of Escherichia coli population by respiratory activity and membrane potential of cells during growth and long-term starvation. Microbiol. Res. 2011, 166, 129–135. [Google Scholar] [CrossRef]
- Posch, T.; Pernthaler, J.; Alfreider, A.; Psenner, R. Cell-specific respiratory activity of aquatic bacteria studied with the tetrazolium reduction method, cyto-clear slides, and image analysis. Appl. Environ. Microbiol. 1997, 63, 867–873. [Google Scholar] [CrossRef]
- Kenarova, A.; Encheva, M.; Chipeva, V. Physiological diversity of bacterial communities from different soil locations on Livingston Island, South Shetland archipelago, Antarctica. Pol. Biol. 2013, 36, 223–233. [Google Scholar] [CrossRef]
- Loferer-Krössbacher, M.; Klima, J.; Psenner, R. Determination of bacterial cell dry mass by transmission electron microscopy and densitometric image analysis. Appl. Environ. Microbiol. 1998, 64, 688–694. [Google Scholar] [CrossRef]
- Deming, J.W. Psychrophiles and polar regions. Curr. Opin. Microbiol. 2002, 5, 301–309. [Google Scholar] [CrossRef]
- Wagner, D.; Kobabe, S.; Liebner, S. Bacterial community structure and carbon turnover in permafrost-affected soils of the Lena Delta, North Eastern Siberia. Can. J. Microbiol. 2009, 55, 73–83. [Google Scholar] [CrossRef]
- Laybourn-Parry, J.; Pearce, D.A. The biodiversity and ecology of Antarctic lakes: Models for evolution. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 2273–2289. [Google Scholar] [CrossRef] [PubMed]
- Huston, A.L.; Methe, B.; Deming, J.W. Purification, characterization, and sequencing of an extracellular cold-active aminopeptidase produced by marine psychrophile Colwellia psychrerythraea strain 34H. Appl. Environ. Microbiol. 2004, 70, 3321–3328. [Google Scholar] [CrossRef] [PubMed]
- Ortega, G.; Lain, A.; Tadeo, X.; Lopez-Mendez, B.; Castano, D.; Millet, O. Halophilic enzyme activation induced by salts. Sci. Rep. 2011, 1, 6. [Google Scholar] [CrossRef]
- Karan, R.; Capes, M.D.; DasSarma, S. Function and biotechnology of extremophilic enzymes. Biochemistry 2011, 76, 686–693. [Google Scholar]
- Lei, F.; Zhao, Q.; Sun-Waterhouse, D.; Zhao, M. Characterization of a salt-tolerant aminopeptidase from marine Bacillus licheniformis SWJS33 that improves hydrolysis and debittering efficiency for soy protein isolate. Food Chem. 2016, 214, 347–353. [Google Scholar] [CrossRef]
- Salwan, R.; Sharma, V.; Chand Kasan, R.; Gulati, A. Bioprospecting psychrotrophic bacteria for serine-type proteases from the cold areas of the Western Himalayas. Curr. Microbiol. 2020, 77, 795–806. [Google Scholar] [CrossRef]
- Qin, Y.; Huang, Z.; Liu, Z. A novel cold-active and salt-tolerant α-amylase from marine bacterium Zunongwangia profunda: Molecular cloning, heterologous expression and biochemical characterization. Extremophiles 2014, 18, 271–281. [Google Scholar] [CrossRef]
- Wang, Q.F.; Miao, J.L.; Hou, Y.H.; Ding, Y.; Wang, G.D.; Li, G.Y. Purification and characterization of an extracellular cold-active serine protease from the psychrophilic bacterium Colwellia sp. NJ341. Biotechnol. Lett. 2005, 27, 1195–1198. [Google Scholar] [CrossRef]
- Tchigvintsev, A.; Tran, H.; Popovic, A.; Kovacic, F.; Brown, G.; Flick, R.; Hajighasemi, M.; Egorova, O.; Somody, J.C.; Tchigvintsev, D.; et al. The environment shapes microbial enzymes: Five cold-active and salt-resistant carboxylesterases from marine metagenomes. Appl. Microbiol. Biotechnol. 2015, 99, 2165–2178. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.F.N.; Weeks, W.F. CRREL Report 82-30, Equations for determining the gas and brine volumes in sea ice samples. J. Glaciol. 1983, 29, 306–316. [Google Scholar] [CrossRef]
- Hoppe, H.G.; Arnosti, C.; Herndl, G.F. Ecological significance of bacterial enzymes in marine environment. In Microbial Enzymes in the Environment Activity, Ecology and Applications; Burns, R.C., Dick, R.P., Eds.; Marcel Dekker Inc.: New York, NY, USA, 2002; pp. 73–107. [Google Scholar]
- Maxwell, S.G. Acquisition, Degradation, and Cycling of Organic Matter within Sea-Ice Brines by Bacteria and Their Viruses. Ph.D. Thesis, University of Washington, Washington, DC, USA, 2020; pp. 1–168. [Google Scholar]
- Stedmon, C.A.; Thomas, D.N.; Papadimitriou, S.; Granskog, M.A.; Dieckmann, G.S. Using fluorescence to characterize dissolved organic matter in Antarctic sea ice brines. J. Geophys. Res. Biogeosci. 2011, 116, G03027. [Google Scholar] [CrossRef]
- Cox, M.; Lehninger, A.L.; Nelson, D.R. Lehninger Principles of Biochemistry; Worth Publishers: New York, NY, USA, 2000; pp. 306–308. [Google Scholar]
- Sun, S.; Jones, R.B.; Fodor, A.A. Inference-based accuracy of metagenome prediction tools varies across sample types and functional categories. Microbiome 2020, 8, 46. [Google Scholar] [CrossRef]
- Chistoserdova, L.; Laukel, M.; Portais, J.C.; Vorholt, J.A.; Lidstrom, M.E. Multiple formate dehydrogenase enzymes in the facultative methylotroph Methylobacterium extorquens AM1 are dispensable for growth on methanol. J. Bacteriol. 2004, 186, 22–28. [Google Scholar] [CrossRef]
- Stibal, M.; Wadham, J.L.; Lis, G.P.; Telling, J.; Pancost, R.D.; Dubnick, A.; Sharp, M.J.; Lawson, E.C.; Butler, C.E.H.; Hasan, F.; et al. Methanogenic potential of Arctic and Antarctic subglacial environments with contrasting organic carbon sources. Glob. Chang. Biol. 2012, 18, 3332–3345. [Google Scholar] [CrossRef]
- Ortiz, M.; Bosch, J.; Coclet, C.; Johnson, J.; Lebre, P.; Salawu-Rotimi, A.; Vikram, S.; Makhalanyane, T.; Cowan, D. Microbial nitrogen cycling in Antarctic soils. Microorganisms 2020, 8, 1442. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.A.; Venceslau, S.S.; Grein, F.; Leavitt, W.D.; Dahl, C.; Johnston, D.T.; Pereira, I.A. A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science 2015, 350, 1541–1545. [Google Scholar] [CrossRef] [PubMed]
- Griesbeck, C.; Hauska, G.; Schütz, M. Biological Sulfide-Oxidation: Sulfide-Quinone Reductase (SQR), the Primary Reaction. In Recent Research Developments in Microbiology; Pandalai, S.G., Ed.; Research Signpost: Trivadrum, India, 2000; Volume 4, pp. 129–203. [Google Scholar]
- Karr, E.A.; Sattley, W.M.; Jung, D.O.; Madigan, M.T.; Achenbach, L.A. Remarkable diversity of phototrophic purple bacteria in a permanently frozen Antarctic lake. Appl. Environ. Microbiol. 2003, 69, 4910–4914. [Google Scholar] [CrossRef]
- Karr, E.A.; Sattley, W.M.; Rice, M.R.; Jung, D.O.; Madigan, M.T.; Achenbach, L.A. Diversity and distribution of sulfate-reducing bacteria in permanently frozen Lake Fryxell, McMurdo Dry Valleys, Antarctica. Appl. Environ. Microbiol. 2005, 71, 6353–6359. [Google Scholar] [CrossRef]
- Christner, B.C.; Priscu, J.C.; Achberger, A.M.; Barbante, C.; Carter, S.P.; Christianson, K.; Michaud, A.B.; Mikucki, J.A.; Mitchell, A.C.; Skidmore, M.L.; et al. WISSARD Science Team. Subglacial Lake Whillans: A microbial ecosysytem beneath the West Antarctic Ice Sheet. Nature 2014, 512, 310–313. [Google Scholar] [CrossRef]
- Mikucki, J.; Pearson, A.; Johnston, D.T.; Turchyn, A.V.; Farquhar, J.; Schrag, D.P.; Anbar, A.D.; Priscu, J.C.; Lee, P.A. A contemporary microbially maintained subglacial ferrous ocean. Science 2009, 324, 397–400. [Google Scholar] [CrossRef]
- Sattley, W.M.; Madigan, M.T. Isolation, characterization, and ecology of cold-active, chemolithotrophic, sulfur-oxidizing bacteria from perennially ice-covered Lake Fryxell, Antarctica. Appl. Environ. Microbiol. 2006, 72, 5562–5568. [Google Scholar] [CrossRef]
- Sattley, W.M.; Madigan, M.T. Temperature and nutrient induced responses of Lake Fryxell sulfate-reducing prokaryotes and description of Desulfovibrio lacusfryxellense sp. nov., a pervasive, cold-active, sulfate-reducing bacterium from Lake Fryxell, Antarctica. Extremophiles 2010, 14, 357–366. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).