An Assessment of Applicability of SNP Chip Developed for Domestic Goats in Genetic Studies of Caucasian Tur (Capra caucasica) †
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weinberg, P.J.; Akkiev, M.I.; Buchukuri, R.G. Tur of the Caucasus: Morphology, taxonomy and conservation strategy. Status and Protecyion of Globally Threatened Species in the Caucasus. In CEPF Biodiversity Investments in the Caucasus Hotspot 2004–2009; Zazanashvili, N., Mallon, D., Eds.; Contour Ltd.: Tbilisi, Georgia, 2009; pp. 83–91. [Google Scholar]
- Lydekker, R. Wild Oxen, Sheep, and Goats of All Lands; Rowland Ward: London, UK, 1898; p. 318. [Google Scholar]
- Tsalkin, V.I. Variability and systematics of tur of the West Caucasus (Caprinae, Artiodactyla). Byulleten Mosk. Obs. Ispyt. Prir. Otd. Biol. 1955, 60, 17–33. (In Russian) [Google Scholar]
- Geptner, V.G.; Nasimovich, A.A.; Bannikov, A.G. Mammals of the Soviet Union. Ungulates and Not Ungulates; Higher School: Moscow, Russia, 1961; Volume 1, p. 776. (In Russia) [Google Scholar]
- Damm, G.R.; Franco, N. CIC Caprinae Atlas of the World; CIC International Council for Game and Wildlife Conservation: Budakeszi, Hungary; Rowland Ward Publications: Johannesburg, South Africa, 2014. [Google Scholar]
- Gromov, I.M.; Gureev, A.A.; Novikov, G.A.; Sokolov, I.I.; Strelkov, P.P.; Chapskii, K.K. Mammal. Fauna of the USSR V.2.; Academy of Sciences of the USSR: Moscow-Leningrad, Russia, 1963; pp. 1047–1050. (In Russian) [Google Scholar]
- Lortkipanidze, B.; Weinberg, P. Capra cylindricornis. The IUCN Red List of Threatened Species 2020: E.T3795A91287260. [CrossRef]
- Weinberg, P. Capra caucasica. The IUCN Red List of Threatened Species 2020: E.T3794A22143809. [CrossRef]
- Manceau, V.; Després, L.; Bouvet, J.; Taberlet, P. Systematics of the genus Capra inferred from mitochondrial DNA sequence data. Mol. Phylogenetics Evol. 1999, 13, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Kazanskaya, E.Y.; Kuznetsova, M.V.; Danilkin, A.A. Phylogenetic reconstructions in the genus Capra (Bovidae, Artiodactyla) based on the mitochondrial DNA analysis. Russ. J. Genet. 2009, 43, 245–253. (In Russian) [Google Scholar] [CrossRef]
- Zvychaynaya, E.Y. Genetic differentiation of the wild goats (genus Capra) based on the analysis of mitochondrial gene cytochrome b and fragment of nuclear gene SRY. Galemys 2010, 22, 255–276. [Google Scholar]
- Kashinina, N.V.; Kholodova, M.V. Currunt genetic structure of Caucasian turs Capra caucasica and C. cylindricornis. In Conference Proceedings: Topical Issues of Modern Zoology and Ecology of Animals; Penza, Russia, 2016; p. 46. [Google Scholar]
- Coates, B.S.; Sumerford, D.V.; Miller, N.J.; Kim, K.S.; Sappington, T.W.; Siegfried, B.D.; Lewis, L.C. Comparative performance of single nucleotide polymorphism and microsatellite markers for population genetic analysis. J. Hered. 2009, 100, 556–564. [Google Scholar] [CrossRef] [Green Version]
- Kijas, J.W.; Townley, D.; Dalrymple, B.P.; Heaton, M.P.; Maddox, J.F.; McGrath, A. The International Sheep Genomics Consortium. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS ONE 2009, 4, e4668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tokarska, M.; Marshall, T.; Kowalczyk, R.; Wójcik, J.M.; Pertoldi, C.; Kristensen, T.N.; Loeschcke, V.; Gregersen, V.R.; Bendixen, C. Effectiveness of microsatellite and SNP markers for parentage and identity analysis in species with low genetic diversity: The case of European bison. Heredity 2009, 103, 326–332. [Google Scholar] [CrossRef] [Green Version]
- Kasarda, R.; Moravcikova, N.; Sidlova, V.; Trakovicka, A.; Kadlecik, O.; Pokoradi, J.; Zidek, R. Suitability of BovineSNP50 BeadChip for the evaluation of the Cervidae family diversity. Czech. J. Anim. Sci. 2015, 60, 391–399. [Google Scholar] [CrossRef] [Green Version]
- Kharzinova, V.R.; Sermyagin, A.A.; Gladyr, E.A.; Okhlopkov, I.M.; Brem, G.; Zinovieva, N.A. A study of applicability of SNP chips developed for Bovine and Ovine species to whole-genome analysis of Reindeer Rangifer tarandus. J. Hered. 2015, 106, 758–761. [Google Scholar] [CrossRef]
- Kharzinova, V.; Dotsev, A.; Solovieva, A.; Sergeeva, O.; Bryzgalov, G.; Reyer, H.; Wimmers, K.; Brem, G.; Zinovieva, N. Insight into the current genetic diversity and population structure of domestic reindeer (Rangifer tarandus) in Russia. Animals 2020, 10, 1309. [Google Scholar] [CrossRef]
- Miller, J.M.; Poissant, J.; Kijas, J.W.; Coltman, D.W. The International Sheep Genomics Consortium. A genome-wide set of SNPs detects population substructure and long range linkage disequilibrium in wild sheep. Mol. Ecol. Resour. 2011, 11, 314–322. [Google Scholar] [CrossRef]
- Miller, J.M.; Kijas, J.W.; Heaton, M.P.; McEwan, J.C.; Coltman, D.W. Consistent divergence times and allele sharing measured from cross-species application of SNP chips developed for three domestic species. Mol. Ecol. Resour. 2012, 12, 1145–1150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dotsev, A.V.; Deniskova, T.E.; Okhlopkov, I.M.; Mészáros, G.; Sölkner, J.; Reyer, H.; Wimmers, K.; Brem, G.; Zinovieva, N.A. Genome-wide SNP analysis unveils genetic structure and phylogeographic history of snow sheep (Ovis nivicola) populations inhabiting the Verkhoyansk Mountains and Momsky Ridge (Northeastern Siberia). Ecol. Evol. 2018, 8, 8000–8010. [Google Scholar] [CrossRef] [PubMed]
- Tosser-Klopp, G.; Bardou, P.; Bouchez, O.; Cabau, C.; Crooijmans, R.; Dong, Y.; Donnadieu-Tonon, C.; Eggen, A.; Heuven, H.C.; Jamli, S.; et al. Design and characterization of a 52 K SNP chip for goats. PLoS ONE 2014, 9, e86227. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, F.; Servin, B.; Talenti, A.; Rochat, E.; Kim, E.S.; Oget, C.; Palhière, I.; Crisà, A.; Catillo, G.; Steri, R.; et al. Signatures of selection and environmental adaptation across the goat genome post-domestication. Genet. Sel. Evol. 2018, 50, 57. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 83, 583–590. [Google Scholar] [CrossRef]
- Keenan, K.; McGinnity, P.; Cross, T.F.; Crozier, W.W.; Prodöhl, P.A. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 2013, 4, 782–788. [Google Scholar] [CrossRef] [Green Version]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Pembleton, L.W.; Cogan, N.O.; Forster, J.W. StAMPP: An R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol. Ecol. Resour. 2013, 13, 946–952. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Stoffel, M.A.; Esser, M.; Kardos, M.; Humble, E.; Nichols, H.; David, P.; Hoffman, J.I. inbreedR: An R package for the analysis of inbreeding based on genetic markers. Methods Ecol. Evol. 2016, 7, 1331–1339. [Google Scholar] [CrossRef]
- Chen, H.; VennDiagram: Generate High-Resolution Venn and Euler Plots. R Package Version 1.6.20. 2018. Available online: https://CRAN.R-project.org/package=VennDiagram (accessed on 22 December 2020).
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francis, R.M. pophelper: An R package and web app to analyze and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reich, D.; Thangaraj, K.; Patterson, N.; Price, A.L.; Singh, L. Reconstructing Indian population history. Nature 2009, 461, 489–494. [Google Scholar] [CrossRef] [Green Version]
- Patterson, N.; Moorjani, P.; Luo, Y.; Mallick, S.; Rohland, N.; Zhan, Y.; Genschoreck, T.; Webster, T.; Reich, D. Ancient admixture in human history. Genetics 2012, 192, 1065–1093. [Google Scholar] [CrossRef] [Green Version]
- Petr, M.; Vernot, B.; Kelso, J. admixr—R package for reproducible analyses using ADMIXTOOLS. Bioinformatics 2019, 35, 3194–3195. [Google Scholar] [CrossRef] [Green Version]
- Becker, R.A.; Wilks, A.R.; Brownrigg, R.; Minka, T.P.; Deckmyn, A.; Maps: Draw Geographical Maps. R Package Version 3.3.0. 2018. Available online: https://CRAN.R-project.org/package=maps (accessed on 22 December 2020).
- Wright, S. Evolution and the Genetics of Population, Variability Within and Among Natural Populations; The University of Chicago Press: Chicago, IL, USA, 1978. [Google Scholar]
- Hartl, D.L.; Clark, A.G. Principles of Population Genetics, 3rd ed.; Sinauer Associates, Inc.: Sunderland, MA, USA, 1997. [Google Scholar]
- Sokolov, I.I. Fauna of the USSR: Mammals: Ungulates (Orders Perissodactyla and Artiodactyla); Publishing house of the Academy of Sciences of the USSR: Leningrad, Moscow, Russia, 1959; p. 265. [Google Scholar]
- Tembotov, A.K. Geography of North. Caucasian Mammals; Nalchik: Elbrus, Russia, 1972; p. 151. [Google Scholar]
- Couturier, M.A.J. Le Bouquetin des Alpes; Allier: Grenoble, France, 1962. [Google Scholar]
- Ayunts, K.R. On the systematics of Caucasian tur. Bulletin of the Moscow Society of Naturalists. Biol. Dep. 1992, 97, 19–25. [Google Scholar]
- Ropiquet, A.; Hassanin, A. Molecular evidence for the polyphyly of the genus Hemitragus (Mammalia, Bovidae). Mol. Phylogenetics Evol. 2005, 36, 154–168. [Google Scholar] [CrossRef]
- Tan, S.; Wang, Z.; Jiang, L.; Peng, R.; Zhang, T.; Peng, Q.; Zou, F. Molecular phylogeny and phylogeography of genus Pseudois (Bovidae, Cetartiodactyla): New insights into the contrasting phylogeographic structure. Ecol. Evol. 2017, 7, 7047–7057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dotsev, A.V.; Kunz, E.; Kharzinova, V.R.; Okhlopkov, I.M.; Lv, F.-H.; Li, M.-H.; Rodionov, A.N.; Shakhin, A.V.; Sipko, T.P.; Medvedev, D.G.; et al. Mitochondrial DNA Analysis Clarifies Taxonomic Status of the Northernmost Snow Sheep (Ovis nivicola) Population. Life 2021, 11, 252. [Google Scholar] [CrossRef] [PubMed]
- Huxley, J. Clines: An Auxiliary Taxonomic Principle. Nature 1938, 142, 19–220. [Google Scholar] [CrossRef]

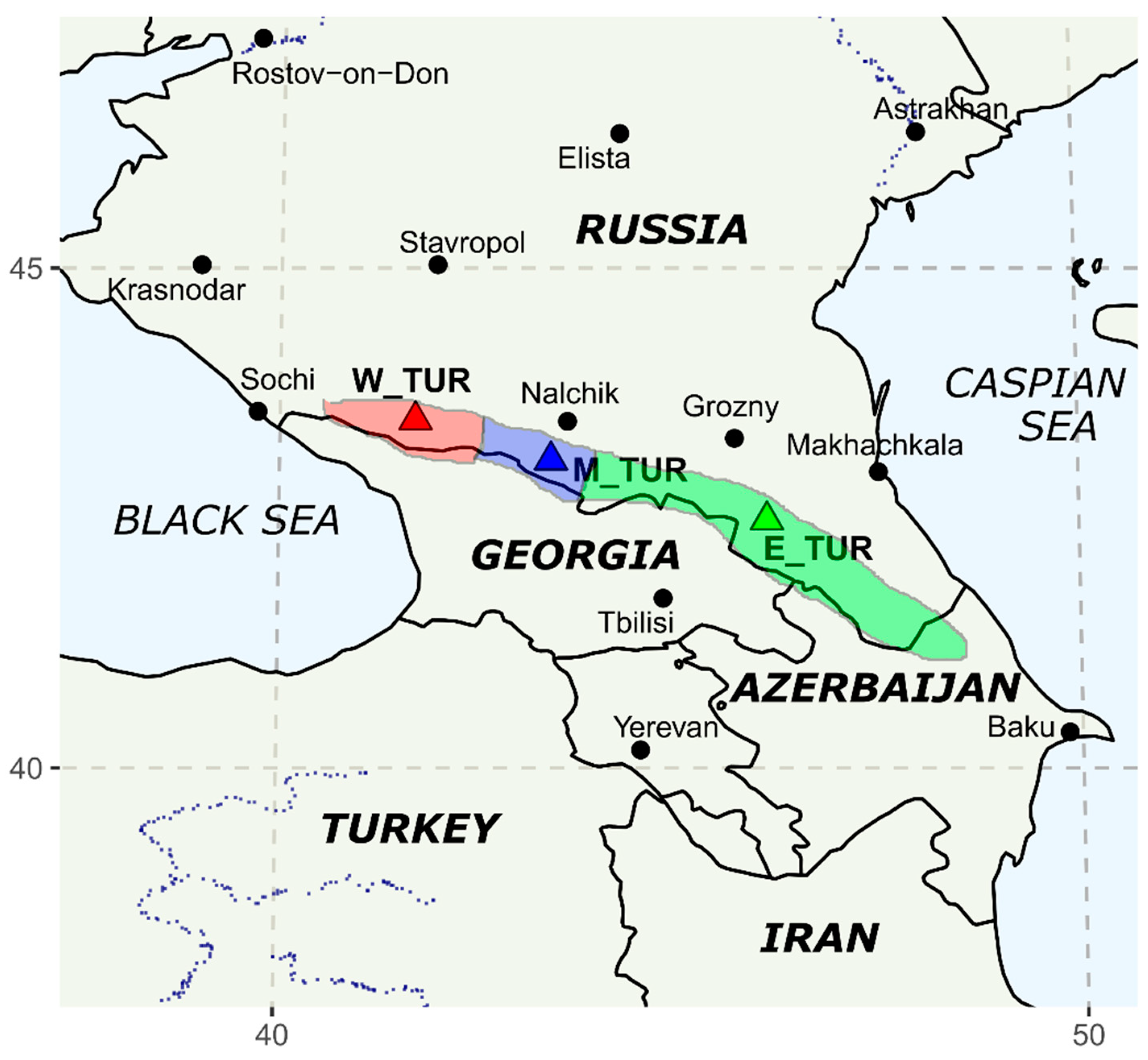

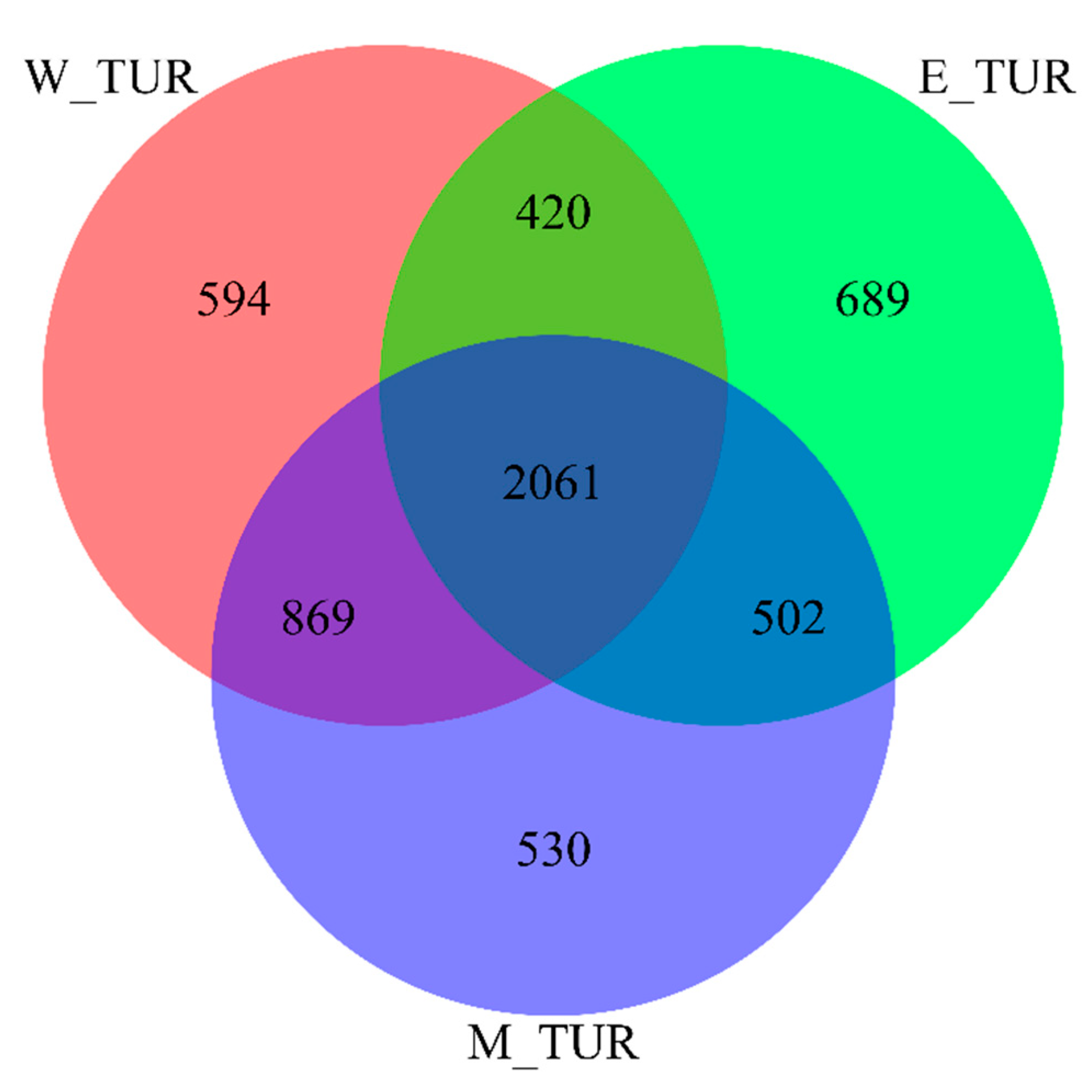


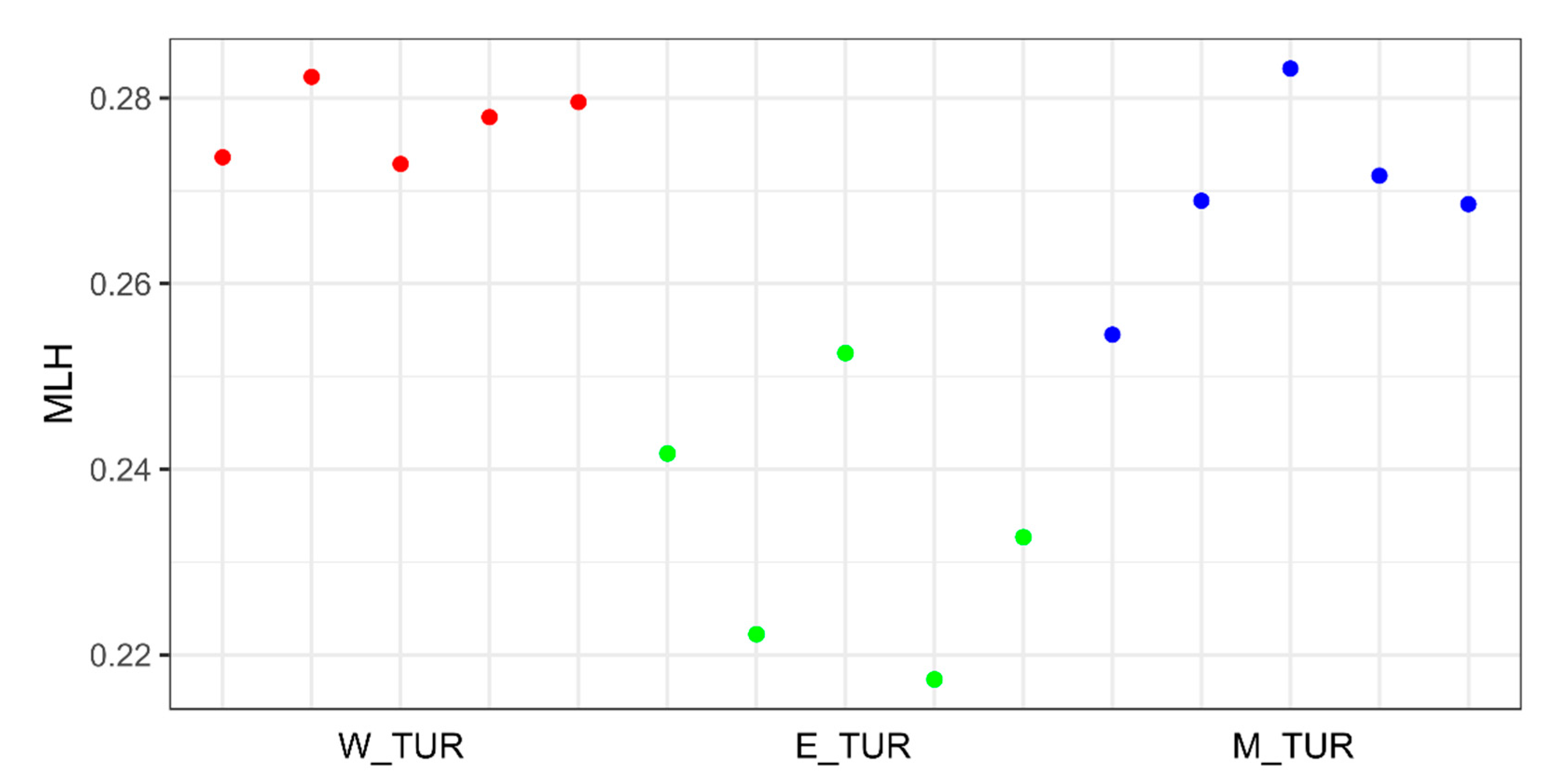

| Group | W_TUR | E_TUR | M_TUR |
|---|---|---|---|
| W_TUR | 0.000 | ||
| E_TUR | 0.161 | 0.000 | |
| M_TUR | 0.099 | 0.135 | 0.000 |
| Group | n | HO (±SE) | HE (±SE) | FIS [95% CI] | AR (±SE) |
|---|---|---|---|---|---|
| W_TUR | 5 | 0.308 ± 0.004 | 0.312 ± 0.003 | 0.015 [0.002; 0.028] | 1.775 ± 0.007 |
| E_TUR | 5 | 0.258 ± 0.004 | 0.284 ± 0.003 | 0.074 [0.060; 0.088] | 1.730 ± 0.007 |
| M_TUR | 5 | 0.306 ± 0.004 | 0.316 ± 0.003 | 0.028 [0.015; 0.041] | 1.790 ± 0.007 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dotsev, A.V.; Rodionov, A.N.; Kharzinova, V.R.; Petrov, S.N.; Medvedev, D.G.; Bagirov, V.A.; Brem, G.; Zinovieva, N.A. An Assessment of Applicability of SNP Chip Developed for Domestic Goats in Genetic Studies of Caucasian Tur (Capra caucasica). Diversity 2021, 13, 312. https://doi.org/10.3390/d13070312
Dotsev AV, Rodionov AN, Kharzinova VR, Petrov SN, Medvedev DG, Bagirov VA, Brem G, Zinovieva NA. An Assessment of Applicability of SNP Chip Developed for Domestic Goats in Genetic Studies of Caucasian Tur (Capra caucasica). Diversity. 2021; 13(7):312. https://doi.org/10.3390/d13070312
Chicago/Turabian StyleDotsev, Arsen V., Andrey N. Rodionov, Veronika R. Kharzinova, Sergey N. Petrov, Dmitry G. Medvedev, Vugar A. Bagirov, Gottfried Brem, and Natalia A. Zinovieva. 2021. "An Assessment of Applicability of SNP Chip Developed for Domestic Goats in Genetic Studies of Caucasian Tur (Capra caucasica)" Diversity 13, no. 7: 312. https://doi.org/10.3390/d13070312
APA StyleDotsev, A. V., Rodionov, A. N., Kharzinova, V. R., Petrov, S. N., Medvedev, D. G., Bagirov, V. A., Brem, G., & Zinovieva, N. A. (2021). An Assessment of Applicability of SNP Chip Developed for Domestic Goats in Genetic Studies of Caucasian Tur (Capra caucasica). Diversity, 13(7), 312. https://doi.org/10.3390/d13070312








