Abstract
The geographical pattern of genetic diversity was investigated in the endemic Sicilian pond turtle Emys trinacris across its entire distribution range, using 16 microsatellite loci. Overall, 245 specimens of E. trinacris were studied, showing high polymorphic microsatellite loci, with allele numbers ranging from 7 to 30. STRUCTURE and GENELAND analyses showed a noteworthy, geographically based structuring of the studied populations in five well-characterized clusters, supported by a moderate degree of genetic diversity (FST values between 0.075 and 0.160). Possible explanations for the genetic fragmentation observed are provided, where both natural and human-mediated habitat fragmentation of the Sicilian wetlands played a major role in this process. Finally, some conservation and management suggestions aimed at preventing the loss of genetic variability of the species are briefly reported, stressing the importance of considering the five detected clusters as independent Management Units.
1. Introduction
The western Palaearctic genus Emys Duméril, 1805 contains two species, Emys orbicularis (Linnaeus, 1758) and Emys trinacris Fritz et al., 2005. Emys orbicularis includes a variety of morphologically and genetically defined subspecies [1,2,3,4,5,6,7,8]. Among them, E. orbicularis hellenica (corresponding to mitochondrial lineage IV sensu Stuckas et al. [6]) and E. o. galloitalica (corresponding to mitochondrial lineage V) occur in peninsular Italy and Sardinia [5,6,7]. Emys trinacris, characterized by the mitochondrial lineage III, is endemic to Sicily (Italy) and is the only autochthonous pond turtle occurring there [2,9,10].
The taxonomic status of the Sicilian pond turtle is currently debated. When it was described, species status was inferred by nuclear-genomic fingerprinting [2] and later confirmed using nuclear microsatellite markers [5,7]. However, Speybroeck et al. [11] proposed treating the taxon as a subspecies of E. orbicularis, reflecting the weak genetic divergence of E. trinacris and E. orbicularis compared to other European reptiles. On the other hand, Speybroeck et al. [11] conceded that further distinct species could be currently subsumed under E. orbicularis. Among turtles, several taxa with less genetic divergence are currently recognized as full species (e.g., Graptemys [12,13,14]). To avoid unnecessary nomenclatural changes, we here continue to treat E. trinacris as a distinct species until new genetic evidence becomes available.
Regardless of the taxonomic status, the Sicilian endemic pond turtle is listed, as well as Emys orbicularis, as “EN” (i.e., “Endangered”) in the Italian IUCN Red List of Threatened Species [15]. Moreover, it is included in both Appendix II of the EU Council Directive 92/43/EEC (“Habitats Directive”) and in Appendix II of the “Bern Convention on the Conservation of European Wildlife and Natural Habitats”.
According to several authors, Emys trinacris is widely distributed in Sicily, but the distribution is very fragmented due to its limited dispersal capability [7,16,17]. Moreover, populations of E. trinacris, as well as of the congeneric E. orbicularis, are facing several threats, i.e., habitat loss and fragmentation [2,7,18,19,20,21], invasive alien species [22,23,24], human trading [25], recreational fishing [26], and possibly parasite spill-over [27].
Based on a mitochondrial marker (cytochrome b gene) and 15 microsatellite loci, a strong genetic structuring of the Sicilian pond turtle was found throughout the island by Vamberger et al. [7]. In agreement with these results, attention should be paid when individuals are translocated. Such an approach is indicated as pivotal to avoid the loss of genetic variability through a homogenization process. Even though the sampling was adequate in the study by Vamberger et al. [7], some locations were not sampled or represented only by few individuals. Based on 16 nuclear microsatellite loci and a significantly increased sampling, we here reassess the genetic structuring of E. trinacris populations in Sicily. We provide a sound population genetic database covering the whole distribution of E. trinacris in Sicily for management and conservation actions for this threatened pond turtle.
2. Materials and Methods
2.1. Sampling, DNA Extraction and Selected Loci Amplification
Sicilian pond turtles were collected in permanent water bodies throughout Sicily (Figure S1; Table S1; no precise geographical coordinates are provided since the species is subject to poaching). Turtles were captured using baited hoop traps, immersed in water and equipped with a flotation system, so that turtles and other air-breathing animals that were eventually caught could breathe at the surface [28]. The traps were left fishing for about 48 h per session, and then checked for the possible presence of captured turtles.
Tissue, blood or saliva samples were collected from each turtle. DNA extraction, PCRs and genotyping followed the protocols described in Vamberger et al. [7].
The fragment lengths were determined on an ABI 3130xl Genetic Analyzer (Applied Biosystems, Foster, CA, USA) using the GeneScan–600 LIZ Size Standard (Applied Biosystems, Foster, CA, USA) and the software PEAK SCANNER 1.0 (Life Technologies, Carlsbad, CA, USA).
All methods and experimental protocols on turtles were conducted in strict accordance with the recommendations of the Italian Ministry of the Environment “Ministero dell’Ambiente e della Tutela del Territorio e del Mare” (U. prot. 000884/PNM 08/05/2014; 0023415/PNM 17/11/2014 and 0006880/PNM 5/04/2016).
2.2. Genetic Cluster Analysis and Diversity Indices
Microsatellite genotypes and sample spatial location data were analysed for all loci and samples using two Bayesian clustering software STRUCTURE 2.3.3 [29,30] and GENELAND 4.0.9 [31]. In STRUCTURE, the admixture model and correlated allele frequencies were used, setting an arbitrary upper bond value of K = 10; the most likely number of clusters (K) was determined using the ΔK method [32] with the software STRUCTURE HARVESTER [33] through its online interface (http://taylor0.biology.ucla.edu/structureHarvester/). Calculations for each K were repeated 10 times using a MCMC chain of 750,000 generations for each run, including a burn-in of 250,000 generations. Population structuring and individual admixture were visualized using the software DISTRUCT 1.1 [34]. Following Randi [35], individuals with proportions of cluster membership below 80% were treated as having mixed ancestries. Afterwards, data of admixed individuals were removed to avoid introducing noise from alleles of other clusters.
In GENELAND, the geographic information was used to detect spatial delineation of genetic discontinuities. The number of clusters (K) was inferred using 500,000 MCMC iterations with 100 burn-in generations.
In addition, microsatellite data of the inferred clusters were analysed by calculating population genetic diversity indices using CONVERT 1.31 [36] and GENETIX 4.05.2 [37]. Hardy–Weinberg equilibrium (HWE) for all loci and all clusters was tested in GENEPOP ON THE WEB 4.7 [38,39] and the pairwise FST values were computed through the software ARLEQUIN 3.5.1.2 [40]. Tests for recent bottlenecks were performed with the program BOTTLENECK 1.2.02 [41] using the strict one-step stepwise mutation model (SMM; Ohta and Kimura [42]) as suggested by Piry et al. [41] for microsatellite loci. In order to compare our results, a Discriminant Analysis of Principal Components (DAPCs) was computed using the same dataset, with the package ADEGENET [43,44] for CRAN R 3.6.3.
3. Results
Altogether, 245 samples of the Sicilian pond turtle from 21 different sampling sites were studied (Figure S1; Table S1), including 62 new samples and 183 samples from Vamberger et al. [7], thus covering the whole distribution of the species in Sicily. All studied microsatellite loci were highly polymorphic, with allele numbers ranging from 7 to 30, and a total allele number of 293.
The highest support in both STRUCTURE and GENELAND analyses was found for K = 5, suggesting the split of the Sicilian pond turtles into five different clusters: the Nebrodi mountains (cluster A), the Sicilian south-western area (cluster B); south-eastern Sicily (cluster C); the Sicani mountains (province of Palermo; cluster D); north-western Sicily (cluster E) (Figure 1 and Figure S2). DACP analysis (Figure 2) supported the differentiation of the five clusters recognized by STRUCTURE and GENELAND (Figure S2).
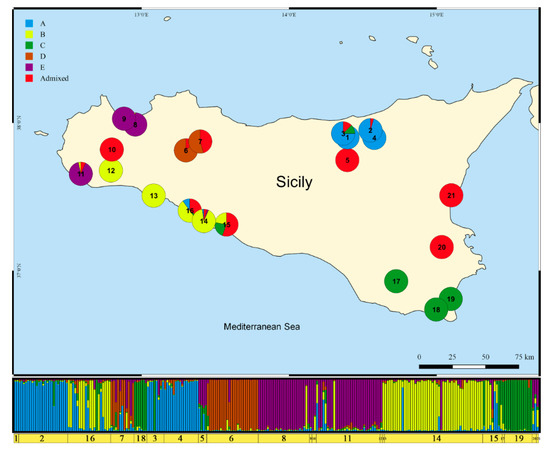
Figure 1.
Genotypic structuring of 245 Sicilian pond turtles (Emys trinacris) from 21 sampling sites for K=5 using 16 microsatellite loci based on the STRUCTURE analysis. Distinct clusters are colour-coded; samples are arranged according to the numbers of the pooled sampling sites (Table S1). Within each cluster, an individual turtle is represented by a vertical segment that reflects its ancestry. Mixed ancestries are indicated by differently coloured sectors corresponding to inferred genetic percentages of the respective cluster. Colours of pooled sampling sites in the map correspond to STRUCTURE clusters; slices represent turtles with mixed ancestries or conflicting cluster assignment (percentages).
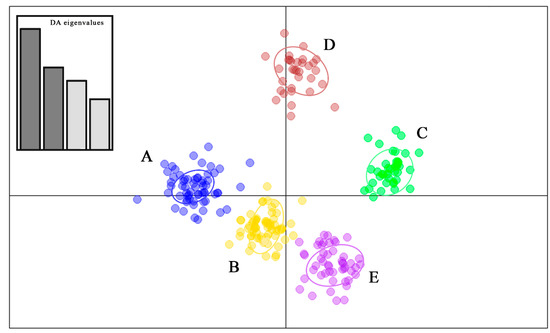
Figure 2.
Discriminant Analysis of Principal Components (DAPC) for microsatellite data of the five clusters (A–E) of Emys trinacris. The oval outlines correspond to 95% confidence intervals. (DA Eigenvalues: 891, 606.9, 455.1, 326).
Observed heterozygosity (Ho) values (range 0.54–0.66) were lower than the expected ones (He, range 0.60–0.76) (Table 1) In addition, all clusters showed evidence of recent bottlenecking (see Table 1).

Table 1.
Genetic diversity indices and BOTTLENECK analysis of STRUCTURE clusters of Emys trinacris. Colours refer to clusters (see Figure 1). The category admixed individuals comprises turtles with admixed genotypes irrespective of their collection sites.
Moreover, all identified clusters of E. trinacris showed a significant deviation from HWE at all loci and within all clusters defined by STRUCTURE and GENELAND (Table 2). Pairwise FST values between the five a priori defined cluster, based on the STRUCTURE results, ranged from 0.075 to 0.160 (Table 3); all FST values were significant with a p-value of 0.05. FST values indicated moderate genetic differentiation among sampled Sicilian pond turtles, where the highest value was observed between the comparison of the clusters “D” and “C”.

Table 2.
Hardy–Weinberg Exact test (Fisher’s method) based on the STRUCTURE clusters of Emys trinacris.

Table 3.
Pairwise FST based on the STRUCTURE clusters of Emys trinacris.
4. Discussion
The results obtained in this study suggest the presence of a clear structuring of Emys trinacris populations, with a moderate degree of genetic differentiation among the five identified clusters (Table 3). The clusters are supported by moderate genetic diversity indices among them (see Table 1); a similar moderate genetic structuring can be also observed in E. orbicularis [45]. Surprisingly however, the genetic structuring only partially agrees with that revealed by Vamberger et al. [7]. While Vamberger et al. [7] found eight well differentiated clusters within E. trinacris, our increased sampling covering some new sampling localities concluded that only five clusters exist. Nevertheless, three of them were in full agreement with the ones from Vamberger et al. [7], i.e., clusters A, C, D. Both studies highlighted a moderate genetic structuring of E. trinacris in Sicily, with a slightly different grouping of the studied samples, possibly due to an increased sampling scheme and the implementation of a further microsatellite marker, suggesting a long-lasting persistence of turtles in the respective areas of Sicily. In contrast, the subspecies of E. orbicularis do not show such a pronounced substructuring (e.g., [7,8]). Stöck et al. [46] found low levels of genetic structuring in the studied Sicilian herpetofauna using two mitochondrial markers, the cytochrome b and 16S rDNA genes, and a nuclear marker, the tropomyosin intron. However, it is well known that the current genetic structuring of many other freshwater taxa in Sicily can be ascribed to paleogeographic and paleoclimatic events [47,48,49]. The pronounced genetic structure in the Sicilian pond turtle could thus be due to the role of Sicily as a “refuge area” during the Plio-Pleistocene climatic fluctuations [50]. The local physiography can maintain genetic diversity (see Vamberger et al. [7]), corresponding to a “refugia within refugia” pattern [51].
In addition, increased habitat fragmentation and the consequent reduction in population size and gene flow levels [52] could also contribute to the current structuring, which shows evidence of recent bottleneck experienced by all the local clusters of the species (Table 1). The paucity of water bodies and habitats suitable for the presence of the species in Sicily prevents the populations from establishing sound metapopulation dynamics, thus precluding significant inter-population gene flow. In some areas, Emys trinacris is locally abundant but these regions are separated from other inhabited areas by large gaps where the species is completely absent (e.g., in the Madonie and Peloritani mountain chains).
In recent centuries, the wetlands in Sicily have been significantly modified due to remediation measures to reclaim agricultural land and to fight against malaria (“Provveditorato alle opere pubbliche per la Sicilia” [53]; see also Naselli-Flores and Marrone [23]). During those years, wetlands across Sicily were altered completely by local authorities by re-shaping the surrounding landscape. Furthermore, according to ISPRA (“Istituto Superiore per la Protezione e la Ricerca Ambientale” [54]), in the last century Sicily has undergone a severe degradation of the wetlands, where 34.5% of all the Sicilian wetlands are in a state of complete degradation and only 5% remain untouched (cf. Table 10 in [54]).
In addition, the presence of invasive species (e.g., Trachemys scripta [24,55]) may also contribute to the pronounced genetic fragmentation of E. trinacris. To date, Trachemys scripta and other non-native emydid taxa are known to occur all over Sicily [23,24,56], and coexist with E. trinacris in some of the sampled localities (e.g., Agrigento: Monteallegro, Diga Gorgo; Agrigento: Imera river; Palermo: Bosco Ficuzza; Catania: Simeto river; pers. obs.). The invasive pond slider is known to have a negative impact on the local turtle populations, i.e., competing for food, basking and nesting sites [18,57,58,59,60,61,62,63,64], and being vectors for parasites and other pathogens (helminth cross-transmission: [65,66,67]; Salmonella: [68,69]; oxyurid and strongylid nematodes: [70]).
In conclusion, to preserve the genetic variability of E. trinacris, protection actions by local management entities are required. According to our improved genetic database, which can now serve in conservation actions, we propose that the five population clusters highlighted by present study should be considered as independent Management Units for Sicily. As already proposed by Vamberger et al. [7], it is necessary to avoid translocating turtles, even when events or activities regarding the restocking or release of treated or seized individuals are carried out. It is thus of outstanding importance to genetically characterize all individuals before any translocation actions can be approved.
Supplementary Materials
The following materials are available online at https://www.mdpi.com/1424-2818/12/9/343/s1, Figure S1: Pooled sampling sites of Emys trinacris. Consecutive numbers of pooled sites refer to Table S1, Figure S2: Genotypic structuring of 245 Sicilian pond turtles (Emys trinacris) from 21 sampling sites for K = 5 using 16 microsatellite loci. Samples are arranged according to the numbers of the pooled sampling sites (see Table S1). Colours of pooled sampling sites in the map correspond to GENELAND clusters; slices represent turtles with mixed ancestries or conflicting cluster assignment (percentages). Table S1: Origin of the studied Emys trinacris samples. Sample sites values refers to number showed in Figure S1.
Author Contributions
Conceptualization, M.V., U.F. and M.A.; Methodology, M.V.; Software, L.V.; Validation, M.V., U.F., M.A. and F.M.; Formal Analysis, L.V.; Investigation, M.V. and L.V.; Resources, M.V. and U.F.; Data Curation, M.V. and L.V.; Writing—Original Draft Preparation, L.V.; Writing—Review and Editing, L.V., M.V., U.F., M.A. and F.M.; Visualization, L.V.; Supervision, M.V.; Project Administration, M.V.; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
New tissue samples were collected using permits of the Italian “Ministero dell’Ambiente e della Tutela del Territorio e del Mare” (U. prot. 000884/PNM 08/05/2014; 0023415/PNM 17/11/2014 and 0006880/PNM 5/04/2016)”. Francesco Sacco and Rita Scardino (Palermo, Italy) are acknowledged for the support they provided with the collection and analysis of some of the studied samples. Laboratory work was conducted at Senckenberg Dresden (SGN-SNSDMol-Lab).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lenk, P.; Fritz, U.; Joger, U.; Wink, M. Mitochondrial phylogeography of the European pond turtle, Emys orbicularis (Linnaeus 1758). Mol. Ecol. 1999, 8, 1911–1922. [Google Scholar] [CrossRef] [PubMed]
- Fritz, U.; Fattizzo, T.; Guicking, D.; Tripepi, S.; Pennisi, M.G.; Lenk, P.; Joger, U.; Wink, M. A new cryptic species of pond turtle from southern Italy, the hottest spot in the range of the genus Emys (Reptilia, Testudines, Emydidae). Zool. Scr. 2005, 34, 351–371. [Google Scholar] [CrossRef]
- Fritz, U.; Guicking, D.; Kami, H.; Arakelyan, M.; Auer, M.; Ayaz, D.; Ayres Fernández, C.; Bakiev, A.; Celani, A.; Havaš, P.; et al. Mitochondrial phylogeography of European pond turtles (Emys orbicularis, Emys trinacris)—An update. Amphibia-Reptilia 2007, 28, 418–426. [Google Scholar] [CrossRef]
- Fritz, U.; Ayaz, D.; Hundsdörfer, A.K.; Kotenko, T.; Guicking, D.; Wink, M.; Tok, C.V.; Çiçek, K.; Buschbom, J. Mitochondrial diversity of European pond turtles (Emys orbicularis) in Anatolia and the Ponto-Caspian Region: Multiple old refuges, hotspot of extant diversification and critically endangered endemics. Org. Divers. Evol. 2009, 9, 100–114. [Google Scholar] [CrossRef]
- Pedall, I.; Fritz, U.; Stuckas, H.; Valdeón, A.; Wink, M. Gene flow across secondary contact zones of the Emys orbicularis complex in the Western Mediterranean and evidence for extinction and re-introduction of pond turtles on Corsica and Sardinia (Testudines: Emydidae). J. Zool. Syst. Evol. Res. 2011, 49, 44–57. [Google Scholar] [CrossRef]
- Stuckas, H.; Velo-Antón, G.; Fahd, S.; Kalboussi, M.; Rouag, R.; Arculeo, M.; Marrone, F.; Sacco, F.; Vamberger, M.; Fritz, U. Where are you from, stranger? The enigmatic biogeography of North African pond turtles (Emys orbicularis). Org. Divers. Evol. 2014, 14, 295–306. [Google Scholar] [CrossRef]
- Vamberger, M.; Stuckas, H.; Sacco, F.; D’Angelo, S.; Arculeo, M.; Cheylan, M.; Corti, C.; Lo Valvo, M.; Marrone, F.; Wink, M.; et al. Differences in gene flow in a twofold secondary contact zone of pond turtles in southern Italy (Testudines: Emydidae: Emys orbicularis galloitalica, E. o. hellenica, E. trinacris). Zool. Scr. 2015, 44, 233–249. [Google Scholar] [CrossRef]
- Pöschel, J.; Heltai, B.; Graciá, E.; Franch Quintana, M.; Velo-Antón, G.; Arribas, O.; Valdeón, A.; Wink, M.; Fritz, U.; Vamberger, M. Complex hybridization patterns in European pond turtles (Emys orbicularis) in the Pyrenean Region. Sci. Rep. 2018, 8, 15925. [Google Scholar] [CrossRef]
- Marrone, F.; Sacco, F.; Arizza, V.; Arculeo, M. Amendment of the type locality of the endemic Sicilian pond turtle Emys trinacris Fritz et al. 2005, with some notes on the highest altitude reached by the species (Testudines, Emydidae). Acta Herpetol. 2016, 11, 59–61. [Google Scholar] [CrossRef]
- Scardino, R.; Mazzoleni, S.; Rovatsos, M.; Vecchioni, L.; Dumas, F. Molecular cytogenetics characterization of the Sicilian endemic pond turtle Emys trinacris (Testudines, Emydidae). Genes 2020, 11, 702. [Google Scholar] [CrossRef]
- Speybroeck, J.; Beukema, W.; Dufresnes, C.; Fritz, U.; Jablonski, D.; Lymberakis, P.; Martínez-Solano, I.; Razzetti, E.; Vamberger, M.; Vences, M.; et al. Species list of the European herpetofauna—2020 Update by the Taxonomic Committee of the Societas Europaea Herpetologica. Amphibia-Reptilia 2020, 41, 139–189. [Google Scholar] [CrossRef]
- Praschag, P.; Ihlow, F.; Flecks, M.; Vamberger, M.; Fritz, U. Diversity of North American map and sawback turtles (Testudines: Emydidae: Graptemys). Zool. Scr. 2017, 46, 675–682. [Google Scholar] [CrossRef]
- TTWG [Turtle Taxonomy Working Group]. Turtles of the World: Annotated Checklist and Atlas of Taxonomy, Synonymy, Distribution, and Conservation Status, 8th ed.; Chelonian Research Monographs: Lunenburg, MA, USA, 2017; Volume 7, 292p. [Google Scholar]
- Thomson, R.C.; Spinks, P.Q.; Shaffer, H.B. Molecular phylogeny and divergence of the map turtles (Emydidae: Graptemys). Mol. Phylogenet. Evol. 2018, 121, 61–70. [Google Scholar] [CrossRef]
- Rondinini, C.; Battistoni, A.; Peronace, V.; Teofili, C. Lista Rossa IUCN dei Vertebrati Italiani; Comitato Italiano IUCN e Ministero dell’Ambiente e della Tutela del Territorio e del Mare: Roma, Italy, 2013. [Google Scholar]
- Marrone, F.; Sacco, F.; Kehlmaier, C.; Arizza, V.; Arculeo, M. Some like it cold: The glossiphoniid parasites of the Sicilian endemic pond turtle Emys trinacris (Testudines, Emydidae), an example of “parasite inertia”? J. Zool. Syst. Evol. Res. 2016, 54, 60–66. [Google Scholar] [CrossRef]
- Iannella, M.; Cerasoli, F.; D’Alessandro, P.; Console, G.; Biondi, M. Coupling GIS spatial analysis and Ensemble Niche Modelling to investigate climate change-related threats to the Sicilian pond turtle Emys trinacris, an endangered species from the Mediterranean. PeerJ 2018, 6, e4969. [Google Scholar] [CrossRef]
- Vamberger, M. European pond turtle (Emys orbicularis) in Slovenia; Rogner, M., Ed.; Edition Chimaira; European Pond Turtle (Emys orbicularis): Frankfurt am Main, Germany, 2009; pp. 191–193. [Google Scholar]
- Vamberger, M.; Kos, I. First observations on some aspects on the natural history of European pond turtles Emys orbicularis in Slovenia. Biologia 2011, 66, 170–174. [Google Scholar] [CrossRef]
- Vamberger, M.; Poboljšaj, K.; Govedič, M.; Debeljak Šabec, N.; Žagar, A. Conservation activities for European pond turtles (Emys orbicularis) in Slovenia. Herpetol. Notes 2013, 6, 123–126. [Google Scholar]
- Vamberger, M.; Lipovšek, G.; Šalamun, A.; Cipot, M.; Fritz, U.; Govedič, M. Distribution and population size of the European pond turtle Emys orbicularis in Ljubljansko barje, Slovenia. Vertebr. Zool. 2017, 67, 223–229. [Google Scholar]
- Standfuss, B.; Lipovšek, G.; Fritz, U.; Vamberger, M. Threat or fiction: Is the pond slider (Trachemys scripta) really invasive in Central Europe? A case study from Slovenia. Conserv. Genet. 2016, 17, 557–563. [Google Scholar] [CrossRef]
- Naselli-Flores, L.; Marrone, F. Different invasibility of permanent and temporary waterbodies in a semiarid Mediterranean Island. Inland Waters 2019, 9, 411–421. [Google Scholar] [CrossRef]
- Liuzzo, M.; Termine, R.; Marrone, F. First evidence of an egg-laying attempt of feral Trachemys scripta (Schoepff, 1792) in Sicily (Lake Pergusa, Italy). Herpetol. Notes 2020, 13, 365–368. [Google Scholar]
- Mărginean, G.I.; Gherman, E.; Sos, T. The illegal internet-based trade in European pond turtle Emys orbicularis (Linnaeus, 1758) in Romania: A threat factor for conservation. North-Western J. Zool. 2018, 14, 64–70. [Google Scholar]
- Vecchioni, L.; Cicerone, A.; Scardino, R.; Arizza, V.; Arculeo, M.; Marrone, F. Sicilians are not easily hooked! First assessment of the impact of recreational fishing on the endemic Sicilian pond turtle Emys trinacris (Testudines, Emydidae). Herpetol. Notes 2020, accepted. [Google Scholar]
- Arizza, V.; Sacco, F.; Russo, D.; Scardino, R.; Arculeo, M.; Vamberger, M.; Marrone, F. The good, the bad and the ugly: Emys trinacris, Placobdella costata and Haemogregarina stepanowi in Sicily (Testudines, Annelida and Apicomplexa). Folia Parasit. 2016, 63, 029. [Google Scholar] [CrossRef]
- Ream, C.; Ream, R. The influence of sampling methods on the estimation of population structure in painted turtles. Am. Midl. Nat. 1966, 325–338. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Res. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Guillot, G.; Mortier, F.; Estoup, A. GENELAND: A computer package for landscape genetics. Mol. Ecol. Notes 2005, 5, 712–715. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Res. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Randi, E. Detecting hybridization between wild species and their domesticated relatives. Mol. Ecol. 2008, 17, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Glaubitz, J.C. CONVERT: A user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol. Ecol. Notes 2004, 4, 309–310. [Google Scholar] [CrossRef]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. GENETIX 4.05, Logiciel Sous Windows TM Pour la Génétique des Populations; Laboratoire Génome, Populations, Interactions, CNRS UMR 5000; Université de Montpellier II: Montpellier, France, 2004. [Google Scholar]
- Raymond, M.; Rousset, F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Rousset, F. GENEPOP’007: A complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. ARLEQUIN suite ver. 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Piry, S.G.; Luikart, G.; Cornuet, J.M. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Ohta, T.; Kimura, M. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet. Res. Cambr. 1973, 22, 201–204. [Google Scholar] [CrossRef]
- Jombart, T. ADEGENET: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef]
- Velo-Antón, G.; García-París, M.; Cordero Rivera, A. Patterns of nuclear and mitochondrial DNA variation in Iberian populations of Emys orbicularis (Emydidae): Conservation implications. Conserv. Genet. 2008, 9, 1263–1274. [Google Scholar] [CrossRef]
- Stöck, M.; Grifoni, G.; Armor, N.; Scheidt, U.; Sicilia, A.; Novarini, N. On the origin of the recent herpetofauna of Sicily: Comparative phylogeography using homologous mitochondrial and nuclear genes. Zool. Anz. 2016, 261, 70–81. [Google Scholar] [CrossRef]
- Ferrito, V.; Pappalardo, A.M.; Canapa, A.; Barucca, M.; Doadrio, I.; Olmo, E.; Tigano, C. Mitochondrial phylogeography of the killifish Aphanius fasciatus (Teleostei, Cyprinodontidae) reveals highly divergent Mediterranean populations. Mar. Biol. 2013, 160, 3193–3208. [Google Scholar] [CrossRef]
- Vecchioni, L.; Deidun, A.; Sciberras, J.; Sciberras, A.; Marrone, F.; Arculeo, M. The late Pleistocene origin of the Italian and Maltese populations of Potamon fluviatile (Malacostraca: Decapoda): Insights from an expanded sampling of molecular data. Eur. Zool. J. 2017, 84, 575–582. [Google Scholar] [CrossRef]
- Vella, A.; Vella, N. First Population genetic structure analysis of the freshwater crab Potamon fluviatile (Brachyura: Potamidae) reveals fragmentation at small geographical scale. Genet. Aquat. Org. 2020, 4, 49–59. [Google Scholar] [CrossRef]
- Hewitt, G.M. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Gómez, A.; Lunt, D.H. Refugia within Refugia: Patterns of Phylogeographic concordance in the Iberian peninsula. In Phylogeography of Southern European Refugia; Weiss, S., Ferrand, N., Eds.; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar] [CrossRef]
- Harrison, S.; Hastings, A. Genetic and evolutionary consequences of metapopulation structure. Trends Ecol. Evol. 1996, 11, 180–183. [Google Scholar] [CrossRef]
- Consoli, N. Gl’interventi di Piccola Bonifica Nella Lotta Contro la Malaria in Sicilia; Provveditorato alle opere pubbliche per la Sicilia. Estratto dalla “Rivista Sanitaria Siciliana”; Sanzo, F., Ed.; C. Industria Tipografica Editrice: Moncalieri, Turin, Italy, 1928; p. 18. [Google Scholar]
- D’Antoni, S.; Battisti, C.; Cenni, M.; Rossi, G.L. (a cura di) Contributi per la tutela della biodiversità delle zone umide. Rapp. ISPRA 2011, 153, 461. [Google Scholar]
- Sperone, E.; Crescente, A.; Brunelli, E.; Paolillo, G.; Tripepi, S. Sightings and successful reproduction of allochthonous reptiles in Calabria. Acta Herpetol. 2010, 5, 265–273. [Google Scholar] [CrossRef]
- Marrone, F.; Naselli-Flores, L. A review on the animal xenodiversity in Sicilian inland waters (Italy). AIOL 2015, 6, 2–12. [Google Scholar] [CrossRef][Green Version]
- Cadi, A.; Joly, P. Competition for basking places between the endangered European pond turtle (Emys orbicularis galloitalica) and the introduced red-eared slider (Trachemys scripta elegans). Can. J. Zool. 2003, 81, 1392–1398. [Google Scholar] [CrossRef]
- Cadi, A.; Delmas, V.; Prévot-Julliard, A.C.; Joly, P.; Pieau, C.; Girondot, M. Successful reproduction of the introduced slider turtle (Trachemys scripta elegans) in the south of France. Aquat. Conserv. 2004, 14, 237–246. [Google Scholar] [CrossRef]
- Pérez-Santigosa, N.; Díaz-Paniagua, C.; Hidalgo-Vila, J. The reproductive ecology of exotic Trachemys scripta elegans in an invaded area of southern Europe. Aquat. Conserv. 2008, 18, 1302–1310. [Google Scholar] [CrossRef]
- Pérez-Santigosa, N.; Florencio, M.; Hidalgo-Vila, J.; Díaz-Paniagua, C. Does the exotic invader turtle, Trachemys scripta elegans, compete for food with coexisting native turtles? Amphibia-Reptilia 2011, 32, 167–175. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Thuiller, W.; Schioppa, E.P. From introduction to the establishment of alien species: Bioclimatic differences between presence and reproduction localities in the slider turtle. Divers. Distrib. 2009, 15, 108–116. [Google Scholar] [CrossRef]
- Vamberger, M.; Lipovšek, G.; Gregorič, M. First reproduction record of Trachemys scripta (Schoepff, 1792), in Slovenia. Herpetozoa 2012, 25, 76–79. [Google Scholar]
- Crescente, A.; Sperone, E.; Paolillo, G.; Bernabo, I.; Brunelli, E.; Tripepi, S. Nesting ecology of the exotic Trachemys scripta elegans in an area of Southern Italy (Angitola Lake, Calabria). Amphibia-Reptilia 2014, 35, 366–370. [Google Scholar] [CrossRef]
- Pearson, S.H.; Arvey, H.W.; Spotila, J.R. Juvenile invasive red eared slider turtles negatively impact the growth of native turtles: Implications for global freshwater turtle populations. Biol. Conserv. 2015, 186, 115–121. [Google Scholar] [CrossRef]
- Moravec, F.; Vargas-Vázquez, J. Some endohelminths from the freshwater turtle Trachemys scripta from Yucatán, México. J. Nat. Hist. 1998, 32, 455–468. [Google Scholar] [CrossRef]
- Hidalgo-Vila, J.; Ribas, A.; Florencio, M. Falcaustra donanaensis sp. nov. (Nematoda: Kathlaniidae) a parasite of Mauremys leprosa (Testudines, Bataguridae) in Spain. Parasitol. Res. 2006, 99, 410–413. [Google Scholar] [CrossRef]
- Meyer, L.; Du Preez, L.; Bonneau, E.; Héritier, L.; Quintana, M.F.; Valdeón, A.; Sadaoui, A.; Kechemir-Issad, N.; Palacios, C.; Verneau, O. Parasite host-switching from the invasive American red-eared slider, Trachemys scripta elegans, to the native Mediterranean pond turtle, Mauremys leprosa, in natural environments. Aquat. Invasions 2015, 10, 79–91. [Google Scholar] [CrossRef]
- Bringsøe, H. NOBANIS—Invasive Alien Species Fact Sheet, Trachemys scripta. Online Database of the North European and Baltic Network on Invasive Alien Species. 2004. Available online: http://www.nobanis.org (accessed on 7 July 2020).
- Shen, L.; Shi, H.; Wang, R.; Liu, D.; Pang, X. An invasive species red-eared slider (Trachemys scripta elegans) carrying Salmonella pathogens in Hainan island. Mol. Pathog. 2011, 2, 4. [Google Scholar] [CrossRef]
- Rataj, A.V.; Lindtner-Knific, R.; Vlahović, K.; Mavri, U.; Dovč, A. Parasites in pet reptiles. Acta Vet. Scand. 2011, 53, 33. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).