Seasonal and Agricultural Response of Acidobacteria Present in Two Fynbos Rhizosphere Soils
Abstract
1. Introduction
2. Materials and Methods
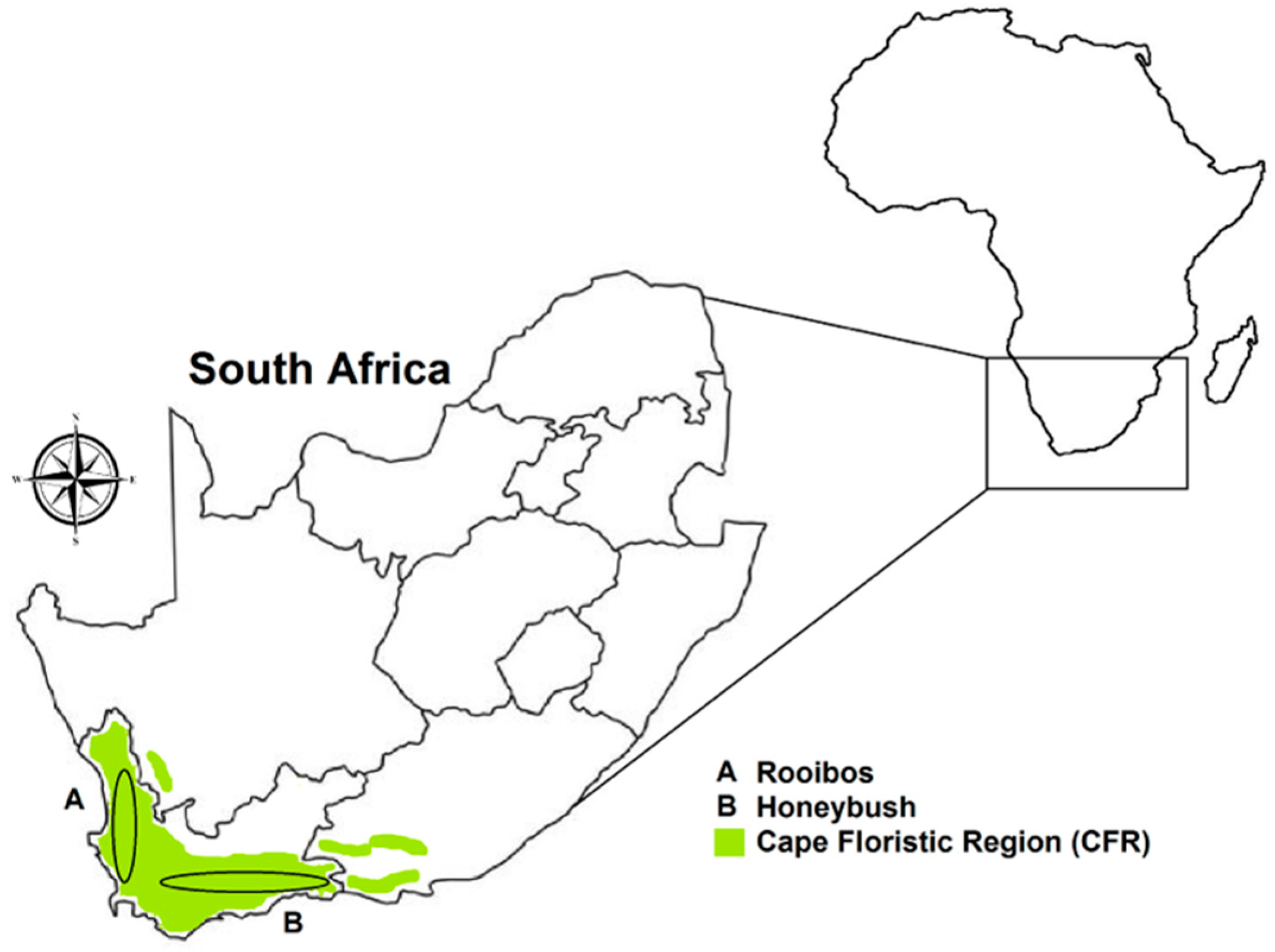
2.1. Experimental Data
2.2. Sequence Processing
2.3. Statistical Analysis
3. Results
3.1. Abiotic Soil Properties
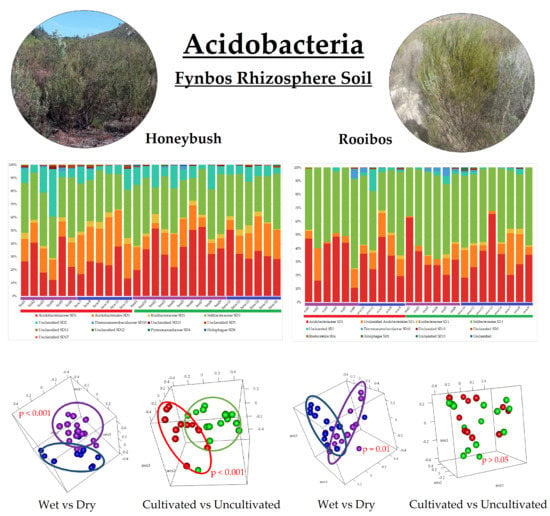
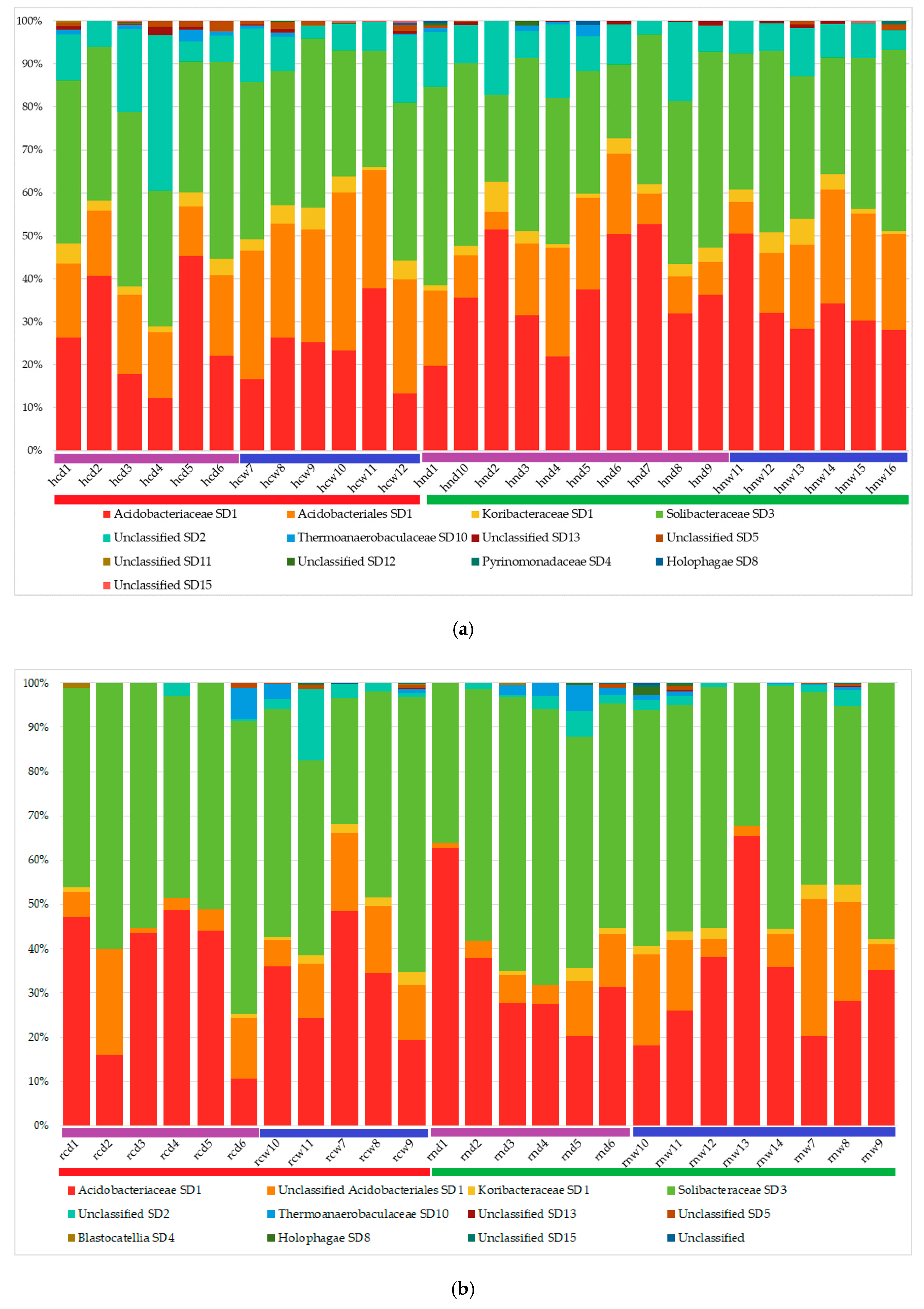
3.2. Acidobacterial Community Composition
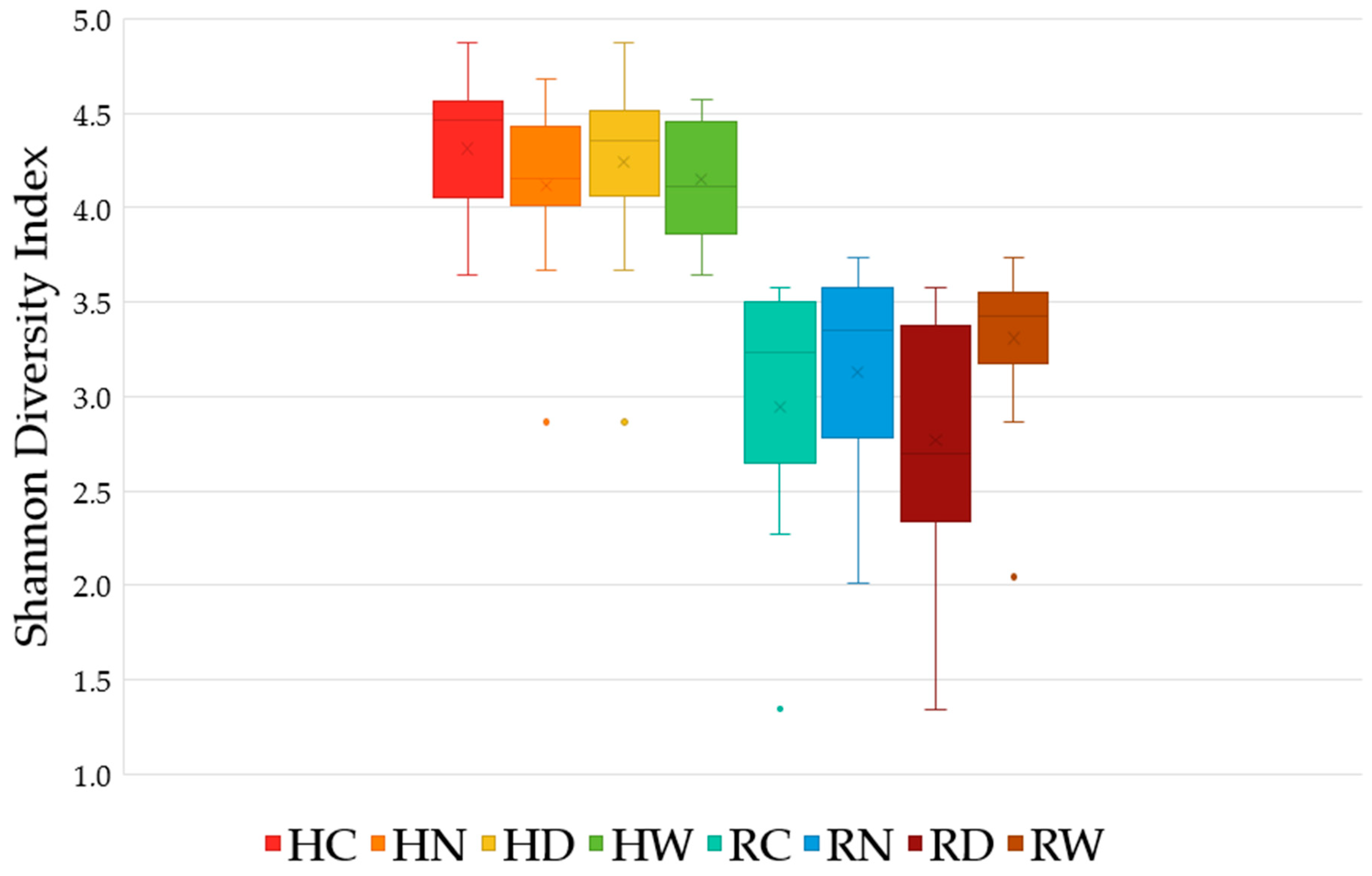
3.3. Alpha-Diversity
3.4. Beta-Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rebelo, A.G.; Boucher, C.; Helme, N.; Mucina, L.; Rutherford, M.C. Fynbos Biome. In The Vegetation of South Africa, Lesotho and Swaziland; South African National Biodiversity Institute: Pretoria, South Africa, 2006; pp. 52–219. [Google Scholar]
- Cowling, R.M.; Rundel, P.W.; Lamont, B.B.; Arroyo, M.K.; Arianoutsou, M. Plant diversity in mediterranean-climate regions. Trends Ecol. Evol. 1996, 11, 362–366. [Google Scholar] [CrossRef]
- Cowling, R.M.; Ojeda, F.; Lamont, B.B.; Rundel, P.W.; Lechmere-Oertel, R. Rainfall reliability, a neglected factor in explaining convergence and divergence of plant traits in fire-prone mediterranean-climate ecosystems. Glob. Ecol. Biogeogr. 2005, 14, 509–519. [Google Scholar] [CrossRef]
- Joubert, E.; Gelderblom, W.C.A.; Louw, A.; de Beer, D. South African herbal teas: Aspalathus linearis, Cyclopia spp. and Athrixia phylicoides-A review. J. Ethnopharmacol. 2008, 119, 376–412. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, A.A.; Kuramae, E.E.; de Hollander, M.; Pijl, A.S.; van Veen, J.A.; Tsai, S.M. Acidobacterial community responses to agricultural management of soybean in Amazon forest soils. FEMS Microbiol. Ecol. 2013, 83, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Montecchia, M.S.; Tosi, M.; Soria, M.A.; Vogrig, J.A.; Sydorenko, O.; Correa, O.S.; Moora, M. Pyrosequencing Reveals Changes in Soil Bacterial Communities after Conversion of Yungas Forests to Agriculture. PLoS ONE 2015, 10, e0119426. [Google Scholar] [CrossRef]
- Trivedi, P.; Delgado-Baquerizo, M.; Anderson, I.C.; Singh, B.K. Response of soil properties and microbial communities to agriculture: Implications for primary productivity and soil health indicators. Front. Plant Sci. 2016, 7, 990. [Google Scholar] [CrossRef]
- Jesus, E.D.C.; Marsh, T.L.; Tiedje, J.M.; Moreira, F.M.D.S. Changes in land use alter the structure of bacterial communities in Western Amazon soils. ISME J. 2009, 3, 1004–1011. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Fierer, N.; Schimel, J.P. Effects of drying-rewetting frequency on soil carbon and nitrogen transformations. Soil Biol. Biochem. 2002, 34, 777–787. [Google Scholar] [CrossRef]
- Postma, A.; Slabbert, E.; Postma, F.; Jacobs, K. Soil bacterial communities associated with natural and commercial Cyclopia spp. FEMS Microbiol. Ecol. 2016, 92, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Brink, C.; Postma, A.; Slabbert, E.; Postma, F.; Muasya, A.; Jacobs, K. Bacterial communities associated with natural and commercially grown rooibos (Aspalathus linearis). Pedosphere 2019, in press. [Google Scholar]
- Aulakh, M.S.; Wassmann, R.; Bueno, C.; Kreuzwieser, J.; Rennenberg, H. Characterization of root exudates at different growth stages of ten rice (Oryza sativa L.) cultivars. Plant Biol. 2001, 3, 139–148. [Google Scholar] [CrossRef]
- Bell, C.W.; Acosta-Martinez, V.; McIntyre, N.E.; Cox, S.; Tissue, D.T.; Zak, J.C. Linking microbial community structure and function to seasonal differences in soil moisture and temperature in a Chihuahuan Desert grassland. Microb. Ecol. 2009, 58, 827–842. [Google Scholar] [CrossRef]
- Pietikäinen, J.; Pettersson, M.; Bååth, E. Comparison of temperature effects on soil respiration and bacterial and fungal growth rates. FEMS Microbiol. Ecol. 2005, 52, 49–58. [Google Scholar] [CrossRef]
- Barns, S.M.; Takala, S.L.; Kuske, C.R. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 1999, 65, 1731–1737. [Google Scholar] [CrossRef]
- Tank, M.; Costas, A.M.G.; Bryant, D.A. Chloracidobacterium. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons Ltd.: New York, NY, USA, 2018; pp. 1–9. ISBN 9781118960608. [Google Scholar]
- Huber, K.J.; Geppert, A.M.; Wanner, G.; Fösel, B.U.; Wüst, P.K.; Overmann, J. The first representative of the globally widespread subdivision 6 Acidobacteria, Vicinamibacter silvestris gen. nov., sp. nov., isolated from subtropical savannah soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 2971–2979. [Google Scholar] [CrossRef]
- Vieira, S.; Luckner, M.; Wanner, G.; Overmann, J. Luteitalea pratensis gen. nov., sp. nov. a new member of subdivision 6 Acidobacteria isolated from temperate grassland soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 1408–1414. [Google Scholar] [CrossRef]
- Quaiser, A.; López-García, P.; Zivanovic, Y.; Henn, M.R.; Rodriguez-Valera, F.; Moreira, D. Comparative analysis of genome fragments of Acidobacteria from deep Mediterranean plankton. Environ. Microbiol. 2008, 10, 2704–2717. [Google Scholar] [CrossRef]
- O’Connor-Sánchez, A.; Rivera-Domínguez, A.J.; de los Santos-Briones, C.; López-Aguiar, L.K.; Peña-Ramírez, Y.J.; Prieto-Davo, A. Acidobacteria appear to dominate the microbiome of two sympatric Caribbean Sponges and one Zoanthid. Biol. Res. 2014, 47, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, J.; Gonzalez, J.M.; Saiz-Jimenez, C.; Ludwig, W. Detection and phylogenetic relationships of highly diverse uncultured acidobacterial communities in Altamira Cave using 23S rRNA sequence analyses. Geomicrobiol. J. 2005, 22, 379–388. [Google Scholar] [CrossRef]
- Hugenholtz, P.; Pitulle, C.; Hershberger, K.L.; Pace, N.R. Novel Division Level Bacterial Diversity in a Yellowstone Hot Spring Novel Division Level Bacterial Diversity in a Yellowstone Hot Spring. J. Bacteriol. 1998, 180, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Kleinsteuber, S.; Müller, F.D.; Chatzinotas, A.; Wendt-Potthoff, K.; Harms, H. Diversity and in situ quantification of Acidobacteria subdivision 1 in an acidic mining lake. FEMS Microbiol. Ecol. 2008, 63, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Barns, S.M.; Cain, E.C.; Sommerville, L.; Kuske, C.R. Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl. Environ. Microbiol. 2007, 73, 3113–3116. [Google Scholar] [CrossRef]
- Ludwig, W.; Bauer, S.H.; Bauer, M.; Held, I.; Kirchhof, G.; Huber, I.; Spring, S.; Hartmann, A.; Schleifer, K.H. Detection and in situ identification of representatives o fa widely distributed new bacterial phylum. FEMS Microbiol. Lett. 1997, 153, 181–190. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Yilmaz, P. Refining the taxonomic structure of the phylum Acidobacteria. Int. J. Syst. Evol. Microbiol. 2018, 68, 3796–3806. [Google Scholar] [CrossRef]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef]
- Jones, R.T.; Robeson, M.S.; Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. Int. Soc. Microb. Ecol. 2009, 3, 442–453. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Sinninghe Damsté, J.S. Acidobacteria. In eLS; John Wiley & Sons Ltd.: Chichester, UK, 2018; pp. 1–10. [Google Scholar]
- Rodrigues, J.L.M.; Pellizari, V.H.; Mueller, R.; Baek, K.; Jesus, E.D.C.; Paula, F.S.; Mirza, B.; Hamaou, G.S.; Tsai, S.M.; Feiglf, B.; et al. Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2013, 110, 988–993. [Google Scholar] [CrossRef]
- Sait, M.; Davis, K.E.R.; Janssen, P.H. Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl. Environ. Microbiol. 2006, 72, 1852–1857. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, A.A.; Venturini, A.M.; Meyer, K.M.; Klein, A.M.; Tiedje, J.M.; Brendan, B.J.; Nüsslein, K.; Tsai, S.M.; Rodrigues, J.L.M. Differential response of Acidobacteria subgroups to forest-to-pasture conversion and their biogeographic patterns in the western Brazilian Amazon. Front. Microbiol. 2015, 6, 1443. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Kielak, A.; Pijl, A.S.; Van Veen, J.A.; Kowalchuk, G.A. Phylogenetic diversity of Acidobacteria in a former agricultural soil. ISME J. 2009, 3, 378–382. [Google Scholar] [CrossRef]
- Da Rocha, U.N.; Plugge, C.M.; George, I.; Van Elsas, J.D.; Van Overbeek, L.S. The rhizosphere selects for particular groups of Acidobacteria and Verrucomicrobia. PLoS ONE 2013, 8, e82443. [Google Scholar]
- Maseko, S.T.; Dakora, F.D. Accumulation of mineral elements in the rhizosphere and shoots of Cyclopia and Aspalathus species under different settings of the Cape fynbos. S. Afr. J. Bot. 2017, 110, 103–109. [Google Scholar] [CrossRef]
- Schloss, P.D.; Gevers, D.; Westcott, S.L. Reducing the Effects of PCR Amplification and Sequencing Artifacts on 16S rRNA-Based Studies. PLoS ONE 2011, 6, e27310. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Keet, J.H.; Ellis, A.G.; Hui, C.; Le Roux, J.J. Strong spatial and temporal turnover of soil bacterial communities in South Africa’s hyperdiverse fynbos biome. Soil Biol. Biochem. 2019, 136, 107541. [Google Scholar] [CrossRef]
- Stafford, W.H.L.; Baker, G.C.; Brown, S.A.; Burton, S.G.; Cowan, D.A. Bacterial diversity in the rhizosphere of Proteaceae species. Environ. Microbiol. 2005, 7, 1755–1768. [Google Scholar] [CrossRef] [PubMed]
- Slabbert, E.; Jacobs, S.M.; Jacobs, K. The soil bacterial communities of South African fynbos riparian ecosystems invaded by Australian Acacia species. PLoS ONE 2014, 9, e86560. [Google Scholar] [CrossRef] [PubMed]
- Ward, N.L.; Challacombe, J.F.; Janssen, P.H.; Henrissat, B.; Coutinho, P.M.; Wu, M.; Xie, G.; Haft, D.H.; Sait, M.; Badger, J.; et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 2009, 75, 2046–2056. [Google Scholar] [CrossRef]
- Männistö, M.K.; Kurhela, E.; Tiirola, M.; Häggblom, M.M. Acidobacteria dominate the active bacterial communities of Arctic tundra with widely divergent winter-time snow accumulation and soil temperatures. FEMS Microbiol. Ecol. 2013, 84, 47–59. [Google Scholar] [CrossRef]
- Lee, S.H.; Cho, J.C. Distribution patterns of the members of phylum Acidobacteria in global soil samples. J. Microbiol. Biotechnol. 2009, 19, 1281–1287. [Google Scholar] [CrossRef]
- Mikha, M.M.; Rice, C.W.; Milliken, G.A. Carbon and nitrogen mineralization as affected by drying and wetting cycles. Soil Biol. Biochem. 2005, 37, 339–347. [Google Scholar] [CrossRef]
- Van Gestel, M.; Ladd, J.N.; Amato, M. Microbial biomass responses to seasonal change and imposed drying regimes at increasing depths of undisturbed topsoil profiles. Soil Biol. Biochem. 1992, 24, 103–111. [Google Scholar] [CrossRef]
- Castro, H.F.; Classen, A.T.; Austin, E.E.; Norby, R.J.; Schadt, C.W. Soil Microbial Community Responses to Multiple Experimental Climate Change Drivers. Appl. Environ. Microbiol. 2010, 76, 999–1007. [Google Scholar] [CrossRef]
- Lauber, C.L.; Ramirez, K.S.; Aanderud, Z.; Lennon, J.; Fierer, N. Temporal variability in soil microbial communities across land-use types. ISME J. 2013, 7, 1641–1650. [Google Scholar] [CrossRef]
- de Menezes, A.B.; Müller, C.; Clipson, N.; Doyle, E. The soil microbiome at the Gi-FACE experiment responds to a moisture gradient but not to CO2 enrichment. Microbiology 2016, 162, 1572–1582. [Google Scholar] [CrossRef]
- Lee, K.C.; Caruso, T.; Archer, S.D.J.; Gillman, L.N.; Lau, M.C.Y.; Cary, S.C.; Lee, C.K.; Pointing, S.B. Stochastic and Deterministic Effects of a Moisture Gradient on Soil Microbial Communities in the McMurdo Dry Valleys of Antarctica. Front. Microbiol. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bardgett, R.D.; Freeman, C.; Ostle, N.J. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2008, 2, 805–814. [Google Scholar] [CrossRef]
- Parton, W.; Silver, W.L.; Burke, I.C.; Grassens, L.; Harmon, M.E.; Currie, W.S.; King, J.Y.; Carol Adair, E.; Brandt, L.A.; Hart, S.C.; et al. Global-Scale Similarities in Nitrogen Release Patterns During Long-Term Decomposition. Science 2007, 315, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Pankratov, T.A.; Dedysh, S.N. Granulicella paludicola gen. nov., sp. nov., Granulicella pectinivorans sp. nov., Granulicella aggregans sp. nov. and Granulicella rosea sp. nov., acidophilic, polymer-degrading acidobacteria from Sphagnum peat bogs. Int. J. Syst. Evol. Microbiol. 2010, 60, 2951–2959. [Google Scholar] [CrossRef] [PubMed]
- Männistö, M.K.; Rawat, S.; Starovoytov, V.; Häggblom, M.M. Terriglobus saanensis sp. nov., an acidobacterium isolated from tundra soil. Int. J. Syst. Evol. Microbiol. 2011, 61, 1823–1828. [Google Scholar] [CrossRef] [PubMed]
- García-Fraile, P.; Benada, O.; Cajthaml, T.; Baldrian, P.; Llado, S. Terracidiphilus gabretensis gen. nov., sp. nov., an Abundant and Active Forest Soil Acidobacterium Important in Organic Matter Transformation. Appl. Environ. Microbiol. 2016, 82, 560–569. [Google Scholar] [CrossRef]
- Denef, K.; Six, J.; Bossuyt, H.; Frey, S.D.; Elliott, E.T.; Merckx, R.; Paustian, K. Influence of dry-wet cycles on the interrelationship between aggregate, particulate organic matter, and microbial community dynamics. Soil Biol. Biochem. 2001, 33, 1599–1611. [Google Scholar] [CrossRef]
- Halverson, L.J.; Jones, T.M.; Firestone, M.K. Release of Intracellular Solutes by Four Soil Bacteria Exposed to Dilution Stress. Soil Sci. Soc. Am. J. 2000, 64, 1630. [Google Scholar] [CrossRef]
- Pereira de Castro, A.; Sartori da Silva, M.R.S.; Quirino, B.F.; da Cunha Bustamante, M.M.; Krüger, R.H. Microbial Diversity in Cerrado Biome (Neotropical Savanna) Soils. PLoS ONE 2016, 11, e0148785. [Google Scholar] [CrossRef]
- Catão, E.C.P.P.; Lopes, F.A.C.C.; Araújo, J.F.; De Castro, A.P.; Barreto, C.C.; Bustamante, M.M.C.C.; Quirino, B.F.; Krüger, R.H.; Barreto, C.C.; Krüger, R.H.; et al. Soil Acidobacterial 16S rRNA Gene Sequences Reveal Subgroup Level Differences between Savanna-Like Cerrado and Atlantic Forest Brazilian Biomes. Int. J. Microbiol. 2014, 2014, 1–12. [Google Scholar] [CrossRef]
- Challacombe, J.F.; Eichorst, S.A.; Hauser, L.; Land, M.; Xie, G. Biological Consequences of Ancient Gene Acquisition and Duplication in the Large Genome of Candidatus Solibacter usitatus Ellin6076. PLoS ONE 2011, 6, 24882. [Google Scholar] [CrossRef] [PubMed]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; van Veen, J.A.; Kuramae, E.E. The ecology of Acidobacteria: Moving beyond genes and genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef]
- Singh, B.K.; Munro, S.; Potts, J.M.; Millard, P. Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl. Soil Ecol. 2007, 36, 147–155. [Google Scholar] [CrossRef]
- Lee, S.H.; Ka, J.O.; Cho, J.C. Members of the phylum Acidobacteria are dominant and metabolically active in rhizosphere soil. FEMS Microbiol. Lett. 2008, 285, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Kalam, S.; Das, S.N.; Basu, A.; Podile, A.R. Population densities of indigenous Acidobacteria change in the presence of plant growth promoting rhizobacteria (PGPR) in rhizosphere. J. Basic Microbiol. 2017, 57, 376–385. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Diamond, S.; Butterfield, C.N.; Thomas, B.C.; Banfield, J.F. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 2018, 558, 440–444. [Google Scholar] [CrossRef]
- Liu, J.; Sui, Y.; Yu, Z.; Yao, Q.; Shi, Y.; Chu, H.; Jin, J.; Liu, X.; Wang, G. Diversity and distribution patterns of acidobacterial communities in the black soil zone of northeast China. Soil Biol. Biochem. 2016, 95, 212–222. [Google Scholar] [CrossRef]
- Li, W.; Liu, X.; Niu, S. Differential responses of the acidobacterial community in the topsoil and subsoil to fire disturbance in Pinus tabulaeformis stands. PeerJ 2019, 7, e8047. [Google Scholar] [CrossRef]
- Sui, X.; Zhang, R.; Frey, B.; Yang, L.; Li, M.H.; Ni, H. Land use change effects on diversity of soil bacterial, Acidobacterial and fungal communities in wetlands of the Sanjiang Plain, northeastern China. Sci. Rep. 2019, 9, 18535. [Google Scholar] [CrossRef]
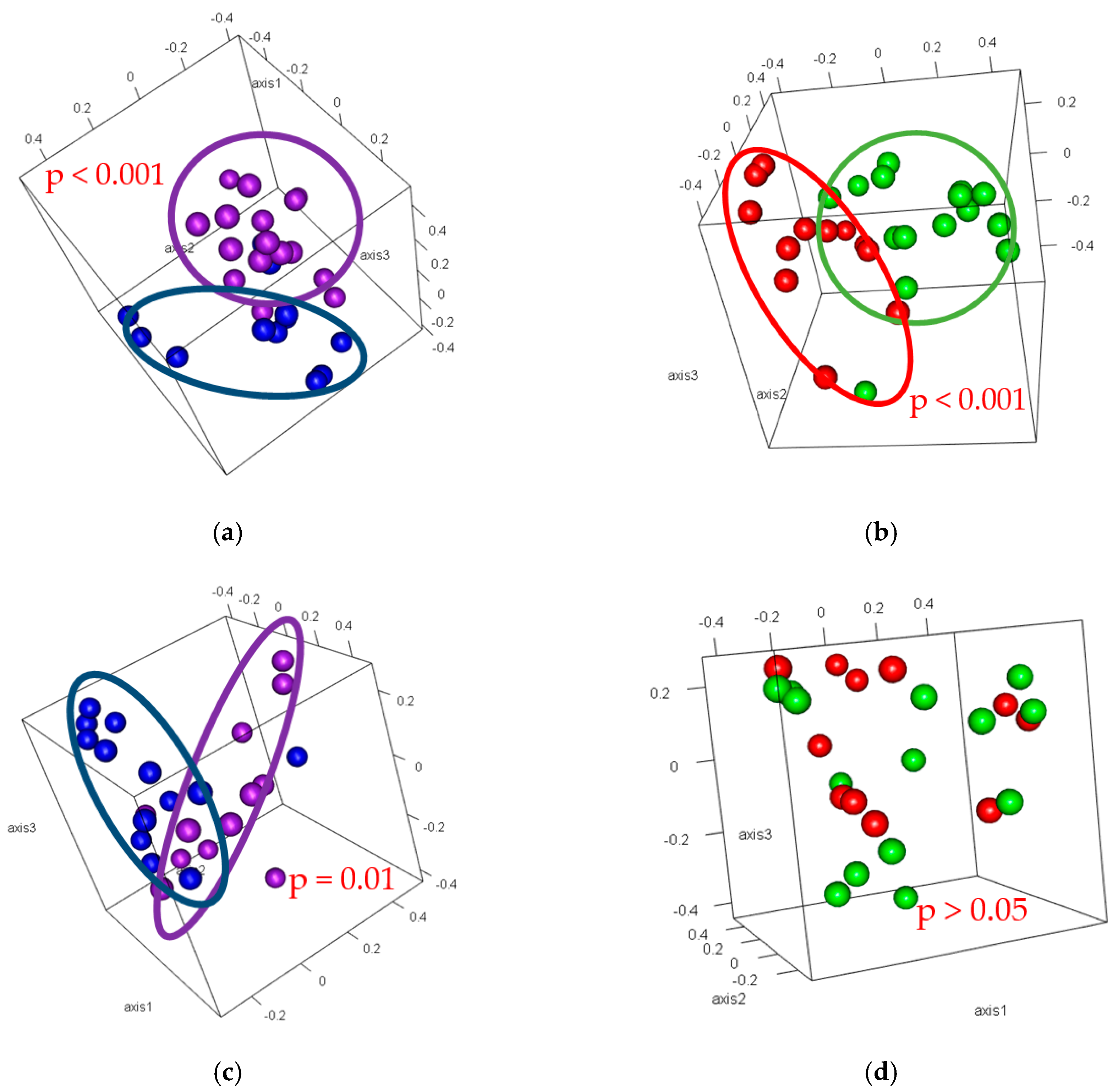
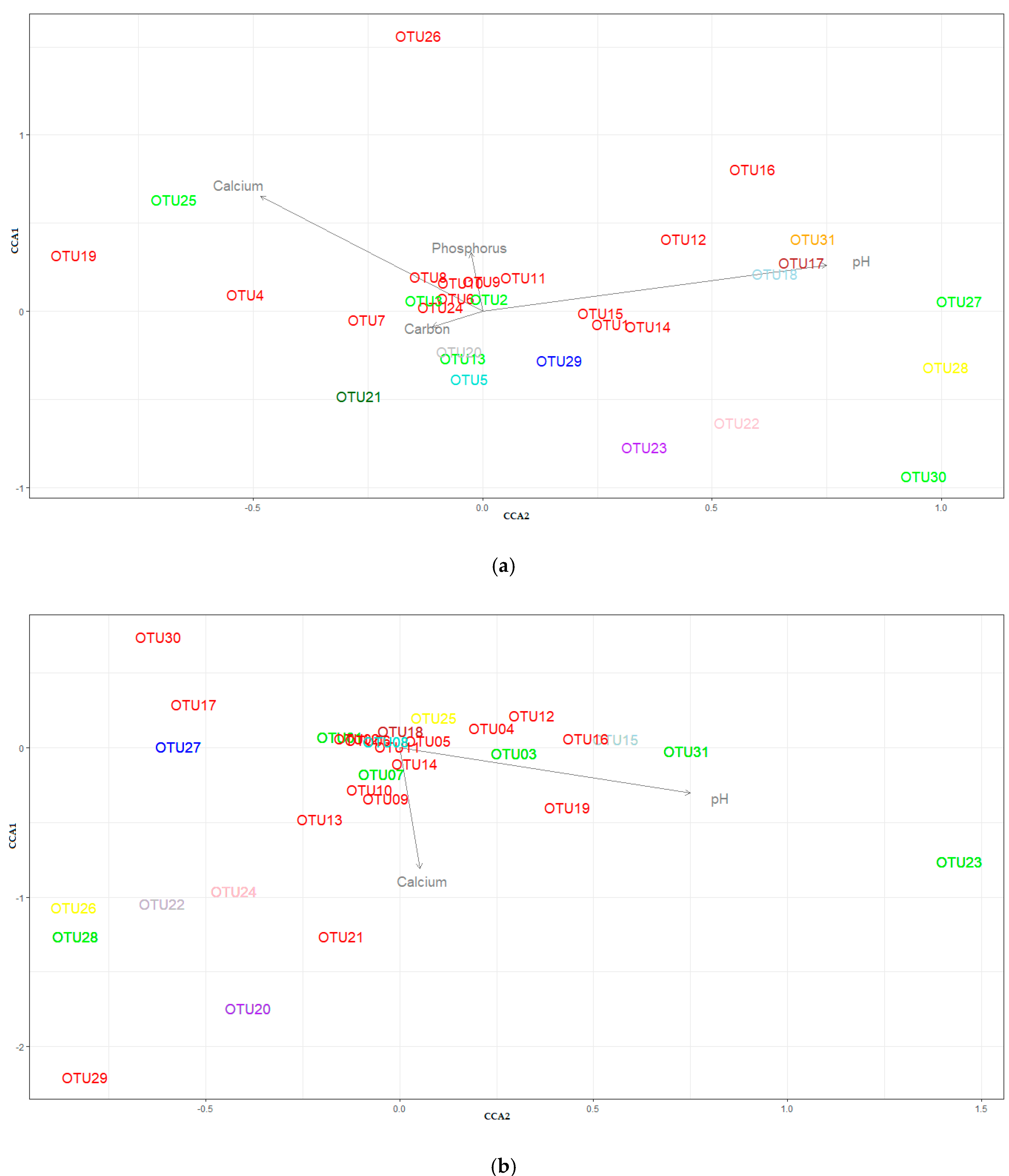
| Soil Properties | Dry 1 | Wet 1 | Cultivated 2 | Uncultivated 2 |
|---|---|---|---|---|
| pH | 4.15 ± 0.67 | 4.37 ± 0.61 | 4.75 ± 0.42 * | 3.83 ± 0.46 * |
| H+ | 3.03 ± 2.61 | 2.31 ± 2.11 | 1.42 ± 1.40 | 3.80 ± 2.55 |
| P | 3.76 ± 2.63 | 5.00 ± 2.63 | 5.23 ± 2.83 | 3.50 ± 2.31 |
| K+ | 52.18 ± 14.23 | 69.17 ± 33.29 | 56.69 ± 28.75 | 61.25 ± 22.20 |
| Na+ | 0.18 ± 0.07 | 0.17 ± 0.09 | 0.12 ± 0.04 | 0.21 ± 0.08 |
| Ca2+ | 1.88 ± 0.87 | 1.51 ± 0.79 | 1.41 ± 0.77 | 1.99 ± 0.84 |
| Mg2+ | 1.27 ± 0.65 | 1.03 ± 0.63 | 0.92 ± 0.58 * | 1.38 ± 0.63 * |
| C | 2.05 ± 0.73 * | 11.31 ± 4.90 * | 6.88 ± 5.74 | 5.08 ± 5.52 |
| NO3-N | 1.28 ± 1.27 | 1.48 ± 1.78 | 2.26 ± 1.85 * | 0.63 ± 0.20 * |
| NH4-N | 7.08 ± 1.50 | 7.59 ± 1.38 | 7.28 ± 1.75 | 7.30 ± 1.21 |
| Soil Properties | Dry 1 | Wet 1 | Cultivated 2 | Uncultivated 2 |
|---|---|---|---|---|
| pH | 4.65 ± 0.38 | 4.48 ± 0.29 | 4.29 ± 0.24 * | 4.81 ± 0.19 * |
| H+ | 0.57 ± 0.30 | 0.50 ± 0.22 | 0.58 ± 0.30 | 0.49 ± 0.22 |
| P | 6.67 ± 2.10 | 5.54 ± 5.06 | 8.00 ± 4.13 * | 4.31 ± 2.75 * |
| K+ | 49.58 ± 34.08 | 24.62 ± 9.00 | 48.42 ± 34.26 | 25.69 ± 11.18 |
| Na+ | 0.10 ± 0.07 | 0.05 ± 0.03 | 0.09 ± 0.08 | 0.06 ± 0.03 |
| Ca2+ | 0.65 ± 0.36 | 0.81 ± 0.54 | 0.40 ± 0.20 | 1.04 ± 0.41 |
| Mg2+ | 0.32 ± 0.05 | 0.34 ± 0.24 | 0.25 ± 0.08 | 0.41 ± 0.20 |
| C | 0.31 ± 0.15 * | 0.77 ± 0.60 * | 0.39 ± 0.20 | 0.70 ± 0.64 |
| NO3-N | 1.30 ± 0.75 | 0.84 ± 0.72 | 1.49 ± 0.87 | 0.66 ± 0.29 |
| NH4-N | 7.76 ± 1.83 | 8.01 ± 0.85 | 8.09 ± 1.57 | 7.71 ± 1.21 |
| Group | Group ID | Filtered Reads 1 | Acidobacteria Reads | Average Relative Abundance (%) | p-Value 2 |
|---|---|---|---|---|---|
| Honeybush Cultivated | HC | 238,526 | 22,518 | 9.4 ± 5.1 | 0.09 |
| Honeybush Uncultivated | HN | 405,815 | 28,270 | 7.0 ± 2.4 | |
| Honeybush Dry | HD | 451,358 | 31,591 | 7.0 ± 2.5 | 0.19 |
| Honeybush Wet | HW | 192,983 | 19,197 | 9.9 ± 4.9 | |
| Rooibos Cultivated | RC | 514,844 | 21,691 | 4.2 ± 5.4 | 0.14 |
| Rooibos Uncultivated | RN | 521,043 | 23,498 | 4.5 ± 3.9 | |
| Rooibos Dry | RD | 717,981 | 14,811 | 2.1 ± 2.0 | 0.006 * |
| Rooibos Wet | RW | 317,906 | 30,378 | 9.6 ± 2.8 |
| Taxonomic Classification | HC | HN | HW | HD | RC | RN | RD | RW | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subdivision | Family | Genus | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD |
| SD1 | Acidobacteriaceae | Acidicapsa | 1.3 | 1.5 | 2.2 | 1.8 | 1.2 | 1.5 | 2.2 | 1.7 | 0.5 | 0.6 | 0.3 | 1.0 | 0.5 | 1.1 | 0.3 | 0.6 |
| Acidipila | 3.0 * | 2.0 | 9.4 * | 5.0 | 4.6 * | 5.0 | 7.5 * | 5.3 | 8.3 | 9.0 | 7.7 | 5.2 | 9.1 | 8.7 | 7.5 | 5.1 | ||
| Unclassified | 0.1 | 0.2 | 0.1 | 0.2 | 0.1 | 0.2 | 0.1 | 0.2 | 0.1 | 0.1 | 0.2 | 0.4 | 0.2 | 0.2 | 0.1 | 0.4 | ||
| Unclassified | 10.1 | 5.5 | 13.8 | 6.0 | 11.5 | 5.2 | 12.5 | 6.3 | 12.8 | 6.3 | 11.6 | 8.9 | 9.6 * | 5.4 | 13.1 * | 9.2 | ||
| Bryocella | 0.1 * | 0.1 | 0.4 * | 1.0 | 0.4 | 1.1 | 0.2 | 0.3 | 0.6 | 3.5 | 0.8 | 1.2 | 1.0 | 3.4 | 0.6 | 1.2 | ||
| Edaphobacter | 2.3 | 2.4 | 1.4 | 1.8 | 1.5 | 1.2 | 2.0 | 2.6 | 1.5 | 2.1 | 1.4 | 2.1 | 2.3 | 2.8 | 1.1 | 0.8 | ||
| Granulicella | 2.1 * | 0.9 | 4.4 * | 3.3 | 2.7 * | 1.9 | 3.7 * | 3.5 | 4.3 | 2.6 | 4.1 | 4.3 | 5.1 | 4.3 | 3.8 | 2.8 | ||
| Occallatibacter | 1.3 | 1.0 | 1.4 | 0.9 | 1.5 | 0.8 | 1.2 | 1.0 | 0.6 | 0.7 | 1.0 | 1.8 | 0.9 | 2.0 | 0.8 | 0.6 | ||
| Telmatobacter | - | - | - | - | - | - | <0.1 | 0.1 | - | - | - | - | <0.1 | 0.1 | ||||
| Terracidiphilus | 2.2 | 1.3 | 1.6 | 1.0 | 1.7 | 1.2 | 1.9 | 1.3 | 1.4 | 1.2 | 1.3 | 1.4 | 1.0 | 1.7 | 1.5 | 0.8 | ||
| Terriglobus | 0.6 | 1.7 | 0.3 | 0.5 | 0.4 | 0.4 | 0.5 | 1.5 | 0.2 | 0.3 | 0.2 | 0.4 | <0.1 | 0.4 | 0.3 | 0.3 | ||
| Unclassified | 0.9 | 0.6 | 0.4 | 0.5 | 1.1 * | 0.6 | 0.3 * | 0.4 | 1.0 | 0.6 | 1.4 | 1.1 | 0.6 | 0.5 | 1.4 | 0.9 | ||
| Unclassified | Unclassified | 0.1 | 0.2 | - | - | - | <0.1 | 0.2 | - | - | 0.1 | 0.1 | - | - | <0.1 | 0.1 | ||
| Unclassified | Unclassified | 2.5 | 1.1 | 1.6 | 2.0 | 2.6 | 1.9 | 1.7 | 1.5 | 3.0 | 1.7 | 5.3 | 4.0 | 2.3 | 1.7 | 5.0 | 3.8 | |
| Koribacteraceae | Candidatus Koribacter | 2.9 | 1.4 | 2.7 | 1.9 | 3.3 | 1.8 | 2.5 | 1.6 | 1.6 | 1.0 | 2.0 | 1.3 | 1.1 | 0.9 | 2.1 | 1.0 | |
| Unclassified | Unclassified | 20.3 | 7.2 | 13.7 | 6.7 | 23.2 * | 6.9 | 13.1 * | 5.4 | 8.9 | 5.6 | 8.9 | 5.1 | 6.8 | 5.4 | 9.7 | 5.0 | |
| Unclassified | Unclassified | 0.1 | 0.1 | - | - | 0.1 | 0.1 | - | - | 0.1 | 0.1 | 0.1 | 0.1 | <0.1 | 0.1 | 0.1 | 0.1 | |
| SD2 | Unclassified | Unclassified | 13.2 | 9.2 | 9.7 | 4.6 | 9.0 * | 3.5 | 12.7 * | 8.3 | 3.9 | 4.7 | 1.9 | 1.7 | 1.7 | 1.8 | 3.2 | 4.2 |
| SD3 | Solibacteraceae | Unclassified | 0.1 | 0.3 | - | - | 0.1 | 0.3 | - | - | - | - | - | - | - | - | - | |
| Bryobacter | 13.3 | 1.8 | 14.8 | 4.2 | 14.2 | 3.9 | 14.0 | 3.1 | 23.2 | 10.4 | 21.0 | 7.3 | 23.8 | 10.6 | 21.3 | 6.8 | ||
| Candidatus Solibacter | 18.0 | 4.7 | 16.4 | 6.7 | 16.5 | 4.0 | 17.5 | 7.2 | 16.6 | 8.4 | 16.8 | 10.8 | 19.8 | 13.4 | 15.5 | 5.4 | ||
| Unclassified | <0.1 | 0.1 | - | - | <0.1 | 0.1 | - | - | - | - | <0.1 | 0.1 | - | - | <0.1 | 0.1 | ||
| Paludibaculum | <0.1 | 0.1 | <0.1 | 0.1 | <0.1 | 0.1 | <0.1 | 0.1 | - | - | 0.1 | 0.2 | 0.1 | 0.2 | - | - | ||
| Unclassified | 3.4 | 1.5 | 4.7 | 3.0 | 3.0 | 2.1 | 4.8 | 2.7 | 9.5 | 5.9 | 12.0 | 4.9 | 10.4 | 5.7 | 11.1 | 4.9 | ||
| Unclassified | <0.1 | 0.1 | - | - | - | - | <0.1 | 0.1 | <0.1 | 0.0 | <0.1 | 0.1 | - | - | <0.1 | 0.1 | ||
| SD4 | Pyrinomonadaceae | Pyrinomonas | - | - | <0.1 | 0.2 | <0.1 | 0.2 | - | - | - | - | - | - | - | - | - | - |
| Unclassified | Unclassified | - | - | - | - | - | - | - | - | 0.1 | 0.3 | <0.1 | 0.1 | 0.1 | 0.3 | <0.1 | 0.1 | |
| SD5 | Unclassified | Unclassified | 0.9 * | 0.8 | 0.2 * | 0.4 | 0.7 | 0.7 | 0.4 | 0.7 | 0.4 | 0.4 | 0.3 | 0.3 | 0.4 | 0.4 | 0.3 | 0.3 |
| SD8 | Unclassified | Unclassified | 0.1 | 0.2 | 0.1 | 0.3 | 0.1 | 0.2 | 0.1 | 0.3 | <0.1 | 0.1 | 0.3 | 0.5 | 0.1 | 0.1 | 0.2 | 0.5 |
| SD10 | Thermoanaerobaculaceae | Unclassified | 0.6 | 0.8 | 0.3 | 0.7 | 0.2 * | 0.3 | 0.5 * | 0.9 | 1.3 | 2.3 | 1.2 | 1.6 | 2.6 | 2.5 | 0.7 | 1.0 |
| SD11 | Unclassified | Unclassified | <0.1 | 0.1 | - | - | - | - | <0.1 | 0.1 | - | - | - | - | - | - | - | - |
| SD12 | Unclassified | Unclassified | 0.1 | 0.1 | 0.1 | 0.3 | 0.1 | 0.1 | 0.1 | 0.3 | - | - | - | - | - | - | - | - |
| SD13 | Unclassified | Unclassified | 0.5 | 0.6 | 0.3 | 0.4 | 0.4 | 0.4 | 0.4 | 0.5 | <0.1 | 0.1 | 0.1 | 0.1 | - | - | 0.1 | 0.2 |
| SD15 | Unclassified | Unclassified | 0.1 | 0.2 | <0.1 | 0.1 | 0.1 | 0.2 | <0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | <0.1 | 0.0 | 0.1 | 0.2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conradie, T.; Jacobs, K. Seasonal and Agricultural Response of Acidobacteria Present in Two Fynbos Rhizosphere Soils. Diversity 2020, 12, 277. https://doi.org/10.3390/d12070277
Conradie T, Jacobs K. Seasonal and Agricultural Response of Acidobacteria Present in Two Fynbos Rhizosphere Soils. Diversity. 2020; 12(7):277. https://doi.org/10.3390/d12070277
Chicago/Turabian StyleConradie, Tersia, and Karin Jacobs. 2020. "Seasonal and Agricultural Response of Acidobacteria Present in Two Fynbos Rhizosphere Soils" Diversity 12, no. 7: 277. https://doi.org/10.3390/d12070277
APA StyleConradie, T., & Jacobs, K. (2020). Seasonal and Agricultural Response of Acidobacteria Present in Two Fynbos Rhizosphere Soils. Diversity, 12(7), 277. https://doi.org/10.3390/d12070277