Oral Microbiome Metabarcoding in Two Invasive Small Mammals from New Zealand
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Ethics Statements
2.2. Sample Collection and Genomic Library Preparation
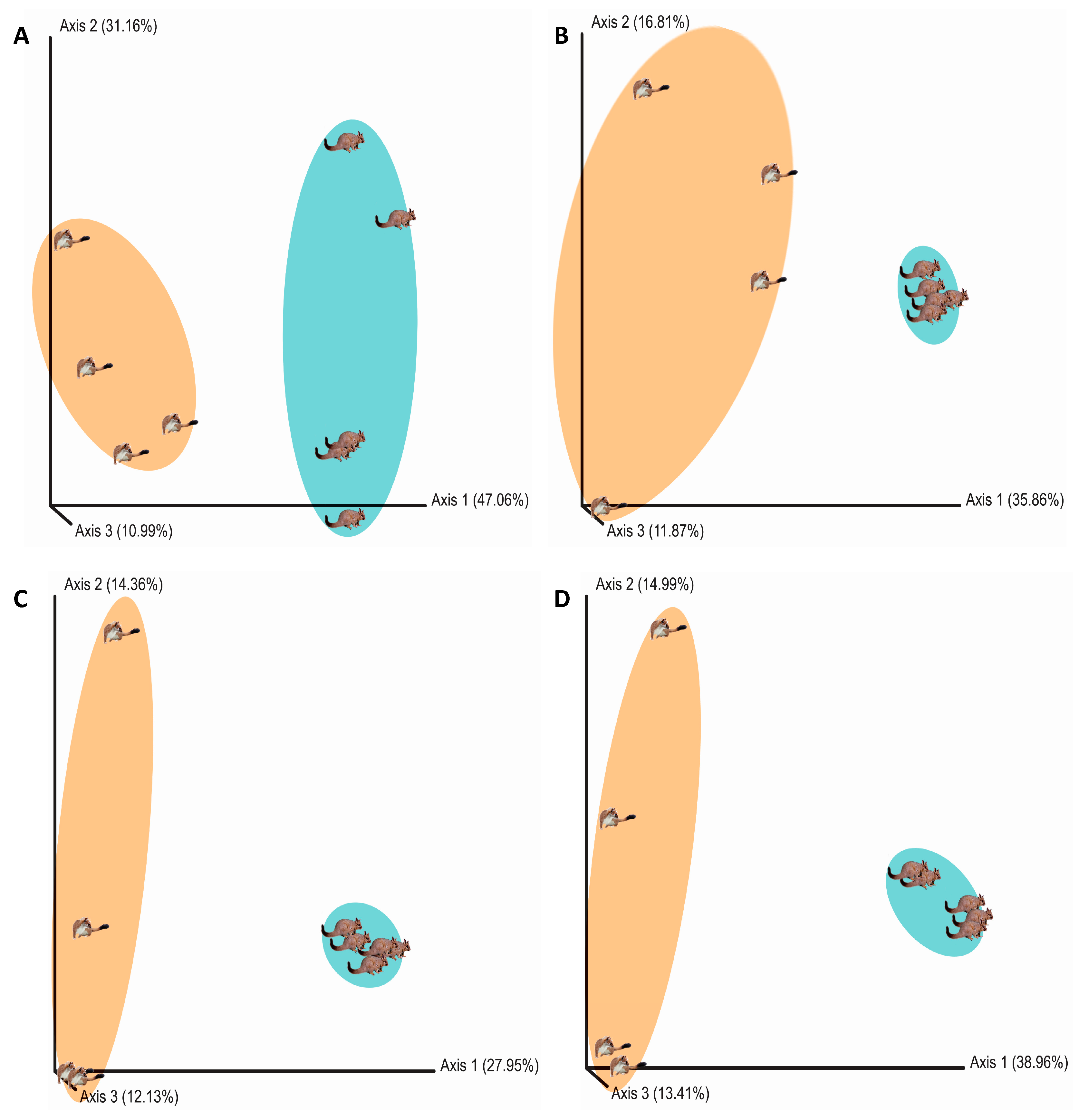
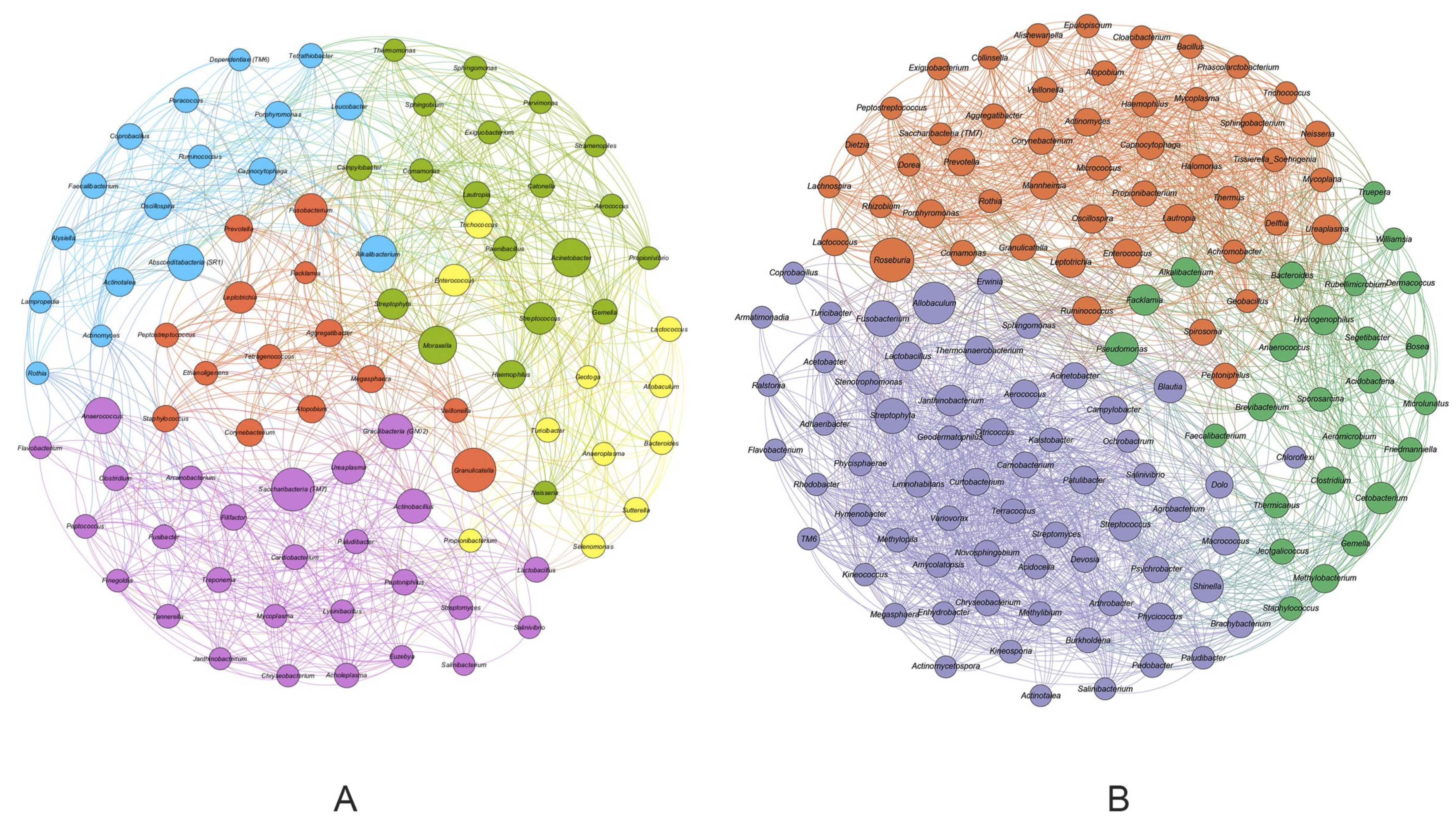
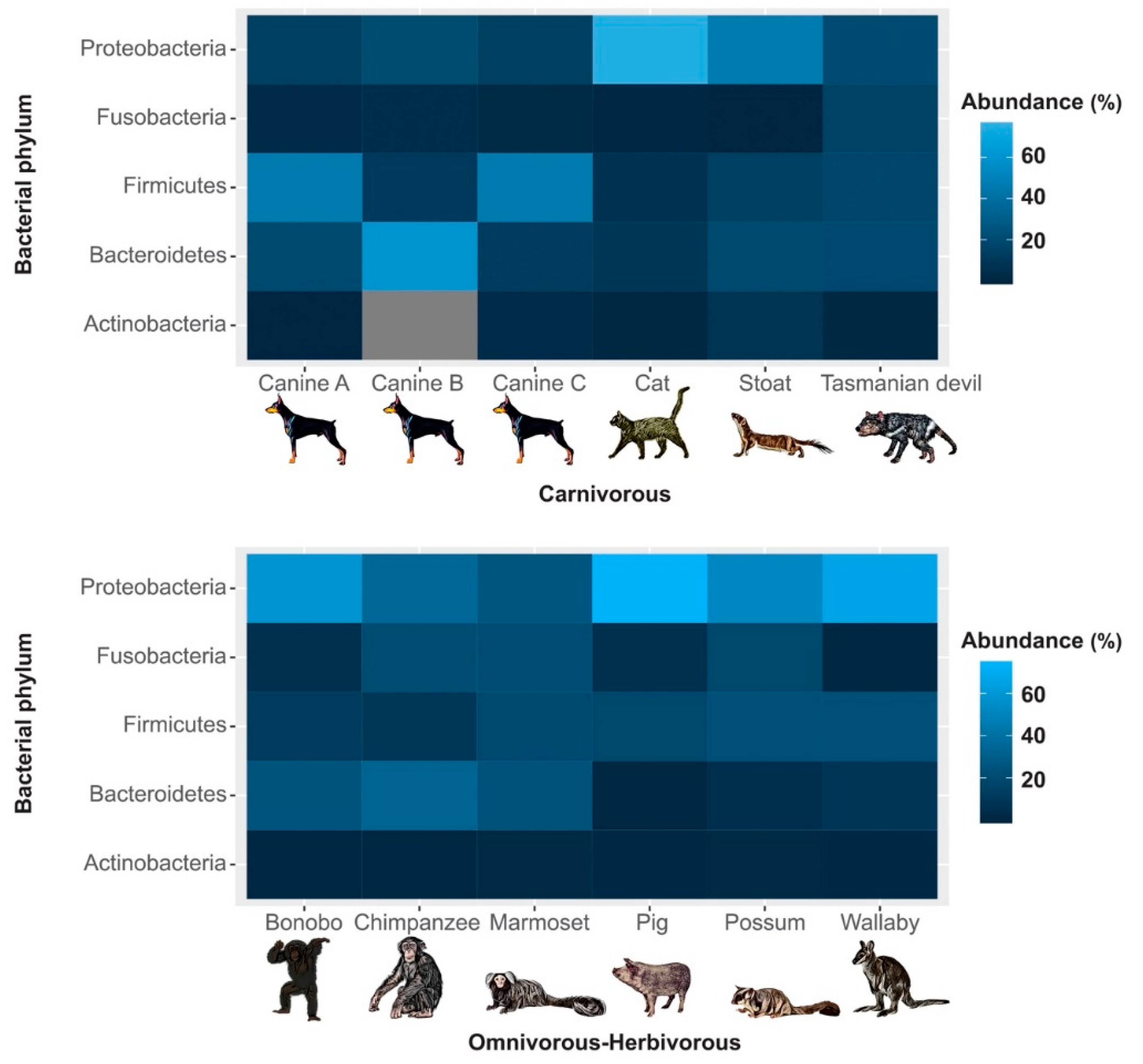
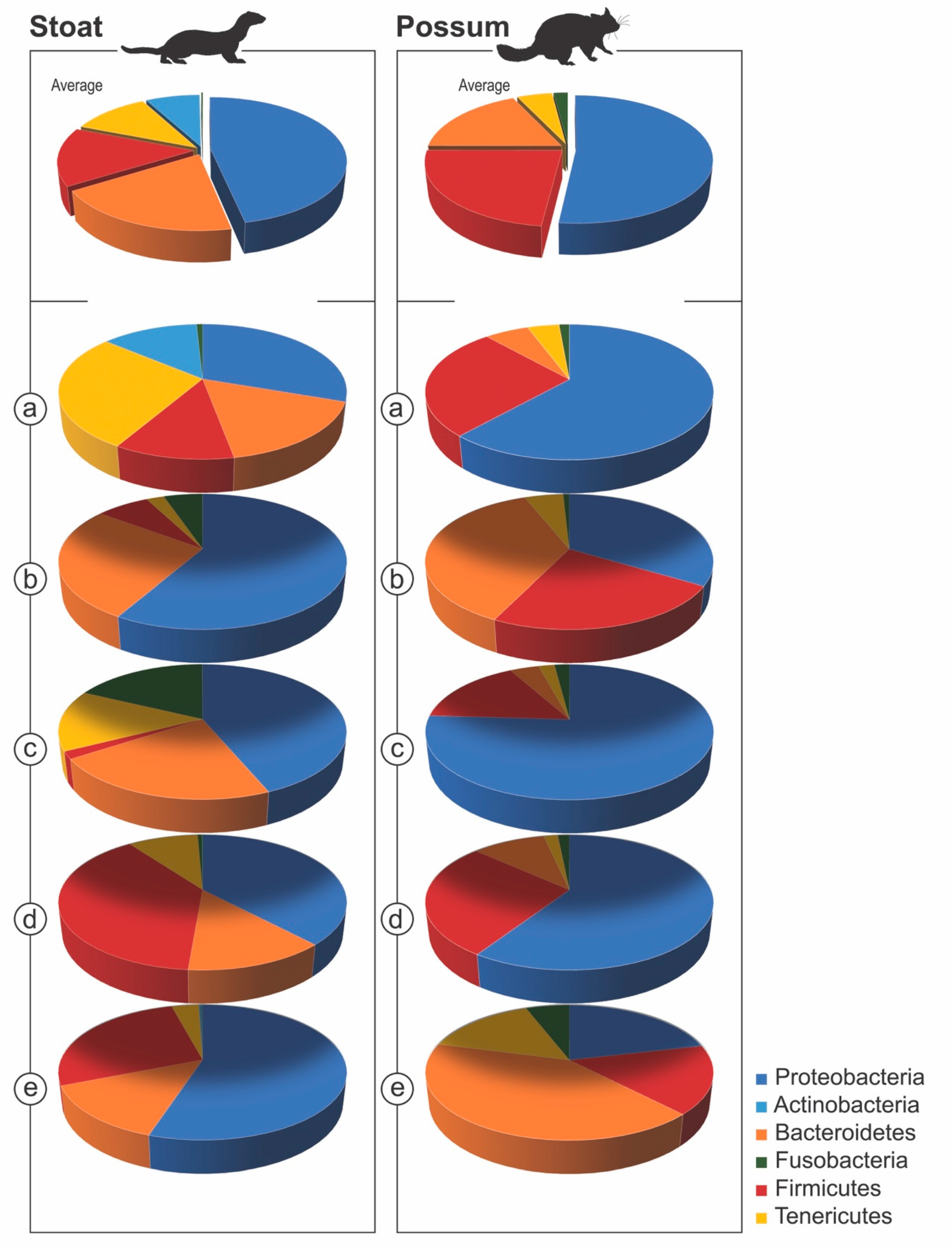
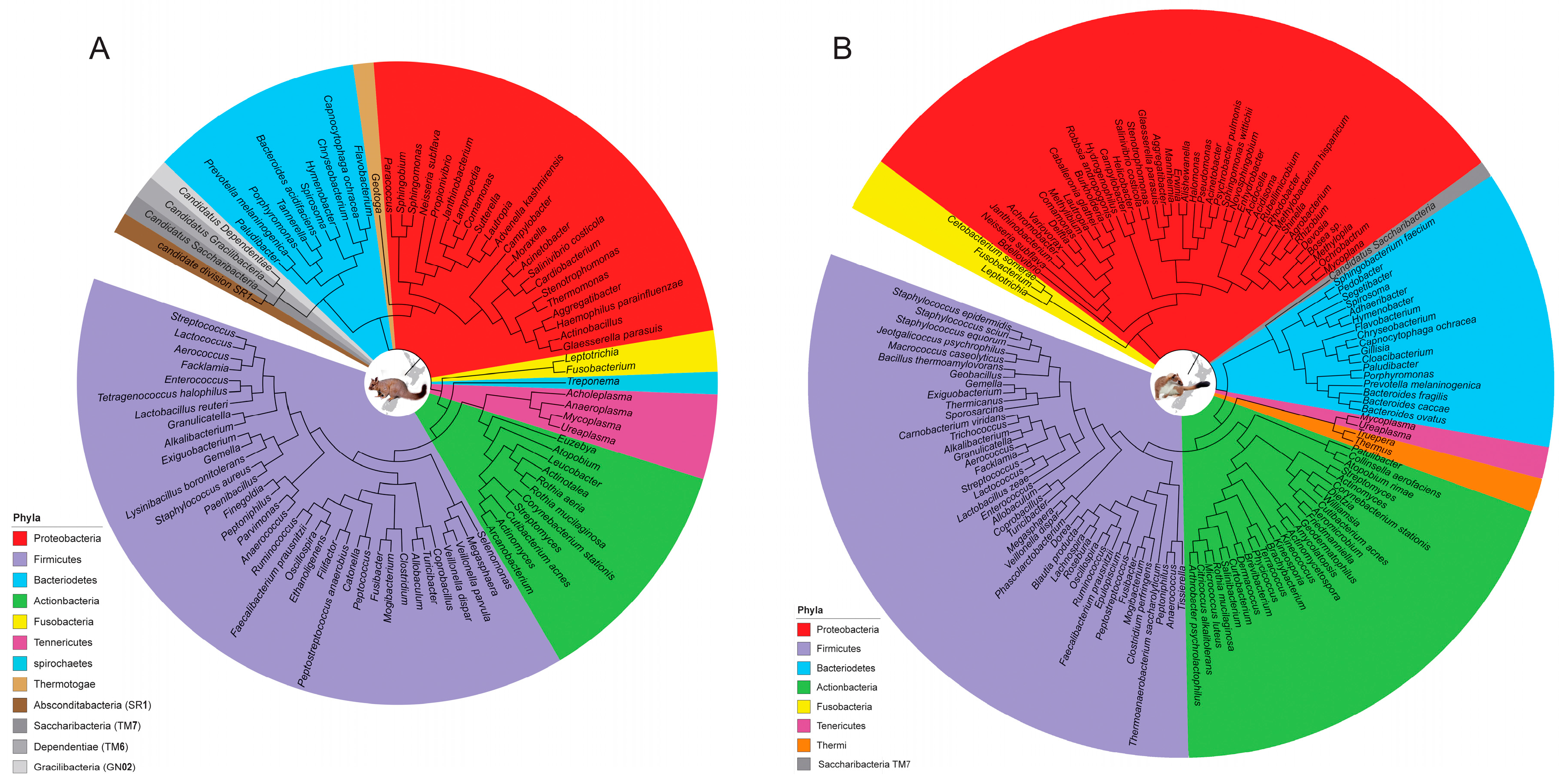
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dyall, S.D.; Brown, M.T.; Johnson, P.J. Ancient Invasions: From Endosymbionts to Organelles. Science 2004, 304, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Horiike, T.; Hamada, K.; Miyata, D.; Shinozawa, T. The Origin of Eukaryotes Is Suggested as the Symbiosis of Pyrococcus into Γ-Proteobacteria by Phylogenetic Tree Based on Gene Content. J. Mol. Evol. 2004, 59, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.; Zilber-Rosenberg, I. Microbes Drive Evolution of Animals and Plants: The Hologenome Concept. MBio 2016, 7, e01395-15. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Apprill, A. Marine Animal Microbiomes: Toward Understanding Host–Microbiome Interactions in a Changing Ocean. Front. Mar. Sci. 2017, 4, 222. [Google Scholar] [CrossRef]
- Dethlefsen, L.; Eckburg, P.B.; Bik, E.M.; Relman, D.A. Assembly of the Human Intestinal Microbiota. Trends Ecol. Evol. 2006, 21, 517–523. [Google Scholar] [CrossRef]
- Lee, Y.K.; Mazmanian, S.K. Has the Microbiota Played a Critical Role in the Evolution of the Adaptive Immune System? Science 2010, 330, 1768–1773. [Google Scholar] [CrossRef]
- Archie, E.A.; Theis, K.R. Animal Behaviour Meets Microbial Ecology. Anim. Behav. 2011, 82, 425–436. [Google Scholar] [CrossRef]
- McCutcheon, J.P.; Von Dohlen, C.D. An Interdependent Metabolic Patchwork in the Nested Symbiosis of Mealybugs. Curr. Biol. 2011, 21, 1366–1372. [Google Scholar] [CrossRef]
- Brucker, R.M.; Bordenstein, S.R. The Capacious Hologenome. Zoology 2013, 116, 260–261. [Google Scholar] [CrossRef]
- Wang, J.; Kalyan, S.; Steck, N.; Turner, L.M.; Harr, B.; Künzel, S.; Vallier, M.; Häsler, R.; Franke, A.; Oberg, H.-H. Analysis of Intestinal Microbiota in Hybrid House Mice Reveals Evolutionary Divergence in a Vertebrate Hologenome. Nat. Commun. 2015, 6, 6440. [Google Scholar] [CrossRef]
- Cheng, D.; Chen, S.; Huang, Y.; Pierce, N.E.; Riegler, M.; Yang, F.; Zeng, L.; Lu, Y.; Liang, G.; Xu, Y. Symbiotic Microbiota May Reflect Host Adaptation by Resident to Invasive Ant Species. PLoS Pathog. 2019, 15, e1007942. [Google Scholar] [CrossRef] [PubMed]
- Siebert, J.C.; Görg, C.; Palmer, B.; Lozupone, C. Visualizing Microbiome–Immune System Interplay. Fut. Med. 2019, 11, 2. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Romero, C.; Guo, K.; Murdock, B.J.; Paez-Colasante, X.; Bassis, C.M.; Mikhail, K.A.; Raue, K.D.; Evans, M.C.; Taubman, G.F.; McDermott, A.J. Temporal Evolution of the Microbiome, Immune System and Epigenome with Disease Progression in Als Mice. Dis. Models Mech. 2020, 13, dmm041947. [Google Scholar] [CrossRef]
- Rawls, J.F.; Mahowald, M.A.; Ley, R.E.; Gordon, J.I. Reciprocal Gut Microbiota Transplants from Zebrafish and Mice to Germ-Free Recipients Reveal Host Habitat Selection. Cell 2006, 127, 423–433. [Google Scholar] [CrossRef]
- Rosenberg, E.; Zilber-Rosenberg, I. The Hologenome Concept: Human, Animal and Plant Microbiota; Springer: New York, NY, USA, 2014; pp. 1–178. [Google Scholar]
- Zilber-Rosenberg, I.; Rosenberg, E. Role of Microorganisms in the Evolution of Animals and Plants: The Hologenome Theory of Evolution. FEMS Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef]
- Theis, K.R.; Dheilly, N.M.; Klassen, J.L.; Brucker, R.M.; Baines, J.F.; Bosch, T.C.; Cryan, J.F.; Gilbert, S.F.; Goodnight, C.J.; Lloyd, E.A. Getting the Hologenome Concept Right: An Eco-Evolutionary Framework for Hosts and Their Microbiomes. Msystems 2016, 1, e00028-16. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.R.; Pop, M.; DeBoy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic Analysis of the Human Distal Gut Microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- Barker, C.J.; Gillett, A.; Polkinghorne, A.; Timms, P. Investigation of the Koala (Phascolarctos cinereus) Hindgut Microbiome Via 16s Pyrosequencing. Vet. Microbiol. 2013, 167, 554–564. [Google Scholar] [CrossRef]
- Moeller, A.H. The Shrinking Human Gut Microbiome. Curr. Opin. Microbiol. 2017, 38, 30–35. [Google Scholar] [CrossRef]
- Raymann, K.; Moeller, A.H.; Goodman, A.L.; Ochman, H. Unexplored Archaeal Diversity in the Great Ape Gut Microbiome. MSphere 2017, 2, e00017–e00026. [Google Scholar] [CrossRef]
- Cheng, Y.; Fox, S.; Pemberton, D.; Hogg, C.; Papenfuss, A.T.; Belov, K. The Tasmanian Devil Microbiome—Implications for Conservation and Management. Microbiome 2015, 3, 76. [Google Scholar] [CrossRef] [PubMed]
- Chhour, K.-L.; Hinds, L.A.; Jacques, N.A.; Deane, E.M. An Observational Study of the Microbiome of the Maternal Pouch and Saliva of the Tammar Wallaby, Macropus Eugenii, and of the Gastrointestinal Tract of the Pouch Young. Microbiology 2010, 156, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.M. Gene Sequence Analyses of the Healthy Oral Microbiome in Humans and Companion Animals: A Comparative Review. J. Vet. Dent. 2016, 33, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Klein, E.A.; Thompson, E.C.; Blanton, J.M.; Chen, T.; Milella, L.; Buckley, C.M.; Davis, I.J.; Bennett, M.-L.; Marshall-Jones, Z.V. The Canine Oral Microbiome. PLoS ONE 2012, 7, e36067. [Google Scholar] [CrossRef]
- Li, J.; Nasidze, I.; Quinque, D.; Li, M.; Horz, H.-P.; André, C.; Garriga, R.M.; Halbwax, M.; Fischer, A.; Stoneking, M. The Saliva Microbiome of Pan and Homo. BMC Microbiol. 2013, 13, 204. [Google Scholar] [CrossRef]
- Lowe, B.A.; Marsh, T.L.; Isaacs-Cosgrove, N.; Kirkwood, R.N.; Kiupel, M.; Mulks, M.H. Defining the “Core Microbiome” of the Microbial Communities in the Tonsils of Healthy Pigs. BMC Microbiol. 2012, 12, 20. [Google Scholar] [CrossRef]
- Sturgeon, A.; Pinder, S.; Costa, M.; Weese, J. Characterization of the Oral Microbiota of Healthy Cats Using Next-Generation Sequencing. Vet. J. 2014, 201, 223–229. [Google Scholar] [CrossRef]
- Takehara, S.; Zeredo, J.L.; Kumei, Y.; Kagiyama, K.; Fukasawa, K.; Oshiro, A.; Ueno, M.; Kojimahara, N.; Minakuchi, S.; Kawaguchi, Y. Characterization of Oral Microbiota in Marmosets: Feasibility of Using the Marmoset as a Human Oral Disease Model. PLoS ONE 2019, 14, e0207560. [Google Scholar] [CrossRef]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.; Yu, W.-H.; Lakshmanan, A.; Wade, W.G. The Human Oral Microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef]
- Gomez, A.; Espinoza, J.L.; Harkins, D.M.; Leong, P.; Saffery, R.; Bockmann, M.; Torralba, M.; Kuelbs, C.; Kodukula, R.; Inman, J. Host Genetic Control of the Oral Microbiome in Health and Disease. Cell Host Microbe 2017, 22, 269–278.e3. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.D.; Slade, G.; Offenbacher, S. Oral Disease, Cardiovascular Disease and Systemic Inflammation. Periodontology 2000 2000, 23, 110–120. [Google Scholar] [CrossRef]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A. Fusobacterium Nucleatum Infection Is Prevalent in Human Colorectal Carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Seymour, G.; Ford, P.; Cullinan, M.; Leishman, S.; Yamazaki, K. Relationship between Periodontal Infections and Systemic Disease. Clin. Microbiol. Infect. 2007, 13, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Zarco, M.; Vess, T.; Ginsburg, G. The Oral Microbiome in Health and Disease and the Potential Impact on Personalized Dental Medicine. Oral Dis. 2012, 18, 109–120. [Google Scholar] [CrossRef]
- Wilmshurst, J.M.; Anderson, A.J.; Higham, T.F.; Worthy, T.H. Dating the Late Prehistoric Dispersal of Polynesians to New Zealand Using the Commensal Pacific Rat. Proc. Natl. Acad. Sci. USA 2008, 105, 7676–7680. [Google Scholar] [CrossRef]
- Emami-Khoyi, A.; Paterson, A.M.; Hartley, D.A.; Boren, L.J.; Cruickshank, R.H.; Ross, J.G.; Murphy, E.C.; Else, T.-A. Mitogenomics Data Reveal Effective Population Size, Historical Bottlenecks, and the Effects of Hunting on New Zealand Fur Seals (Arctocephalus forsteri). Mitochondrial DNA Part A 2018, 29, 567–580. [Google Scholar] [CrossRef]
- Murphy, E.C.; Russell, J.C.; Broome, K.G.; Ryan, G.J.; Dowding, J.E. Conserving New Zealand’s Native Fauna: A Review of Tools Being Developed for the Predator Free 2050 Programme. J. Ornithol. 2019, 160, 1–10. [Google Scholar] [CrossRef]
- Emami-Khoyi, A.; Hartley, D.A.; Ross, J.G.; Murphy, E.C.; Paterson, A.M.; Cruickshank, R.H.; Else, T.-A. Complete Mitochondrial Genome of the Stoat (Mustela erminea) and New Zealand Fur Seal (Arctocephalus forsteri) and their Significance for Mammalian Phylogeny. Mitochondrial DNA Part A 2016, 27, 4597–4599. [Google Scholar] [CrossRef]
- Emami-Khoyi, A.; Parbhu, S.P.; Ross, J.G.; Murphy, E.C.; Bothwell, J.; Monsanto, D.M.; Vuuren, B.J.V.; Teske, P.R.; Paterson, A.M. De Novo Transcriptome Assembly and Annotation of Liver and Brain Tissues of Common Brushtail Possums (Trichosurusvulpecula) in New Zealand: Transcriptome Diversity after Decades of Population Control. Genes 2020, 11, 436. [Google Scholar] [CrossRef]
- Shanmuganandam, S.; Hu, Y.; Strive, T.; Schwessinger, B.; Hall, R.N. Uncovering the Microbiome of Invasive Sympatric European Brown Hares and European Rabbits in Australia. BioRxiv 2019, 832477. [Google Scholar] [CrossRef]
- Blanchong, J.A.; Robinson, S.J.; Samuel, M.D.; Foster, J.T. Application of Genetics and Genomics to Wildlife Epidemiology. J. Wildl. Manag. 2016, 80, 593–608. [Google Scholar] [CrossRef]
- Flecknell, P. Laboratory Animal Anaesthesia; Academic Press: New York NY, USA, 2015. [Google Scholar]
- Hall, L.; Clarke, K.; Trim, C. Veterinary Anaesthesia, 10th ed.; Harcourt Publishers Limited: London, UK, 2001; p. 225. [Google Scholar]
- Morgan, D.; Scobie, S.; Arthur, D. Evaluation of Zoletil and Other Injectable Anaesthetics for Field Sedation of Brushtail Possums (Trichosurusvulpecula). Anim. Welf. UFAW J. 2012, 21, 457. [Google Scholar] [CrossRef][Green Version]
- Emami-Khoyi, A.; Benmazouz, I.; Ross, J.G.; Boren, L.J.; Murphy, E.C.; Jansen van Vuuren, B.; Teske, P.R.; Paterson, A.M. A Survey of the Oral Cavity Microbiome of New Zealand Fur Seal Pups (Arctocephalus forsteri). Mar. Mamm. Sci. 2019, 36, 334–343. [Google Scholar] [CrossRef]
- Lane, D. 16s/23s Rrna Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991. [Google Scholar]
- Forney, L.J.; Gajer, P.; Williams, C.J.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Karlebach, S.; Brotman, R.M.; Davis, C.C.; Ault, K. Comparison of Self-Collected and Physician-Collected Vaginal Swabs for Microbiome Analysis. J. Clin. Microbiol. 2010, 48, 1741–1748. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Qiime 2: Reproducible, Interactive, Scalable, and Extensible Microbiome Data Science. PeerJ 2018, 2167–9843. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. Dada2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16s Rrna Gene Database and Workbench Compatible with Arb. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Letunic, I. Phylot: Phylogenetic Tree Generator. Available online: Phylot.biobyte.de (accessed on 31 May 2019).
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. Fasttree 2–Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation Evaluation and Phylogenetic Diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Pielou, E.C. The Measurement of Diversity in Different Types of Biological Collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Urbana, IL, USA, 1949; p. 117. [Google Scholar]
- Sørensen, T.J. A Method of Establishing Groups of Equal Amplitude in Plant Sociology Based on Similarity of Species Content and Its Application to Analyses of the Vegetation on Danish Commons; Kommission, I., Hos, E., Eds.; Munksgaard: Copenhagen, Denmark, 1948. [Google Scholar]
- Lozupone, C.; Knight, R. Unifrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and Qualitative Β Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Gower, J.C. Some Distance Properties of Latent Root and Vector Methods Used in Multivariate Analysis. Biometrika 1966, 53, 325–338. [Google Scholar] [CrossRef]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. Emperor: A Tool for Visualizing High-Throughput Microbial Community Data. Gigascience 2013, 2, 16. [Google Scholar] [CrossRef]
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (Permanova). In Wiley StatsRef: Statistics Reference Online; Wiley Online Library: Hoboken, NJ, USA, 2014; pp. 1–15. [Google Scholar]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. In Proceedings of the Third International AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Morton, J.T.; Sanders, J.; Quinn, R.A.; McDonald, D.; Gonzalez, A.; Vázquez-Baeza, Y.; Navas-Molina, J.A.; Song, S.J.; Metcalf, J.L.; Hyde, E.R. Balance Trees Reveal Microbial Niche Differentiation. MSystems 2017, 2, e00162-16. [Google Scholar] [CrossRef]
- Ward, J.H., Jr. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Edwards, Y.D.; Allenby, G.M. Multivariate Analysis of Multiple Response Data. J. Mark. Res. 2003, 40, 321–334. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G. Picrust2: An Improved and Extensible Approach for Metagenome Inference. BioRxiv 2019, 672295. [Google Scholar]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q. The Biocyc Collection of Microbial Genomes and Metabolic Pathways. Brief. Bioinform. 2017, 20, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. Microbiomeanalyst: A Web-Based Tool for Comprehensive Statistical, Visual and Meta-Analysis of Microbiome Data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Gao, L.; Xu, T.; Huang, G.; Jiang, S.; Gu, Y.; Chen, F. Oral Microbiomes: More and More Importance in Oral Cavity and Whole Body. Protein Cell 2018, 9, 488–500. [Google Scholar] [CrossRef] [PubMed]
- Adler, C.J.; Malik, R.; Browne, G.V.; Norris, J.M. Diet May Influence the Oral Microbiome Composition in Cats. Microbiome 2016, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Kato, I.; Vasquez, A.; Moyerbrailean, G.; Land, S.; Djuric, Z.; Sun, J.; Lin, H.-S.; Ram, J.L. Nutritional Correlates of Human Oral Microbiome. J. Am. Coll. Nutr. 2017, 36, 88–98. [Google Scholar] [CrossRef]
- Gogarten, J.F.; Davies, T.J.; Benjamino, J.; Gogarten, J.P.; Graf, J.; Mielke, A.; Mundry, R.; Nelson, M.C.; Wittig, R.M.; Leendertz, F.H. Factors Influencing Bacterial Microbiome Composition in a Wild Non-Human Primate Community in Taï National Park, Côte D’ivoire. ISME J. 2018, 12, 2559–2574. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.A.; Azcarate-Peril, M.A.; Cadenas, M.B.; Butz, N.; Paster, B.J.; Chen, T.; Bair, E.; Arnold, R.R. The Oral Bacterial Microbiome of Occlusal Surfaces in Children and Its Association with Diet and Caries. PLoS ONE 2017, 12, e0180621. [Google Scholar] [CrossRef] [PubMed]
- Ruparell, A.; Inui, T.; Staunton, R.; Wallis, C.; Deusch, O.; Holcombe, L.J. The Canine Oral Microbiome: Variation in Bacterial Populations across Different Niches. BMC Microbiol. 2020, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hume, I.D. Digestive Physiology and Nutrition of Marsupials; CUP Archive: New York, NY, USA, 1982. [Google Scholar]
- Delafont, V.; Samba-Louaka, A.; Bouchon, D.; Moulin, L.; Héchard, Y. Shedding Light on Microbial Dark Matter: A Tm 6 Bacterium as Natural Endosymbiont of a Free-Living Amoeba. Environ. Microbiol. Rep. 2015, 7, 970–978. [Google Scholar] [CrossRef]
- Khan, I.; Azhar, E.I.; Abbas, A.T.; Kumosani, T.; Barbour, E.K.; Raoult, D.; Yasir, M. Metagenomic Analysis of Antibiotic-Induced Changes in Gut Microbiota in a Pregnant Rat Model. Front. Pharmacol. 2016, 7, 104. [Google Scholar] [CrossRef]
- Wade, W.G. Has the Use of Molecular Methods for the Characterization of the Human Oral Microbiome Changed Our Understanding of the Role of Bacteria in the Pathogenesis of Periodontal Disease? J. Clin. Periodontol. 2011, 38, 7–16. [Google Scholar] [CrossRef]
- Chang, C.-W.; Huang, B.-H.; Lin, S.-M.; Huang, C.-L.; Liao, P.-C. Changes of Diet and Dominant Intestinal Microbes in Farmland Frogs. BMC Microbiol. 2016, 16, 33. [Google Scholar] [CrossRef]
- Ulloa, P.C.; Van Der Veen, M.H.; Krom, B.P. Modulation of the Oral Microbiome by the Host to Promote Ecological Balance. Odontology 2019, 107, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Pearce, D.S. Physiology and Genetics Shape the Microbiome of a Seabird Species (Oceanodromaleucorhoa) More than Environmental and Social Factors. Master’s Thesis, Western Michigan University, Kalamazoo, MI, USA, 2016. [Google Scholar]
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The Oral Microbiota: Dynamic Communities and Host Interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Compo, N.R.; Gomez, D.E.; Tapscott, B.; Weese, J.S.; Turner, P.V. Fecal Bacterial Microbiota of Canadian Commercial Mink (Neovison vison): Yearly, Life Stage, and Seasonal Comparisons. PLoS ONE 2018, 13, e0207111. [Google Scholar] [CrossRef] [PubMed]
- Blekhman, R.; Goodrich, J.K.; Huang, K.; Sun, Q.; Bukowski, R.; Bell, J.T.; Spector, T.D.; Keinan, A.; Ley, R.E.; Gevers, D. Host Genetic Variation Impacts Microbiome Composition across Human Body Sites. Genome Biol. 2015, 16, 191. [Google Scholar] [CrossRef] [PubMed]
- Davenport, E.R.; Cusanovich, D.A.; Michelini, K.; Barreiro, L.B.; Ober, C.; Gilad, Y. Genome-Wide Association Studies of the Human Gut Microbiota. PLoS ONE 2015, 10, e0140301. [Google Scholar] [CrossRef] [PubMed]
- Folseraas, T.; Melum, E.; Rausch, P.; Juran, B.D.; Ellinghaus, E.; Shiryaev, A.; Laerdahl, J.K.; Ellinghaus, D.; Schramm, C.; Weismüller, T.J. Extended Analysis of a Genome-Wide Association Study in Primary Sclerosing Cholangitis Detects Multiple Novel Risk Loci. J. Hepatol. 2012, 57, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Davenport, E.R.; Beaumont, M.; Jackson, M.A.; Knight, R.; Ober, C.; Spector, T.D.; Bell, J.T.; Clark, A.G.; Ley, R.E. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016, 19, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T. Human Genetics Shape the Gut Microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef]
- Knights, D.; Silverberg, M.S.; Weersma, R.K.; Gevers, D.; Dijkstra, G.; Huang, H.; Tyler, A.D.; Van Sommeren, S.; Imhann, F.; Stempak, J.M. Complex Host Genetics Influence the Microbiome in Inflammatory Bowel Disease. Genome Med. 2014, 6, 107. [Google Scholar] [CrossRef]
- Suzuki, T.A.; Phifer-Rixey, M.; Mack, K.L.; Sheehan, M.J.; Lin, D.; Bi, K.; Nachman, M.W. Host Genetic Determinants of the Gut Microbiota of Wild Mice. Mol. Ecol. 2019, 28, 3197–3207. [Google Scholar] [CrossRef]
- Awany, D.; Allali, I.; Dalvie, S.; Hemmings, S.; Mwaikono, K.S.; Thomford, N.E.; Gomez, A.; Mulder, N.; Chimusa, E.R. Host and Microbiome Genome-Wide Association Studies: Current State and Challenges. Front. Genet. 2019, 9, 637. [Google Scholar] [CrossRef]
- Beilsmith, K.; Thoen, M.P.; Brachi, B.; Gloss, A.D.; Khan, M.H.; Bergelson, J. Genome-Wide Association Studies on the Phyllosphere Microbiome: Embracing Complexity in Host–Microbe Interactions. Plant J. 2019, 97, 164–181. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, Stability and Resilience of the Human Gut Microbiota. Nature 2012, 489, 220. [Google Scholar] [CrossRef]
- Taxis, T.M.; Wolff, S.; Gregg, S.J.; Minton, N.O.; Zhang, C.; Dai, J.; Schnabel, R.D.; Taylor, J.F.; Kerley, M.S.; Pires, J.C. The Players May Change but the Game Remains: Network Analyses of Ruminal Microbiomes Suggest Taxonomic Differences Mask Functional Similarity. Nucleic Acids Res. 2015, 43, 9600–9612. [Google Scholar] [CrossRef] [PubMed]
- Bradley, P.H.; Pollard, K.S. Proteobacteria Explain Significant Functional Variability in the Human Gut Microbiome. Microbiome 2017, 5, 36. [Google Scholar] [CrossRef] [PubMed]
- Sharpton, T.J. Role of the Gut Microbiome in Vertebrate Evolution. MSystems 2018, 3, e00174-17. [Google Scholar] [CrossRef] [PubMed]
- Camboim, E.K.; Almeida, A.P.; Tadra-Sfeir, M.Z.; Junior, F.G.; Andrade, P.P.; McSweeney, C.S.; Melo, M.A.; Riet-Correa, F. Isolation and Identification of Sodium Fluoroacetate Degrading Bacteria from Caprine Rumen in Brazil. Sci. World J. 2012, 2012, 178254. [Google Scholar] [CrossRef] [PubMed][Green Version]
| Phylum | Family | Possum Specimens | Stoat Specimens | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | d | e | a | b | c | d | e | ||
| Actinobacteria | Acidobacteriaceae | 2 | 0 | 0 | 9 | 6 | 0 | 0 | 0 | 0 | 0 |
| C111 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | |
| Actinomycetaceae | 6671 | 4399 | 6709 | 0 | 0 | 890 | 324 | 1155 | 238 | 4265 | |
| Brevibacteriaceae | 5 | 7 | 0 | 11 | 7 | 0 | 0 | 0 | 0 | 0 | |
| Cellulomonadaceae | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 4 | |
| Corynebacteriaceae | 226 | 949 | 397 | 50 | 23 | 150 | 604 | 392 | 26 | 79 | |
| Dermabacteraceae | 0 | 0 | 0 | 9 | 13 | 0 | 0 | 0 | 0 | 11 | |
| Dermacoccaceae | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Dietziaceae | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Geodermatophilaceae | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | |
| Intrasporangiaceae | 0 | 0 | 0 | 9 | 37 | 0 | 0 | 0 | 0 | 0 | |
| Kineosporiaceae | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 0 | |
| Microbacteriaceae | 0 | 0 | 0 | 9 | 93 | 744 | 4 | 26 | 43 | 202 | |
| Micrococcaceae | 26 | 18 | 0 | 5 | 36 | 0 | 65 | 90 | 1855 | 12 | |
| Nakamurellaceae | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Nocardioidaceae | 0 | 0 | 0 | 16 | 36 | 0 | 0 | 0 | 0 | 0 | |
| Propionibacteriaceae | 6064 | 30 | 8846 | 123 | 42 | 113 | 6 | 257 | 120 | 115 | |
| Pseudonocardiaceae | 0 | 0 | 0 | 3 | 17 | 0 | 0 | 0 | 0 | 0 | |
| Sporichthyaceae | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | |
| Streptomycetaceae | 0 | 0 | 0 | 4 | 29 | 0 | 0 | 3 | 0 | 0 | |
| Williamsiaceae | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Coriobacteriaceae | 0 | 2 | 9 | 0 | 6 | 0 | 9 | 0 | 2 | 0 | |
| Euzebyaceae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | |
| Gaiellaceae | 0 | 0 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | |
| Patulibacteraceae | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | |
| Solirubrobacteraceae | 0 | 0 | 0 | 7 | 11 | 0 | 0 | 0 | 0 | 0 | |
| Bacteroidetes | Bacteroidaceae | 31 | 9 | 5 | 18 | 4 | 6 | 0 | 3 | 165 | 0 |
| Porphyromonadaceae | 73 | 1286 | 42 | 29 | 20 | 3092 | 2093 | 466 | 482 | 5361 | |
| Prevotellaceae | 38 | 63 | 53 | 35 | 91 | 156 | 572 | 62 | 157 | 494 | |
| S24-7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 7 | 4 | |
| Barnesiellaceae | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | |
| Paraprevotellaceae | 3 | 9 | 0 | 0 | 7 | 4 | 0 | 0 | 6 | 0 | |
| Cyclobacteriaceae | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cytophagaceae | 4 | 0 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 0 | |
| Flavobacteriaceae | 4887 | 23,828 | 17,570 | 2099 | 17 | 2238 | 2809 | 937 | 1856 | 5121 | |
| Weeksellaceae | 11,660 | 7945 | 2085 | 4249 | 14,979 | 405 | 945 | 477 | 111 | 1068 | |
| Sphingobacteriaceae | 7 | 0 | 0 | 15 | 58 | 0 | 4 | 0 | 0 | 0 | |
| Chitinophagaceae | 0 | 0 | 0 | 9 | 21 | 0 | 0 | 0 | 0 | 4 | |
| Chloroflexi | Dolo_23 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| Firmicutes | Bacillaceae | 13 | 4 | 6 | 5 | 12 | 5 | 0 | 0 | 0 | 0 |
| Paenibacillaceae | 0 | 0 | 0 | 0 | 0 | 5575 | 1657 | 0 | 2645 | 61 | |
| Planococcaceae | 0 | 0 | 2 | 14 | 8 | 0 | 0 | 6 | 0 | 0 | |
| Staphylococcaceae | 86 | 7 | 0 | 246 | 300 | 0 | 3 | 3 | 0 | 3 | |
| Exiguobacteraceae | 0 | 4 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | |
| Thermicanaceae | 5 | 0 | 0 | 4 | 9 | 0 | 0 | 0 | 0 | 0 | |
| Gemellaceae | 10,310 | 1821 | 1049 | 12,727 | 15,233 | 3438 | 829 | 84 | 2015 | 0 | |
| Aerococcaceae | 429 | 4295 | 218 | 1718 | 747 | 1226 | 4145 | 113 | 1258 | 1590 | |
| Carnobacteriaceae | 10 | 11 | 5 | 2 | 46 | 80 | 7 | 11 | 130 | 98 | |
| Enterococcaceae | 7 | 0 | 5 | 0 | 8 | 0 | 7 | 0 | 6 | 5 | |
| Lactobacillaceae | 4 | 4 | 0 | 0 | 28 | 0 | 2 | 14 | 12 | 0 | |
| Leuconostocaceae | 0 | 3 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | |
| Streptococcaceae | 51 | 840 | 132 | 3196 | 12,656 | 26,169 | 19,442 | 14,569 | 32,630 | 11,017 | |
| Turicibacteraceae | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 14 | 0 | |
| Clostridiaceae | 170 | 13 | 52 | 458 | 190 | 0 | 0 | 7 | 67 | 13 | |
| Lachnospiraceae | 24 | 31 | 22 | 2 | 41 | 511 | 1153 | 175 | 814 | 191 | |
| Peptococcaceae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 4 | |
| Peptostreptococcaceae | 37 | 6 | 0 | 166 | 53 | 0 | 28 | 26 | 0 | 2 | |
| Ruminococcaceae | 20 | 12 | 6 | 7 | 42 | 4 | 5 | 0 | 0 | 34 | |
| Veillonellaceae | 45 | 62 | 57 | 19 | 35 | 20 | 1666 | 97 | 454 | 57 | |
| Acidaminobacteraceae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 73 | 0 | 12 | |
| Mogibacteriaceae | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 3 | |
| Tissierellaceae | 45 | 0 | 7 | 9 | 6 | 4 | 0 | 15 | 0 | 8 | |
| Erysipelotrichaceae | 0 | 1622 | 6 | 0 | 11 | 15 | 11 | 23 | 145 | 50 | |
| Fusobacteria | Fusobacteriaceae | 21 | 20 | 19 | 54 | 66 | 5151 | 17,078 | 1478 | 7176 | 15,051 |
| Leptotrichiaceae | 784 | 26 | 7 | 0 | 55 | 3050 | 28,266 | 2224 | 7229 | 17,960 | |
| Proteobacteria | Caulobacteraceae | 6 | 0 | 0 | 5 | 10 | 0 | 0 | 0 | 0 | 0 |
| Bradyrhizobiaceae | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Brucellaceae | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | |
| Hyphomicrobiaceae | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | |
| Methylobacteriaceae | 0 | 0 | 0 | 14 | 72 | 0 | 0 | 0 | 0 | 0 | |
| Methylocystaceae | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | |
| Rhizobiaceae | 0 | 11 | 0 | 14 | 30 | 0 | 0 | 0 | 0 | 0 | |
| Rhodobacteraceae | 0 | 0 | 0 | 3 | 11 | 0 | 0 | 0 | 0 | 4 | |
| Acetobacteraceae | 5 | 0 | 2 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | |
| Rhodospirillaceae | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | |
| mitochondria | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 2 | 0 | |
| Erythrobacteraceae | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | |
| Sphingomonadaceae | 6 | 0 | 0 | 4 | 91 | 6 | 0 | 0 | 0 | 0 | |
| Alcaligenaceae | 4 | 0 | 0 | 0 | 0 | 3 | 0 | 2 | 23 | 4 | |
| Burkholderiaceae | 6992 | 29,794 | 1774 | 1606 | 66 | 1099 | 239 | 208 | 86 | 83 | |
| Comamonadaceae | 2032 | 39 | 1616 | 7 | 56 | 99 | 0 | 40 | 14 | 158 | |
| Oxalobacteraceae | 0 | 4 | 0 | 10 | 76 | 0 | 0 | 4 | 0 | 0 | |
| Methylophilaceae | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Neisseriaceae | 11,609 | 28,742 | 19,824 | 10,454 | 35,160 | 25,358 | 10,979 | 22,580 | 43,075 | 4938 | |
| Rhodocyclaceae | 9 | 0 | 0 | 7 | 6 | 8 | 0 | 0 | 6 | 0 | |
| Bdellovibrionaceae | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | |
| Polyangiaceae | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Syntrophobacteraceae | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | |
| Campylobacteraceae | 3 | 0 | 0 | 0 | 11 | 71 | 54 | 19 | 29 | 55 | |
| Helicobacteraceae | 71 | 4 | 95 | 128 | 3686 | 0 | 0 | 0 | 0 | 0 | |
| Chromatiaceae | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cardiobacteriaceae | 3223 | 3521 | 6280 | 2 | 0 | 32 | 0 | 185 | 16 | 387 | |
| Enterobacteriaceae | 41 | 40 | 24 | 35 | 140 | 0 | 0 | 0 | 0 | 0 | |
| Halomonadaceae | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Pasteurellaceae | 6193 | 9601 | 9533 | 5981 | 20,350 | 56,748 | 27,617 | 48,242 | 43,199 | 11,382 | |
| Moraxellaceae | 8 | 0 | 0 | 66 | 281 | 1709 | 2623 | 588 | 298 | 59 | |
| Pseudomonadaceae | 5 | 13 | 6 | 13 | 26 | 0 | 0 | 0 | 0 | 0 | |
| Vibrionaceae | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 2 | 0 | 0 | |
| Sinobacteraceae | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | |
| Xanthomonadaceae | 16 | 3 | 0 | 26 | 77 | 2 | 0 | 0 | 0 | 0 | |
| Spirochaetes | Spirochaetaceae | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 36 | 0 | 3 |
| TM7 | F16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 |
| Tenericutes | Acholeplasmataceae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 51 | 0 | 0 |
| Anaeroplasmataceae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | |
| Mycoplasmataceae | 27,690 | 2951 | 13,142 | 4572 | 4017 | 0 | 4 | 14 | 0 | 0 | |
| Thermotogae | Thermotogaceae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| Thermi | Trueperaceae | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Thermaceae | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Pathway | Possums | Stoats | ||
|---|---|---|---|---|
| Total | p-Values | Total | p-Values | |
| Lipopolysaccharide biosynthesis | 17 | 0 | 17 | 0 |
| Biosynthesis of amino acids | 222 | <0.001 | 222 | <0.001 |
| Peptidoglycan biosynthesis | 13 | <0.001 | 13 | <0.001 |
| Terpenoid backbone biosynthesis | 23 | <0.001 | 23 | <0.001 |
| Streptomycin biosynthesis | 12 | <0.001 | 12 | <0.001 |
| Polyketide sugar unit biosynthesis | 4 | <0.05 | 4 | <0.05 |
| Valine, leucine and isoleucine biosynthesis | 15 | <0.05 | - | - |
| Folate biosynthesis | 29 | <0.05 | - | - |
| Porphyrin and chlorophyll metabolism | 69 | <0.001 | 69 | <0.001 |
| Alanine, aspartate and glutamate metabolism | 62 | <0.001 | 62 | <0.001 |
| Arginine and proline metabolism | 115 | <0.01 | 115 | <0.01 |
| Cysteine and methionine metabolism | 71 | <0.01 | 71 | <0.01 |
| Glycine, serine and threonine metabolism | 78 | <0.01 | 78 | <0.01 |
| D-Glutamine and D-glutamate metabolism | 6 | <0.01 | 6 | <0.01 |
| Thiamine metabolism | 23 | <0.01 | 23 | <0.01 |
| Starch and sucrose metabolism | 65 | <0.01 | 65 | <0.05 |
| Glyoxylate and dicarboxylate metabolism | 51 | <0.01 | 51 | <0.05 |
| Biotin metabolism | 19 | <0.05 | 19 | <0.05 |
| Riboflavin metabolism | 22 | <0.05 | 22 | <0.05 |
| Amino sugar and nucleotide sugar metabolism | 64 | <0.05 | 64 | <0.05 |
| Carbon metabolism | 249 | <0.05 | 249 | <0.05 |
| Butanoate metabolism | 61 | <0.05 | - | - |
| Nicotinate and nicotinamide metabolism | 36 | <0.05 | - | - |
| beta-Alanine metabolism | - | - | 36 | <0.05 |
| Vitamin B6 metabolism | - | - | 12 | <0.05 |
| Phenylalanine metabolism | 41 | <0.05 | 41 | <0.01 |
| Xylene degradation | 20 | <0.01 | 20 | <0.01 |
| Steroid degradation | 9 | <0.05 | 9 | <0.05 |
| Lysine degradation | - | - | 30 | <0.05 |
| Synthesis and degradation of ketone bodies | 5 | <0.05 | 5 | <0.05 |
| Pentose and glucuronate interconversions | 41 | <0.05 | 41 | <0,01 |
| Carbon fixation in photosynthetic organisms | 35 | <0.05 | 35 | <0.05 |
| Pentose phosphate pathway | - | - | 63 | <0.01 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Emami-Khoyi, A.; Benmazouz, I.; Paterson, A.M.; Ross, J.G.; Murphy, E.C.; Bothwell, J.; Alizadeh, H.; van Vuuren, B.J.; Teske, P.R. Oral Microbiome Metabarcoding in Two Invasive Small Mammals from New Zealand. Diversity 2020, 12, 278. https://doi.org/10.3390/d12070278
Emami-Khoyi A, Benmazouz I, Paterson AM, Ross JG, Murphy EC, Bothwell J, Alizadeh H, van Vuuren BJ, Teske PR. Oral Microbiome Metabarcoding in Two Invasive Small Mammals from New Zealand. Diversity. 2020; 12(7):278. https://doi.org/10.3390/d12070278
Chicago/Turabian StyleEmami-Khoyi, Arsalan, Isma Benmazouz, Adrian M. Paterson, James G. Ross, Elaine C. Murphy, Jennifer Bothwell, Hossein Alizadeh, Bettine Jansen van Vuuren, and Peter R. Teske. 2020. "Oral Microbiome Metabarcoding in Two Invasive Small Mammals from New Zealand" Diversity 12, no. 7: 278. https://doi.org/10.3390/d12070278
APA StyleEmami-Khoyi, A., Benmazouz, I., Paterson, A. M., Ross, J. G., Murphy, E. C., Bothwell, J., Alizadeh, H., van Vuuren, B. J., & Teske, P. R. (2020). Oral Microbiome Metabarcoding in Two Invasive Small Mammals from New Zealand. Diversity, 12(7), 278. https://doi.org/10.3390/d12070278





