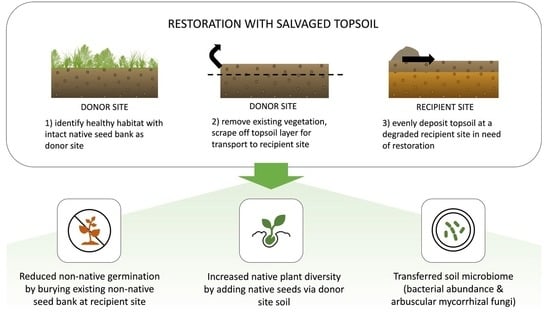
Identifying Mechanisms for Successful Ecological Restoration with Salvaged Topsoil in Coastal Sage Scrub Communities
Abstract
1. Introduction
2. Materials and Methods
2.1. Study System
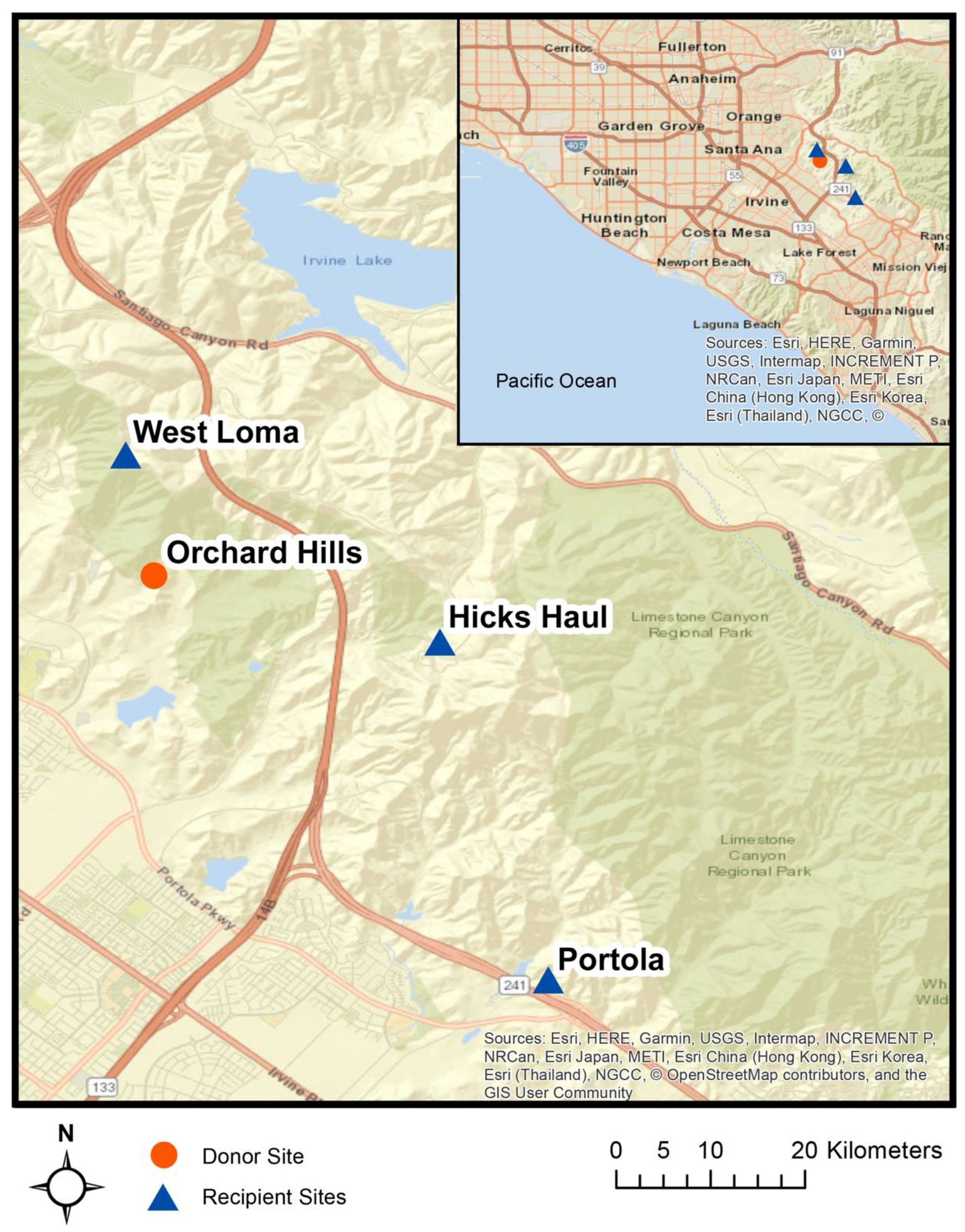
2.2. Site Selection
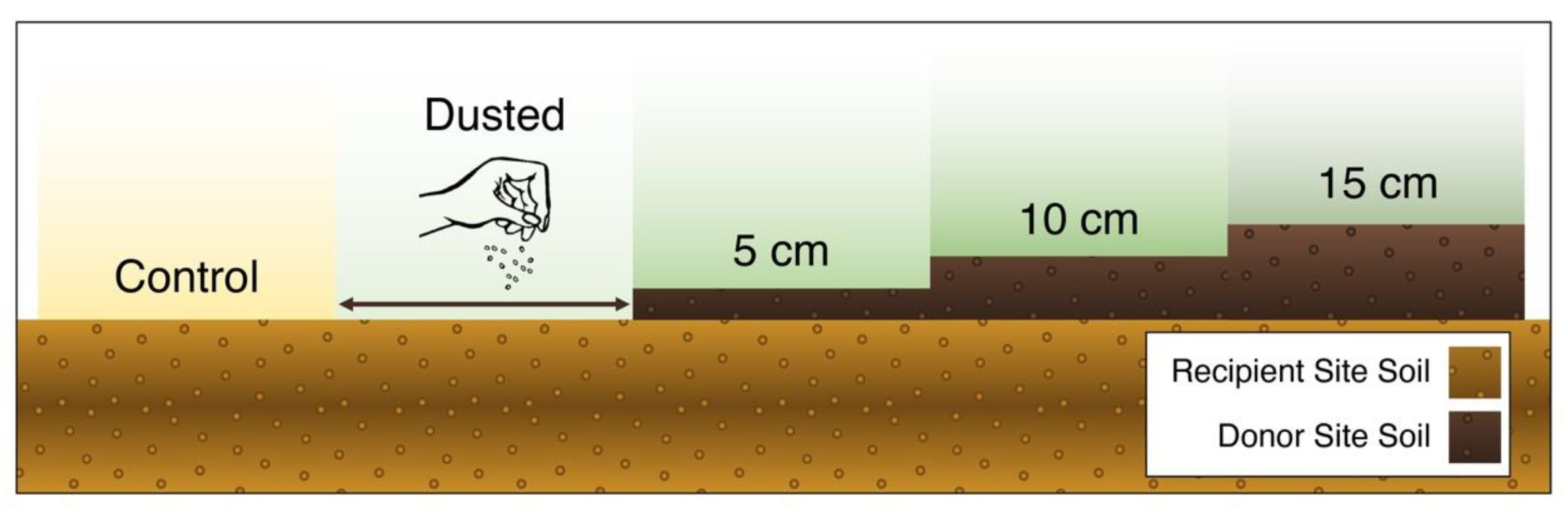
2.3. Experimental design
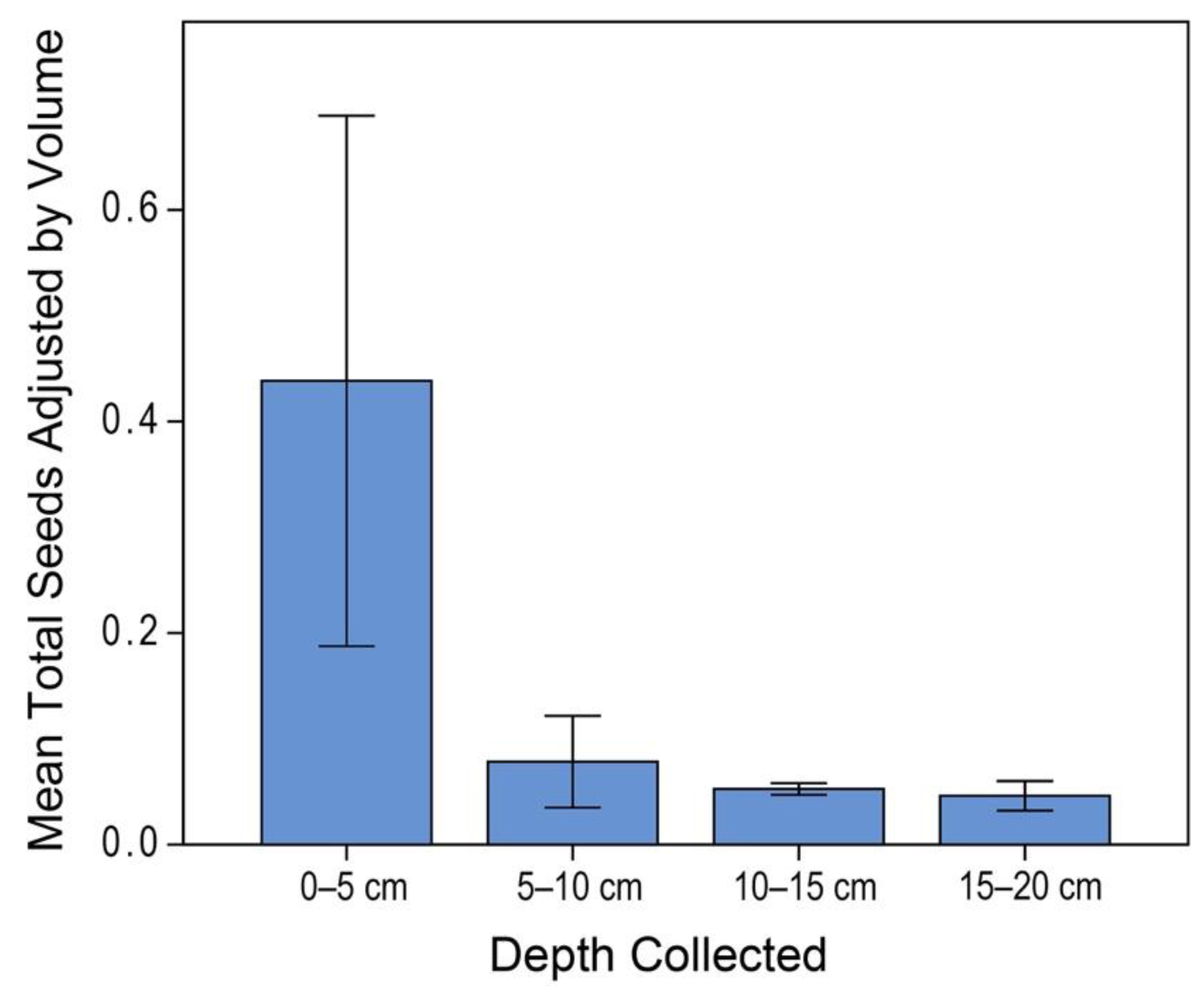
2.4. Donor Site Seed-Bank Characterization
2.5. Plant Communities Data Collection
2.6. Soil Microbes
2.6.1. Bacterial Abundance
2.6.2. Arbuscular Mycorrhizal Fungi
2.7. Fungal Library Construction and Sequencing
2.8. Bioinformatics
2.9. Statistical Analyses
3. Results
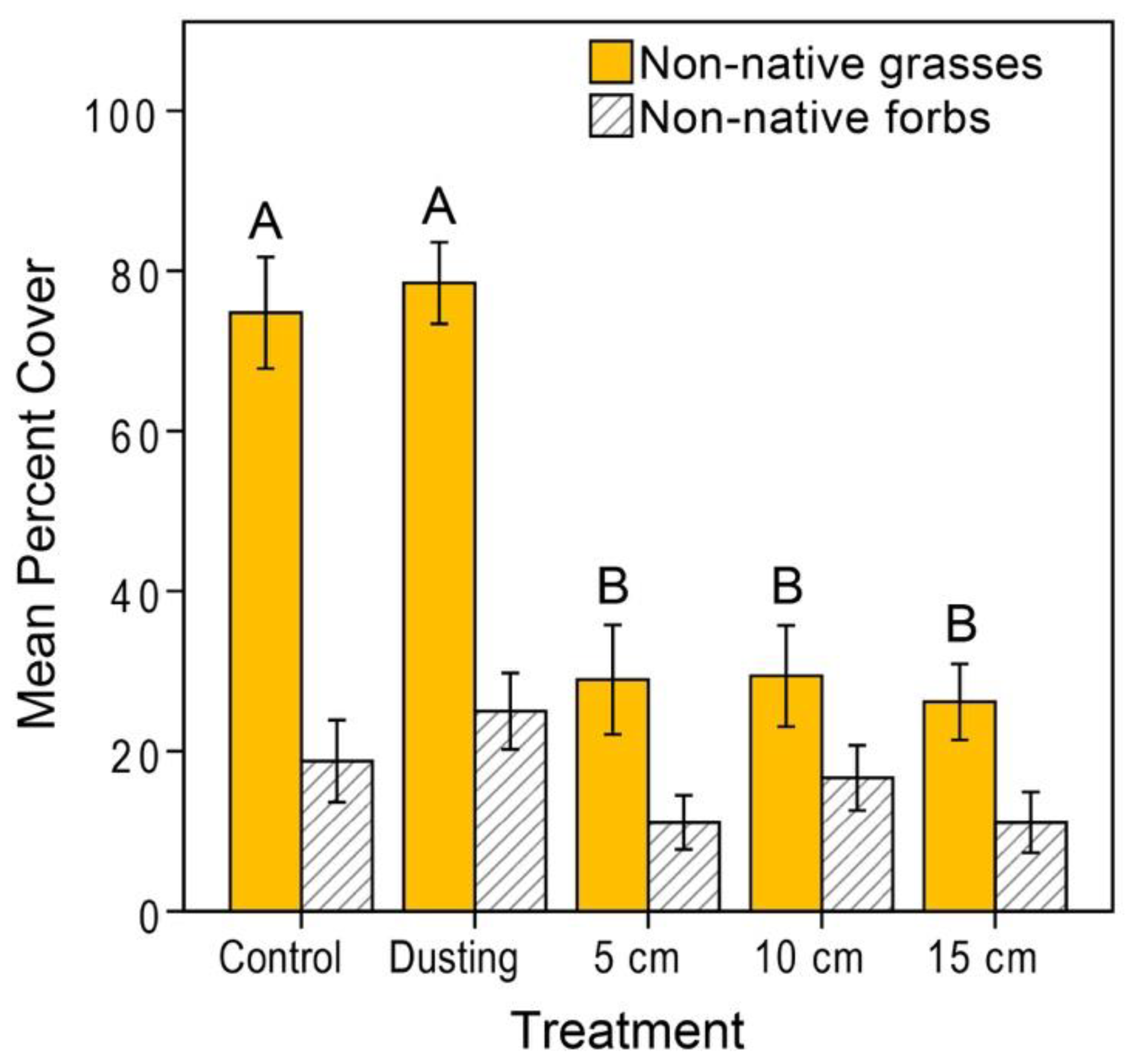
3.1. Donor Site Seed Bank
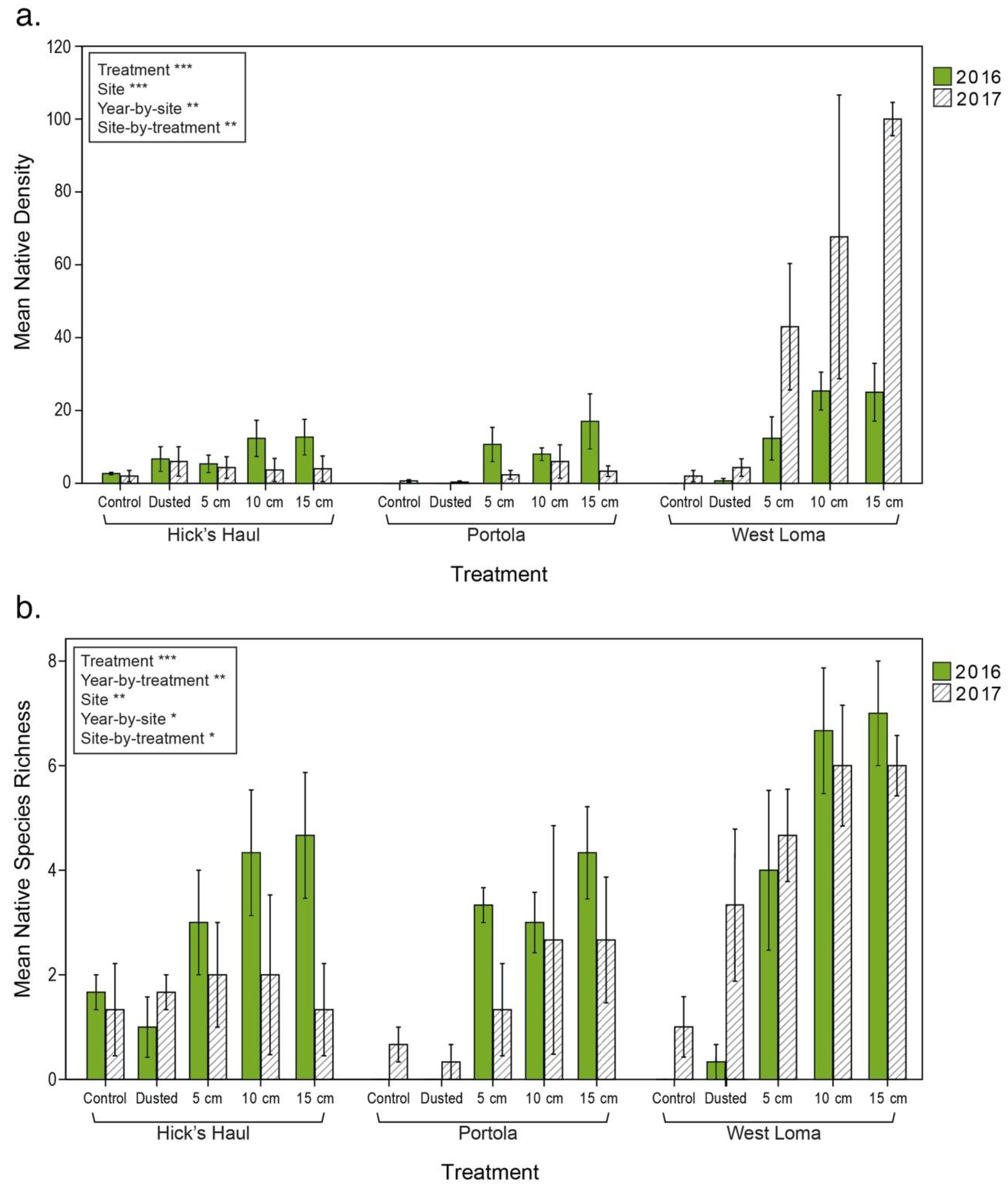
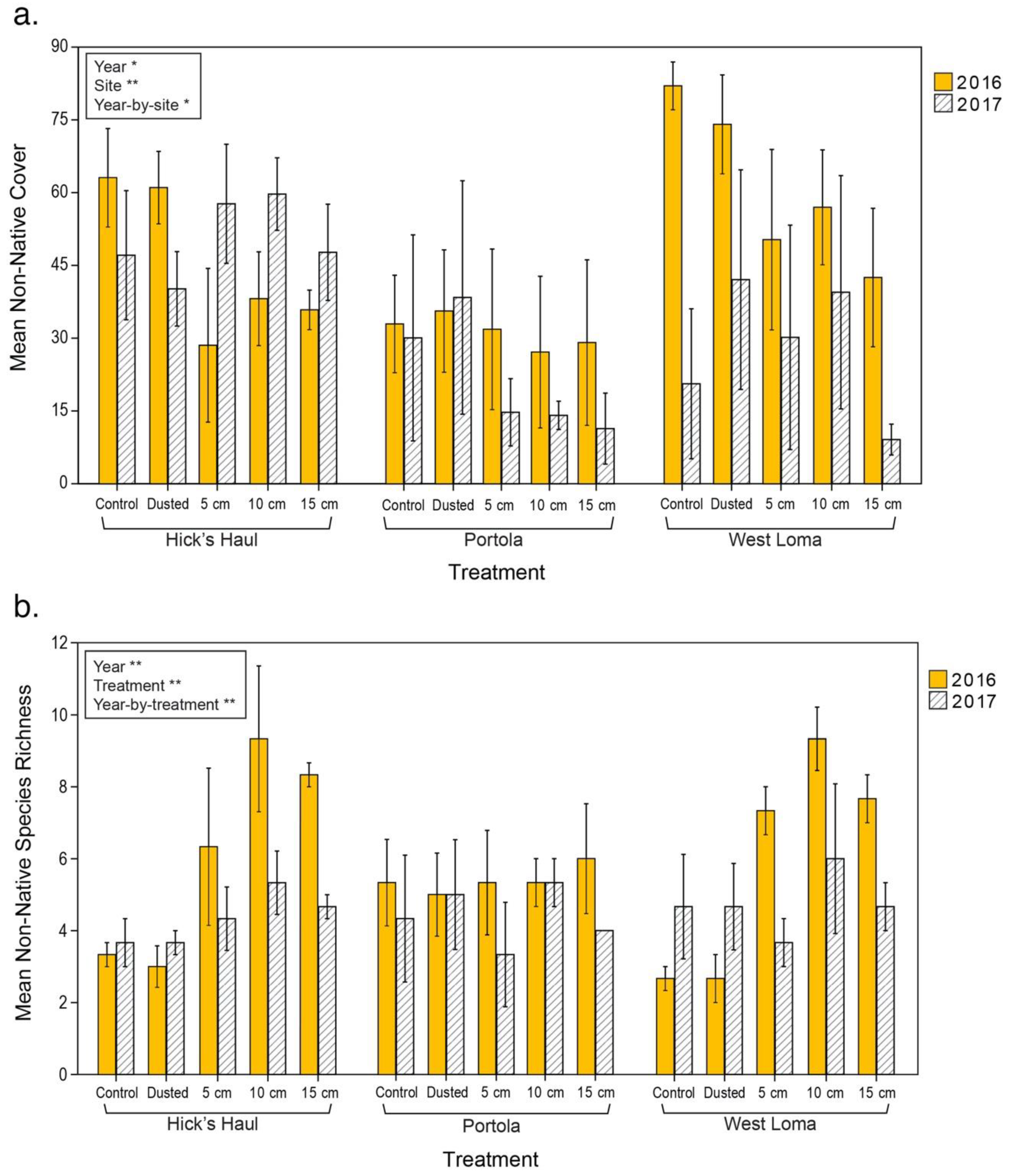
3.2. Vegetation Cover and Species Richness at the Recipient Sites
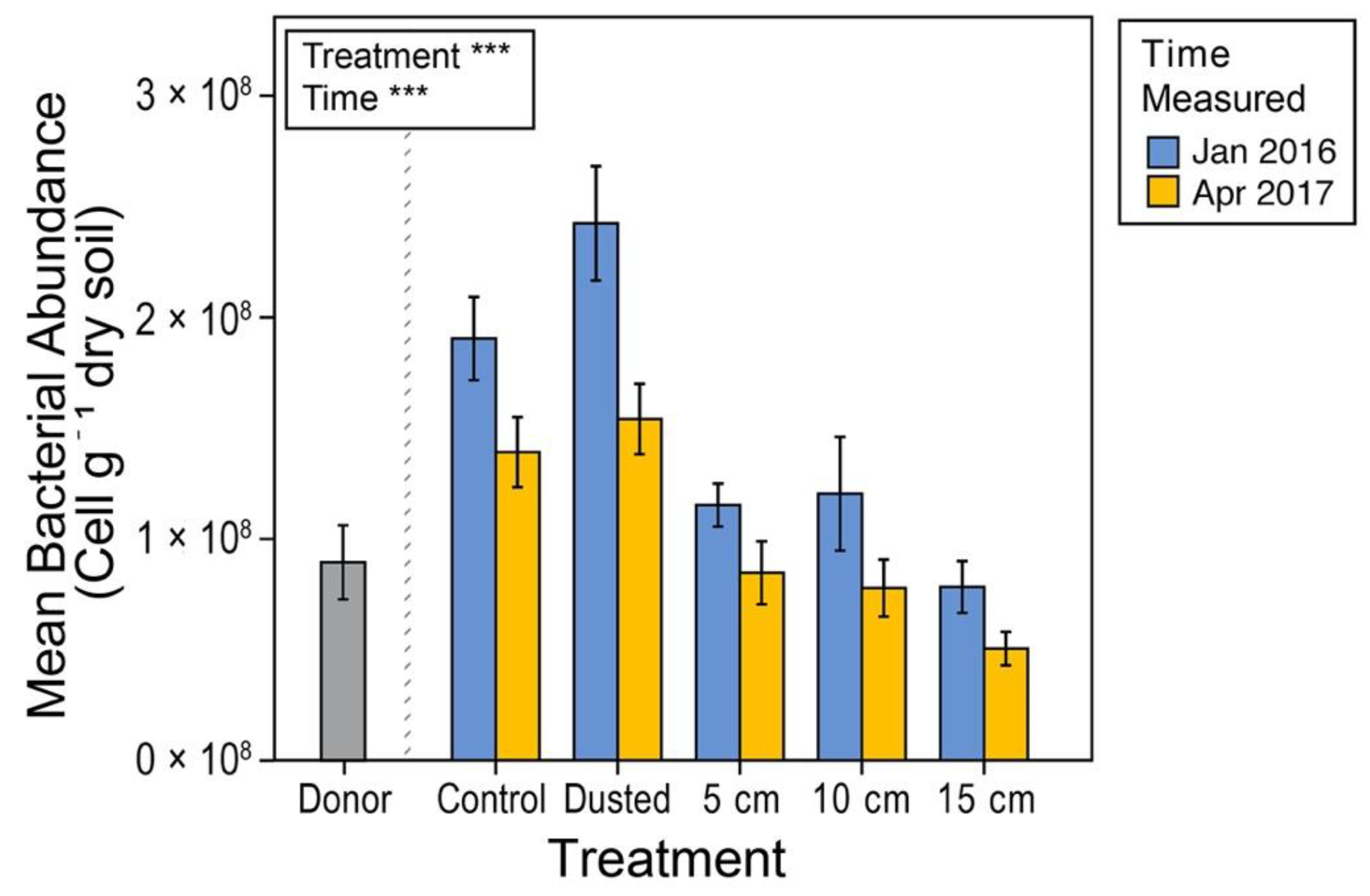
3.3. Soil Microbial Content
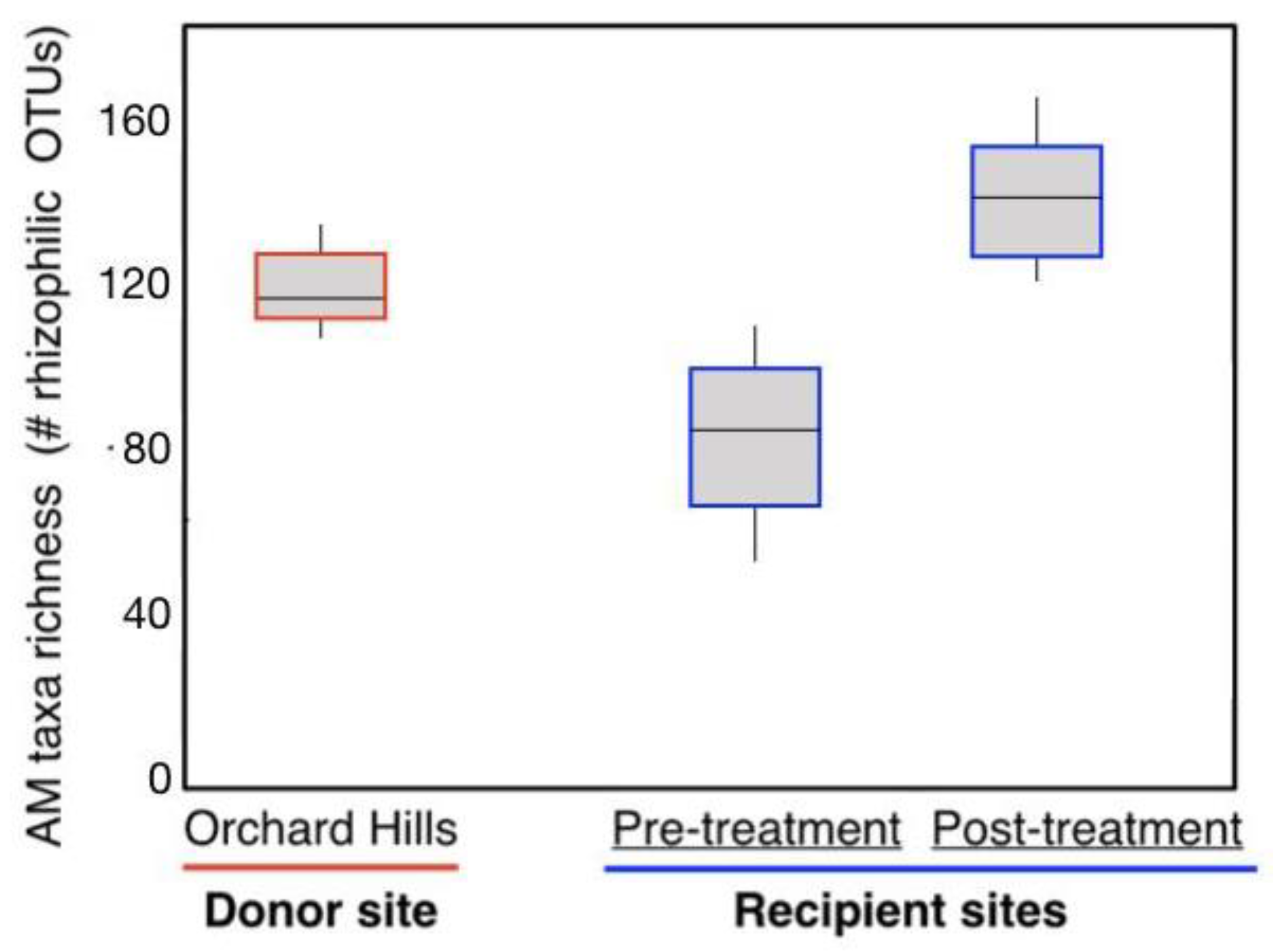
3.4. Arbuscular Mycorrhizal Fungal Communities
4. Discussion
4.1. Response of the Plant Community
4.1.1. Reduction of Competition with Non-Natives via Burial of Invasive Seeds
4.1.2. Addition of Native Plants via Establishment of a Native Seed Bank
4.2. Transference of the Soil Microbiome
4.3. Methodological Implications
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jackson, L.L.; Lopoukhine, N.; Hillyard, D. Ecological Restoration: A Definition and Comments. Restor. Ecol. 1995, 3, 71–75. [Google Scholar] [CrossRef]
- Society for Ecological Restoration International Science & Policy Working Group. The SER International Primer on Ecological Restoration; Society for Ecological Restoration: Washington, DC, USA, 2004. [Google Scholar]
- Lesica, P.; Allendorf, F.W. Ecological Genetics and the Restoration of Plant Communities: Mix or Match? Restor. Ecol. 1999, 7, 42–50. [Google Scholar] [CrossRef]
- McKay, J.K.; Christian, C.E.; Harrison, S.; Rice, K.J. “How Local Is Local?”—A Review of Practical and Conceptual Issues in the Genetics of Restoration. Restor. Ecol. 2005, 13, 432–440. [Google Scholar] [CrossRef]
- Gustafson, D.J.; Halfacre, A.C.; Anderson, R.C. Practical Seed Source Selection for Restoration Projects in an Urban Setting: Tallgrass Prairie, Serpentine Barrens, and Coastal Habitat Examples. Urban Habitats 2008, 5, 18. [Google Scholar]
- Jordan, W.R. The Tallgrass Restoration Handbook: For Prairies, Savannas, and Woodlands; Island Press: Washington, DC, USA, 1997; ISBN 978-1-59726-258-3. [Google Scholar]
- Kimball, S.; Lulow, M.; Sorenson, Q.; Balazs, K.; Fang, Y.-C.; Davis, S.J.; O’Connell, M.; Huxman, T.E. Cost-effective ecological restoration. Restor. Ecol. 2015, 23, 800–810. [Google Scholar] [CrossRef]
- Wardle, D.A.; Bardgett, R.D.; Klironomos, J.N.; Setälä, H.; van der Putten, W.H.; Wall, D.H. Ecological Linkages Between Aboveground and Belowground Biota. Science 2004, 304, 1629–1633. [Google Scholar] [CrossRef]
- Lekberg, Y.; Koide, R.T. Is plant performance limited by abundance of arbuscular mycorrhizal fungi? A meta-analysis of studies published between 1988 and 2003. New Phytol. 2005, 168, 189–204. [Google Scholar] [CrossRef]
- Hoeksema, J.D.; Chaudhary, V.B.; Gehring, C.A.; Johnson, N.C.; Karst, J.; Koide, R.T.; Pringle, A.; Zabinski, C.; Bever, J.D.; Moore, J.C.; et al. A meta-analysis of context-dependency in plant response to inoculation with mycorrhizal fungi. Ecol. Lett. 2010, 13, 394–407. [Google Scholar] [CrossRef]
- van der Putten, W.H.; Bardgett, R.D.; Bever, J.D.; Bezemer, T.M.; Casper, B.B.; Fukami, T.; Kardol, P.; Klironomos, J.N.; Kulmatiski, A.; Schweitzer, J.A.; et al. Plant–soil feedbacks: The past, the present and future challenges. J. Ecol. 2013, 101, 265–276. [Google Scholar] [CrossRef]
- Phillips, M.L.; Aronson, E.L.; Maltz, M.R.; Allen, E.B. Native and invasive inoculation sources modify fungal community assembly and biomass production of a chaparral shrub. Appl. Soil Ecol. 2020, 147, 103370. [Google Scholar] [CrossRef]
- Mosse, B. Growth and Chemical Composition of Mycorrhizal and Non-mycorrhizal Apples. Nature 1957, 179, 922–924. [Google Scholar] [CrossRef]
- Allen, M.F.; Smith, W.K.; Moore, T.S.J.; Christensen, M. Comparative Water Relations and Photosynthesis of Mycorrhizal and Non-Mycorrhizal Bouteloua Gracilis H.b.k. Lag Ex Steud. New Phytol. 1981, 88, 683–693. [Google Scholar] [CrossRef]
- McGonigle, T.P.; Fitter, A.H. Growth and phosphorus inflows of Trifolium repens L. with a range of indigenous vesicular-arbuscular mycorrhizal infection levels under field conditions. New Phytol. 1988, 108, 59–65. [Google Scholar] [CrossRef]
- Maltz, M.R.; Treseder, K.K. Sources of inocula influence mycorrhizal colonization of plants in restoration projects: A meta-analysis. Restor. Ecol. 2015, 23, 625–634. [Google Scholar] [CrossRef]
- Ruiz-Jaen, M.C.; Mitchell Aide, T. Restoration Success: How Is It Being Measured? Restor. Ecol. 2018, 13, 569–577. [Google Scholar] [CrossRef]
- Wortley, L.; Hero, J.-M.; Howes, M. Evaluating Ecological Restoration Success: A Review of the Literature. Restor. Ecol. 2018, 21, 537–543. [Google Scholar] [CrossRef]
- Allen, E.B. The Reconstruction of Disturbed Arid Lands: An Ecological Approach; Routledge: London, UK, 2019; ISBN 978-1-00-030510-4. [Google Scholar]
- Weber, S.E.; Diez, J.M.; Andrews, L.V.; Goulden, M.L.; Aronson, E.L.; Allen, M.F. Responses of arbuscular mycorrhizal fungi to multiple coinciding global change drivers. Fungal Ecol. 2019, 40, 62–71. [Google Scholar] [CrossRef]
- Allen, E.B.; Allen, M.F.; Helm, D.J.; Trappe, J.M.; Molina, R.; Rincon, E. Patterns and regulation of mycorrhizal plant and fungal diversity. Plant Soil 1995, 170, 47–62. [Google Scholar] [CrossRef]
- Stutz, J.C.; Copeman, R.; Martin, C.A.; Morton, J.B. Patterns of species composition and distribution of arbuscular mycorrhizal fungi in arid regions of southwestern North America and Namibia, Africa. Can. J. Bot. 2000, 78, 237–245. [Google Scholar]
- Chaudhary, V.B.; O’Dell, T.E.; Rillig, M.C.; Johnson, N.C. Multiscale patterns of arbuscular mycorrhizal fungal abundance and diversity in semiarid shrublands. Fungal Ecol. 2014, 12, 32–43. [Google Scholar] [CrossRef]
- Stutz, J.C.; Morton, J.B. Successive pot cultures reveal high species richness of arbuscular endomycorrhizal fungi in arid ecosystems. Can. J. Bot. 1996, 74, 1883–1889. [Google Scholar] [CrossRef]
- Maherali, H.; Klironomos, J.N. Influence of Phylogeny on Fungal Community Assembly and Ecosystem Functioning. Science 2007, 316, 1746–1748. [Google Scholar] [CrossRef] [PubMed]
- Cione, N.K.; Padgett, P.E.; Allen, E.B. Restoration of a Native Shrubland Impacted by Exotic Grasses, Frequent Fire, and Nitrogen Deposition in Southern California. Restor. Ecol. 2002, 10, 376–384. [Google Scholar] [CrossRef]
- Tormo, J.; Bochet, E.; García-Fayos, P. Roadfill Revegetation in Semiarid Mediterranean Environments. Part II: Topsoiling, Species Selection, and Hydroseeding. Restor. Ecol. 2007, 15, 97–102. [Google Scholar] [CrossRef]
- Alday, J.G.; Marrs, R.H.; Martínez-Ruiz, C. Vegetation succession on reclaimed coal wastes in Spain: The influence of soil and environmental factors. Appl. Veg. Sci. 2011, 14, 84–94. [Google Scholar] [CrossRef]
- DePuit, E.J. Potential topsoiling strategies for enhancement of vegetation diversity on mined lands. Miner. Environ. 1984, 6, 115–120. [Google Scholar] [CrossRef]
- Abella, S.R.; Chiquoine, L.P.; Newton, A.C.; Vanier, C.H. Restoring a desert ecosystem using soil salvage, revegetation, and irrigation. J. Arid Environ. 2015, 115, 44–52. [Google Scholar] [CrossRef]
- Dixon, P.J. Assessment of Topsoil Salvage and Seed Augmentation in the Restoration of Coastal Sage Scrub on Santa Catalina Island, California. West. North Am. Nat. 2018, 78, 711–721. [Google Scholar] [CrossRef]
- Greipsson, S.; El-Mayas, H. Arbuscular Mycorrhizae of Leymus arenarius on Coastal Sands and Reclamation Sites in Iceland and Response to Inoculation. Restor. Ecol. 2000, 8, 144–150. [Google Scholar] [CrossRef]
- Sheoran, V.; Sheoran, A.S.; Poonia, P. Soil Reclamation of Abandoned Mine Land by Revegetation: A Review. Int. J. Soil Sediment Water 2010, 3, 21. [Google Scholar]
- Hankin, S.L.; Karst, J.; Landhäusser, S.M. Influence of tree species and salvaged soils on the recovery of ectomycorrhizal fungi in upland boreal forest restoration after surface mining. Botany 2015, 93, 267–277. [Google Scholar] [CrossRef]
- Ferris, F.K.; Kleinman, L.H.; Steward, D.G.; Stowe, R.R.; Vicklund, L.E.; Berry, J.D.; Cowan, R.; Dunne, C.G.; Dunne, R.; Fritz, D.M.; et al. Handbook of Western Reclamation Techniques; Office of Technology Transfer: Denver, CO, USA, 1996. [Google Scholar]
- Rundel, P.W. Terrestrial Vegetation of California, 3rd ed.; University of California Press: Berkeley, CA, USA, 2007. [Google Scholar]
- Bowler, P.A. Ecological Restoration of Coastal Sage Scrub and Its Potential Role in Habitat Conservation Plans. Environ. Manag. 2000, 26, S85–S96. [Google Scholar] [CrossRef] [PubMed]
- Kimball, S.; Goulden, M.L.; Suding, K.N.; Parker, S. Altered water and nitrogen input shifts succession in a southern California coastal sage community. Ecol. Appl. 2014, 24, 1390–1404. [Google Scholar] [CrossRef] [PubMed]
- Orange County, California—Historic Data. Available online: http://www.ocwatersheds.com/monitoring/hydrology/historic_data (accessed on 24 November 2019).
- Pake, C.E.; Venable, D.L. Seed Banks in Desert Annuals: Implications for Persistence and Coexistence in Variable Environments. Ecology 1996, 77, 1427–1435. [Google Scholar] [CrossRef]
- Khalili, B.; Weihe, C.; Kimball, S.; Schmidt, K.T.; Martiny, J.B.H. Optimization of a Method To Quantify Soil Bacterial Abundance by Flow Cytometry. mSphere 2019, 4, e00435-19. [Google Scholar] [CrossRef]
- Sylvia, D.M. Techniques for the Study of Mycorrhiza; Academic Press: Cambridge, MA, USA, 1992; ISBN 978-0-08-086053-4. [Google Scholar]
- Brundrett, M.C.; Abbott, L.K. Mycorrhizal fungus propagules in the jarrah forest. New Phytol. 1994, 127, 539–546. [Google Scholar] [CrossRef]
- Koske, R.E.; Tessier, B. A convenient, permanent slide mounting medium. 34(2):59. Mycol. Soc. Am. Newsl. 1983, 34, 59. [Google Scholar]
- Bonfante-Fasolo, P. Anatomy and Morphology of VA Mycorrhizae; CRC Press: Boca Raton, FL, USA, 1984; ISBN 978-1-351-09441-2. [Google Scholar]
- Rohland, N.; Reich, D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012, 22, 939–946. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.; Young, J.P.W. Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol. Ecol. 2008, 65, 339–349. [Google Scholar] [CrossRef]
- Dumbrell, A.J.; Ashton, P.D.; Aziz, N.; Feng, G.; Nelson, M.; Dytham, C.; Fitter, A.H.; Helgason, T. Distinct seasonal assemblages of arbuscular mycorrhizal fungi revealed by massively parallel pyrosequencing. New Phytol. 2011, 190, 794–804. [Google Scholar] [CrossRef]
- Berry, D.; Mahfoudh, K.B.; Wagner, M.; Loy, A. Barcoded Primers Used in Multiplex Amplicon Pyrosequencing Bias Amplification. Appl. Environ. Microbiol. 2012, 78, 612. [Google Scholar] [CrossRef]
- Alvarado, P.; de Teixeira, M.M.; Andrews, L.; Fernandez, A.; Santander, G.; Doyle, A.; Perez, M.; Yegres, F.; Barker, B.M. Detection of Coccidioides posadasii from xerophytic environments in Venezuela reveals risk of naturally acquired coccidioidomycosis infections. Emerg. Microbes Infect. 2018, 7, 1–13. [Google Scholar] [CrossRef]
- Andrews, S. FastQC, A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Mahé, F.; Rognes, T.; Quince, C.; de Vargas, C.; Dunthorn, M. Swarm: Robust and fast clustering method for amplicon-based studies. PeerJ 2014, 2, e593. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Öpik, M.; Vanatoa, A.; Vanatoa, E.; Moora, M.; Davison, J.; Kalwij, J.M.; Reier, Ü.; Zobel, M. The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol. 2010, 188, 223–241. [Google Scholar] [CrossRef] [PubMed]
- Paulson, J.N.; Stine, O.C.; Bravo, H.C.; Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 2013, 10, 1200–1202. [Google Scholar] [CrossRef]
- Phillips, M.L.; Weber, S.E.; Andrews, L.V.; Aronson, E.L.; Allen, M.F.; Allen, E.B. Fungal community assembly in soils and roots under plant invasion and nitrogen deposition. Fungal Ecol. 2019, 40, 107–117. [Google Scholar] [CrossRef]
- Hausmann, N.T.; Hawkes, C.V. Plant neighborhood control of arbuscular mycorrhizal community composition. New Phytol. 2009, 183, 1188–1200. [Google Scholar] [CrossRef]
- Sikes, B.A.; Cottenie, K.; Klironomos, J.N. Plant and fungal identity determines pathogen protection of plant roots by arbuscular mycorrhizas. J. Ecol. 2009, 97, 1274–1280. [Google Scholar] [CrossRef]
- Allen, M.F.; Swenson, W.; Querejeta, J.I.; Egerton-Warburton, L.M.; Treseder, K.K. Ecology of Mycorrhizae: A Conceptual Framework for Complex Interactions Among Plants and Fungi. Annu. Rev. Phytopathol. 2003, 41, 271–303. [Google Scholar] [CrossRef] [PubMed]
- Sikes, B.A.; Powell, J.R.; Rillig, M.C. Deciphering the relative contributions of multiple functions within plant–microbe symbioses. Ecology 2010, 91, 1591–1597. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2009. [Google Scholar]
- Package ‘vegan’; Community Ecology Package, Version 2. 2019. Available online: http://CRAN.R-project.org/package=vegan (accessed on 1 September 2019).
- Balshor, B.J.; Garrambone, M.S.; Austin, P.; Balazs, K.R.; Weihe, C.; Martiny, J.B.H.; Huxman, T.E.; McCollum, J.R.; Kimball, S. The effect of soil inoculants on seed germination of native and invasive species. Botany 2017, 95, 469–480. [Google Scholar] [CrossRef]
- Golos, P.J.; Dixon, K.W.; Erickson, T.E. Plant recruitment from the soil seed bank depends on topsoil stockpile age, height, and storage history in an arid environment. Restor. Ecol. 2016, 24, S53–S61. [Google Scholar] [CrossRef]
- Burke, A. The effect of topsoil treatment on the recovery of rocky plain and outcrop plant communities in Namibia. J. Arid Environ. 2008, 72, 1531–1536. [Google Scholar] [CrossRef]
- Visser, S.; Griffiths, C.L.; Parkinson, D. Topsoil storage effects on primary production and rates of vesicular-arbuscular mycorrhizal development inAgropyron trachycaulum. Plant Soil 1984, 82, 51–60. [Google Scholar] [CrossRef]
- Van Der Heijden, M.G.A.; Bardgett, R.D.; Straalen, N.M.V. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Lankau, E.W.; Lankau, R.A. Plant species capacity to drive soil fungal communities contributes to differential impacts of plant–soil legacies. Ecology 2014, 95, 3221–3228. [Google Scholar] [CrossRef]
- Cox, R.D.; Allen, E.B. Composition of soil seed banks in southern California coastal sage scrub and adjacent exotic grassland. Plant Ecol. 2007, 198, 37. [Google Scholar] [CrossRef]
- Strickland, M.S.; Rousk, J. Considering fungal:bacterial dominance in soils – Methods, controls, and ecosystem implications. Soil Biol. Biochem. 2010, 42, 1385–1395. [Google Scholar] [CrossRef]
- Sylvia, D.M. Nursery Inoculation of Sea Oats with Vesicular-Arbuscular Mycorrhizal Fungi and Outplanting Performance on Florida Beaches. J. Coast. Res. 1989, 5, 9. [Google Scholar]
- Allen, M.F.; Moore, T.S., Jr.; Christensen, M. Phytohormone changes in Bouteloua gracilis infected by vesicular–arbuscular mycorrhizae: I. Cytokinin increases in the host plant. Can. J. Bot. 1980, 58, 371–374. [Google Scholar] [CrossRef]
- Treseder, K.K. The extent of mycorrhizal colonization of roots and its influence on plant growth and phosphorus content. Plant Soil 2013, 371, 1–13. [Google Scholar] [CrossRef]
- Piñeiro, J.; Maestre, F.T.; Bartolomé, L.; Valdecantos, A. Ecotechnology as a tool for restoring degraded drylands: A meta-analysis of field experiments. Ecol. Eng. 2013, 61, 133–144. [Google Scholar] [CrossRef]
- Mummey, D.L.; Rillig, M.C. The invasive plant species Centaurea maculosa alters arbuscular mycorrhizal fungal communities in the field. Plant Soil 2006, 288, 81–90. [Google Scholar] [CrossRef]
- Zhao, H.; Li, X.; Zhang, Z.; Zhao, Y.; Yang, J.; Zhu, Y. Species diversity and drivers of arbuscular mycorrhizal fungal communities in a semi-arid mountain in China. PeerJ 2017, 5, e4155. [Google Scholar] [CrossRef]
- Pringle, A.; Bever, J.D.; Gardes, M.; Parrent, J.L.; Rillig, M.C.; Klironomos, J.N. Mycorrhizal Symbioses and Plant Invasions. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 699–715. [Google Scholar] [CrossRef]
- Lekberg, Y.; Gibbons, S.M.; Rosendahl, S.; Ramsey, P.W. Severe plant invasions can increase mycorrhizal fungal abundance and diversity. ISME J. 2013, 7, 1424–1433. [Google Scholar] [CrossRef]
- Inderjit; van der Putten, W.H. Impacts of soil microbial communities on exotic plant invasions. Trends Ecol. Evol. 2010, 25, 512–519. [Google Scholar] [CrossRef]
- Potthoff, M.; Steenwerth, K.L.; Jackson, L.E.; Drenovsky, R.E.; Scow, K.M.; Joergensen, R.G. Soil microbial community composition as affected by restoration practices in California grassland. Soil Biol. Biochem. 2006, 38, 1851–1860. [Google Scholar] [CrossRef]
- Maltz, M.R.; Bell, C.E.; Mitrovich, M.J.; Iyer, A.R.; Treseder, K.K. Invasive Plant Management Techniques Alter Arbuscular Mycorrhizal Fungi. Ecol. Restor. 2016, 34, 209–215. [Google Scholar] [CrossRef]
- Liddicoat, C.; Weinstein, P.; Bissett, A.; Genie, N.J.C.; Mills, J.G.; Waycott, M.; Breed, M.F. Can bacterial indicators of a grassy woodland restoration inform ecosystem assessment and microbiota-mediated human health? Environ. Int. 2019, 129, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Potthoff, M.; Jackson, L.E.; Steenwerth, K.L.; Ramirez, I.; Stromberg, M.R.; Rolston, D.E. Soil Biological and Chemical Properties in Restored Perennial Grassland in California. Restor. Ecol. 2005, 13, 61–73. [Google Scholar] [CrossRef]
- Brewer, T.E.; Aronson, E.L.; Arogyaswamy, K.; Billings, S.A.; Botthoff, J.K.; Campbell, A.N.; Dove, N.C.; Fairbanks, D.; Gallery, R.E.; Hart, S.C.; et al. Ecological and Genomic Attributes of Novel Bacterial Taxa That Thrive in Subsurface Soil Horizons. mBio 2019, 10, e01318-19. [Google Scholar] [CrossRef]
- Dove, N.C.; Arogyaswamy, K.; Billings, S.; Bothoff, J.; Carey, C.; Cisco, C.; DeForest, J.; Fairbanks, D.; Fierer, N.; Gallery, R.; et al. Continental-scale patterns of extracellular enzyme activity in the subsoil: An overlooked reservoir of microbial activity. Ecol. Lett. 2020. in review. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmidt, K.T.; Maltz, M.; Ta, P.; Khalili, B.; Weihe, C.; Phillips, M.; Aronson, E.; Lulow, M.; Long, J.; Kimball, S. Identifying Mechanisms for Successful Ecological Restoration with Salvaged Topsoil in Coastal Sage Scrub Communities. Diversity 2020, 12, 150. https://doi.org/10.3390/d12040150
Schmidt KT, Maltz M, Ta P, Khalili B, Weihe C, Phillips M, Aronson E, Lulow M, Long J, Kimball S. Identifying Mechanisms for Successful Ecological Restoration with Salvaged Topsoil in Coastal Sage Scrub Communities. Diversity. 2020; 12(4):150. https://doi.org/10.3390/d12040150
Chicago/Turabian StyleSchmidt, Katharina T., Mia Maltz, Priscilla Ta, Banafshe Khalili, Claudia Weihe, Michala Phillips, Emma Aronson, Megan Lulow, Jennifer Long, and Sarah Kimball. 2020. "Identifying Mechanisms for Successful Ecological Restoration with Salvaged Topsoil in Coastal Sage Scrub Communities" Diversity 12, no. 4: 150. https://doi.org/10.3390/d12040150
APA StyleSchmidt, K. T., Maltz, M., Ta, P., Khalili, B., Weihe, C., Phillips, M., Aronson, E., Lulow, M., Long, J., & Kimball, S. (2020). Identifying Mechanisms for Successful Ecological Restoration with Salvaged Topsoil in Coastal Sage Scrub Communities. Diversity, 12(4), 150. https://doi.org/10.3390/d12040150