Abstract
Wolbachia bacteria are widely distributed across invertebrate taxa, including ants, but several aspects of this host-associated interaction are still poorly explored, especially with regard to the ancestral state association, origin, and dispersion patterns of this bacterium. Therefore, in this study, we explored the association of Wolbachia with Formicidae in an evolutionary context. Our data suggest that supergroup F is the ancestral character state for Wolbachia infection in ants, and there is only one transition to supergroup A, and once ants acquired infection with supergroup A, there have been no other strains introduced. Our data also reveal that the origin of Wolbachia in ants likely originated in Asia and spread to the Americas, and then back to Asia. Understanding the processes and mechanisms of dispersion of these bacteria in Formicidae is a crucial step to advance the knowledge of this symbiosis and their implications in an evolutionary context.
1. Introduction
There are many examples of insect–microbe interactions providing benefits to all players involved [1,2,3,4,5], and one of the most well-documented bacteria associated with insects is Wolbachia [6,7]. This bacterium is known for modifying the host’s reproduction for its own benefit, including the induction of cytoplasmic incompatibility, parthenogenesis, and male-killing or feminization [8]. However, it is not known whether these functions are related to a particular Wolbachia strain or host [9]. As this bacterium has been found in association with hundreds of hosts and encompasses an immense diversity, the classification of strains into supergroups has been proposed and about 17 supergroups, to date, have been reported through genotyping a single gene, several genes, or even genomic approaches [10]. These supergroups are called “A to S”, with G and R no longer considered separate supergroups [11,12]. Of these, supergroups found exclusively in arthropods belong to the A, B, E, H, I, K, M, N, O, P, Q, and S supergroups [13,14,15,16,17], with supergroup F being common for nematodes and arthropods [18,19]. Although we know that Wolbachia is associated with several arthropods hosts, we focused on ants, a highly diverse group with more than 13,000 described species and a global distribution [20]. In addition, it is one of the groups of insects best studied in terms of association with Wolbachia. This offers us an excellent opportunity to explore the evolution and biogeography of this association.
Wolbachia has been identified from several ant genera, but few studies have succeeded in identifying the implications for these ant-associated interactions. Monomorium pharaonis appears to have an accelerated colony life cycle [21] and Tapinoma melanocephalum has a nutritional upgrade of vitamin B [22] in the presence of Wolbachia. However, whether these are specific cases within all ant diversity is still unknown. Despite the fact that these bacteria have been identified in several ant genera, little is known about the diversity and evolutionary history of this symbiosis; see [23,24,25,26,27,28,29,30,31,32,33,34,35].
Historically, studying these interactions was conducted by targeted PCR amplification and sequencing of the Wolbachia surface protein (wsp) gene [36]. However, the use of this gene alone has been shown to not be appropriate for diversity studies since it has been reported to have a high rate of recombination [36]. Therefore, the Multilocus Sequence Typing (MLST) approach [37] has been shown to be a more reliable alternative, targeting five different Wolbachia genes, coxA, fbpA, ftsZ, gatB, and hcpA, instead of just one [38]. These five genes are spread across the Wolbachia genome and evolve under purifying selection [38], making them more appropriate for phylogenetic analysis. Once a unique sequence is found with the combination of all of these genes, a sequence type (ST) is assigned and is stored in the Wolbachia MLST database (https://pubmlst.org/wolbachia/).
Many studies have reliably used the MLST approach [39,40,41,42], including some studies of ants [9,24,28,32,33]. However, no studies, to date, in ants have leveraged ancestral state reconstruction and biogeographic range evolution to understand the origin and biogeography of this symbiosis. As a robust phylogenetic tree is necessary to investigate lifestyle transitions in Wolbachia’s evolutionary history, this study aimed to (1) infer the evolutionary history of Wolbachia associated with Formicidae through the MLST approach (five genes), controlling for their different evolutionary models; (2) reconstruct the ancestral state of the different Wolbachia supergroups; (3) investigate the dispersion patterns of these bacteria associated with Formicidae through biogeographic range evolution analyses.
2. Material and Methods
2.1. Phylogenetic Reconstruction
Wolbachia sequences isolated from 70 Formicidae hosts (2079 bp from the five genes) were downloaded from the Wolbachia MLST database (https://pubmlst.org/wolbachia/), as well as metadata information. ST78 from Opistophthalmus chaperi (supergroup F) was added as an outgroup because it has not been found to be associated with ants but shows high similarity with strains associated with ants (Table 1). The sequences were aligned using ClustalW [43] of the BioEdit software [44] and later were used for partitioned phylogenetic reconstruction. PartitionFinder2 (2.1.1) [45] was used to choose the best model of molecular evolution and returned four partitions: charset Subset1 = 1–402 pb; charset Subset2 = 403–831 pb; charset Subset3 = 832–1266 pb and 1636–2079 pb; charset Subset4 = 1267–1635 pb, with GTR + G for Subset1, GTR + I + G for Subset2, GTR + I + G for Subset3, and TIM + I + G for Subset4. Bayesian inference was implemented using MrBayes (3.2.6) [46] on the Cipres Science Gateway [47,48] for phylogenetic reconstruction with the Markov chain Monte Carlo analysis for 1,000,000 generations with sampling every 1000 generations, and discarding the first 25% of trees as burnin. We used the chronos function with the correlated model available in the Ape package [49] with R software [50] to reconstruct a chronogram with a relative time scale [51,52,53].

Table 1.
Wolbachia sequence samples associated with Formicidae included in the present study.
2.2. Reconstruction of the Ancestral State of Wolbachia
To reconstruct the ancestral state of Wolbachia diversity associated with Formicidae, we assigned each supergroup A or F to each tip in our topology. Although there are only a few (three) observations from the supergroup F associated with ants, we included them in the analysis. However, several other studies have already reported that supergroup A is more common in ants [9,28,33], which gives us support that our data are a real representation of what we find in nature. As we only have one observation of Wolbachia belonging to the supergroup B associated with Formicidae (see [9]), we decided to remove this sample from the subsequent analyses. The ‘equal rates’ (ER) model and the ‘all rates different’ (ARD) model were compared to determine which model best explains our data using the likelihood ratio test (LRT) [54,55]. The Ape and Phytools packages [49,56] from the R software [50] were used to reconstruct ancestral character states. We used the MCMC approach to sample character histories from the probability distribution using stochastic character mapping [57]. This method samples character histories in direct proportion to their posterior probability under a model. Using the SIMMAP function [58], we generated 100 stochastic character maps from our dataset. To summarize the set of stochastic maps in a more meaningful way, we estimated the number of changes of each type, the proportion of time spent in each state, and the later probabilities that each internal node is in each state, under the best model.
2.3. Biogeographic Range Evolution Analyses
We used the inferred phylogeny and noted where, geographically, the Wolbachia ST was recovered (Asia, Africa, North America, South America, or Oceania) to implement biogeographic range evolution analyses using the R package BioGeoBEARS v1.1.1 [59] and estimated the ancestral range of Wolbachia associated with Formicidae. We followed the recommendations and parameters available on BioGeoBEARS PhyloWiki (http://phylo.wikidot.com/biogeobears) to test whether the observed biogeographic distribution of Wolbachia associated with Formicidae is best explained with a model that allows for vicariance and long-distance dispersal (DEC + J model) [60] versus a model that allows for only vicariance (DEC model) [61]. In addition, as Wolbachia is widely spread across these five biogeographic areas, we set max_range_size = 5. We used the likelihood ratio test (LRT) and the Akaike information criterion (AIC) to see which model best fits the data.
3. Results
Our Wolbachia phylogeny appears to be fairly robust when examining the results of posterior probability (PP); however, it grouped taxonomically unrelated ants, indicating that there is a lack of specificity of the host. Samples from 35 ant genera were included in this analysis, and as an example, 11 strains of Camponotus recovered from different locations around the world (Brazil, USA, Malaysia, India, and Thailand) were distributed across the phylogeny. This suggests the lack of codivergence of the bacteria and ant host. However, there was a clear and robust distinction between Wolbachia supergroups A and F associated with Formicidae (Figure 1).
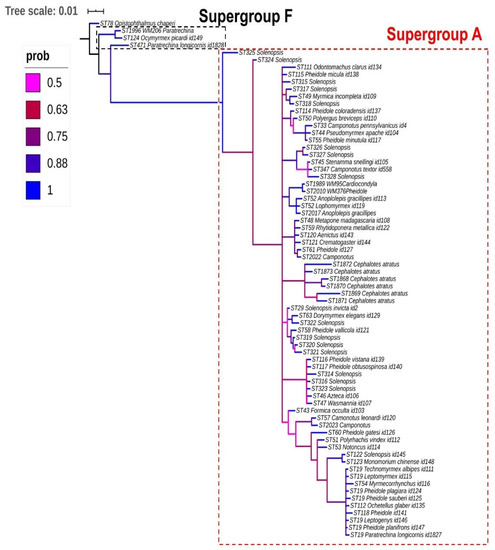
Figure 1.
Bayesian majority-rule consensus tree (MrBayes) from the concatenated dataset (coxA, fbpA, ftsZ, gatB, and hcpA genes) of Wolbachia strains in Formicidae. Note that most strains belong to supergroup A, highlighted by a dotted box red, with the exceptions of three sequence types (STs), which were classified in supergroup F, highlighted by a dotted box black, and ST78 as outgroup.
Based on the likelihood ratio test, the ARD (all rates different) model (LTR= −6.975) was the best fit referring to the transition rates between each state to estimate the ancestral states of the Wolbachia supergroup associated with Formicidae when compared with ER (equal rates) (LTR= −8.530). Our ancestral state reconstruction (ASR) results with all 70 strains of Wolbachia associated with Formicidae show that the ancestor’s supergroup was F and that was consistent across all 100 replicates, which suggests that supergroup F was the ancestor character state (Figure 2A). In addition, the transition from supergroup ancestor F to supergroup A occurred only once within the Formicidae family (Figure 2B). Therefore, once Wolbachia supergroup A is acquired, there is no transition to another type of Wolbachia.
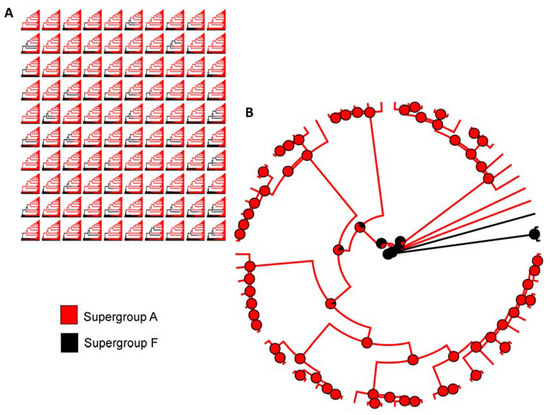
Figure 2.
Ancestral state reconstruction of Wolbachia associated with Formicidae. (A) One hundred stochastic character maps from our dataset. (B) Summary of all stochastic character maps for ancestral state reconstruction. Model = all rates different (ARD).
For the biogeographic analysis, the DEC + J model was returned as the best fit to our data (AIC weight ratio model = 1.28 × 1013), which allows for vicariance and long-distance dispersal of Wolbachia in Formicidae (see Table 2). According to this scenario, the ancestral origin of Wolbachia associated with Formicidae remains ambiguous; however, it is most probable that it originated in Asia. Then, this supergroup F, rarely found among ants, seems to have expanded to Africa. More information about Wolbachia associated with Formicidae in the African region could confirm this trend. Another lineage of Wolbachia, supergroup A, is the most common among Formicidae and appears to have likely expanded to South America. One clade then expanded into North America. In the other major clade, there was a second expansion of Wolbachia back to Asia from South America, introducing supergroup A into this region. Our survey revealed only a few representatives of Wolbachia associated with Formicidae in Oceania and Africa (Madagascar), but our results show that infections in these regions are more recent (Figure 3). Our survey did not recover any data from Europe or Central America.

Table 2.
Likelihood ratio test (LRT) and the Akaike information criterion (AIC) of the DEC model and the DEC + J model to select the optimal model of ancestral areas of Wolbachia associated with Formicidae.
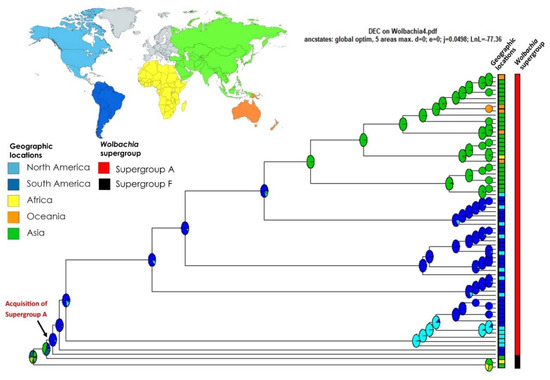
Figure 3.
Ancestral range estimation of Wolbachia associated with Formicidae (MLST approach) with the chronogram using BIOGEOBEARS (DEC + J model). The pies are color-coded with the highest maximum likelihood probability of locations on the continents, and the boxes in the corner are colored according to the geographic locations and Wolbachia supergroup of the tips. The arrow indicates when Wolbachia supergroup A was acquired.
4. Discussion
This is the first study that has sought to investigate the ancestral state of supergroups, as well as to understand the ancestral range of origin in the evolutionary context of Wolbachia associated with ants. In the past, identifying Wolbachia infection was done by genotyping a single gene, wsp only. In general, with this approach, the Wolbachia strains associated with Formicidae belonged to supergroups A and B. With the MLST approach, now with five genes, it was found that the vast majority of Formicidae strains belong to supergroup A, followed by some representatives from supergroup F. There is only a single observation of supergroup B associated with Pheidole sciophila found in Mexico [9] and, therefore, it was not included in the present study. In order to understand if supergroup B, in nature, is atypical as a symbiont of Formicidae or if this is mainly a sampling bias, further studies are needed. In an extensive study, Russell and colleagues [9] used the MLST approach to understand the evolution of this interaction between Wolbachia and its hosts. For this, the authors focused their studies on two insect groups: ants and lycaenid butterflies. In addition to showing the presence of these bacteria in these hosts, the authors also concluded that the Wolbachia bacteria found in each of these groups of insects are different and highly specialized. Furthermore, they concluded that phylogenetic and geographic barriers can influence the evolutionary divergence of these bacteria. The authors found about 41 STs (sequence types) of Wolbachia associated with Formicidae, with few sample sequences included from South America, a hotspot for biodiversity [62,63]. Thus, in the present study, we were able to expand and include samples from this region and add more STs included in the Wolbachia MLST database since 2009 (Table 1).
Including Wolbachia associated with 35 different ant genera, our results indicate a lack of codivergence of Wolbachia with their ant hosts, corroborating what has been found by other studies [9,28,33,64]. The evidence for this conclusion is supported since taxonomically unrelated ants were grouped together in our Wolbachia phylogeny. Surprisingly, supergroup F was the ancestral character state for Wolbachia present in Formicidae, despite being less frequent than supergroup A. As already mentioned above, this supergroup was originally described in nematodes, but with the MLST approach, it has also been found in ants, although less frequently than supergroup A [9,32]. In addition, our results agree with the findings of Comandatore et al. [65] which suggest that supergroup F is a basal branching lineage for all Wolbachia.
This symbiont has already been reported to be transmitted vertically from queens to eggs in several host insects [40,66,67,68], including ants [29,69], and horizontal transmission of Wolbachia in ants is not common but may occasionally occur in related hosts [9,32,70]. In addition, Tseng et al. [32] showed that not all strains of ant Wolbachia have the same potential for being horizontally transferred. Another interesting aspect about these two supergroups being present in ants is that the evolutionary histories seem to be different, with supergroup A being transmitted vertically (maternal) and supergroup F being acquired horizontally [32].
Regarding the potential origin of the ancestral range of Wolbachia in Formicidae, our results seem to explain the dispersions by across continents. Other studies also show that geography can impact Wolbachia diversity on different ant host [9,28,33] and also in other insects, such as butterflies and moths [71]; however, few studies have focused on the origin and pattern of dispersion of these bacteria in different hosts until recently [72]. Our data suggest that the origin of Wolbachia in Formicidae happened in Asia. The supergroup F, although with few samples in ants, seems to have expanded to Africa and Asia. With the evolution of supergroup A associated with ants, Wolbachia then appeared in South America, with one major clade remaining in South America with a single introduction into North America and other South American clades resulting in a jump back to Asia with introductions from Asia into Oceania and Africa (Madagascar). Russell et al. [9] found evidence that Wolbachia strains were grouped according to samples from the Old and New Worlds, and our results also found this grouping. Our work highlights that to fully understand the evolutionary history and mechanisms of Wolbachia dispersion in ants or any other host group, we need much broader sampling of host species and geographic locations. However, our work is shedding light on the evolution and biogeographic history of this symbiotic interaction.
Author Contributions
M.O.R. and C.S.M. contributed equally to this study. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by funding to C.S.M. from the National Science Foundation (NSF DEB 1900357).
Acknowledgments
This manuscript was initiated as part of a class project in the ModelBased Phylogenetics and Hypothesis Testing course in the Department of Entomology at Cornell University. We would like to thank Augusto Santos Rampasso for help in some aspects of the analysis of the data in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kikuchi, Y.; Hosokawa, T.; Fukatsu, T. Insect-microbe mutualism without vertical transmission: A stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 2007, 73, 4308–4316. [Google Scholar] [CrossRef]
- Buchner, P. Endosymbiosis of Animals with Plant Microorganisms.1965.-Google Acadêmico; Buchner, P., Ed.; Interscience Publishers: New York, NY, USA, 1965. [Google Scholar]
- Bourtzis, K.; Miller, T. Insect Symbiosis; CRC Press: Boca Ranton, FL, USA, 2006; Volume 2. [Google Scholar]
- Hu, Y.; Sanders, J.G.; Łukasik, P.; D’Amelio, C.L.; Millar, J.S.; Vann, D.R.; Lan, Y.; Newton, J.A.; Schotanus, M.; Kronauer, D.J.C.; et al. Herbivorous turtle ants obtain essential nutrients from a conserved nitrogen-recycling gut microbiome. Nat. Commun. 2018, 9, 964. [Google Scholar] [CrossRef]
- Saraithong, P.; Li, Y.; Saenphet, K.; Chen, Z.; Chantawannakul, P. Midgut bacterial communities in the giant Asian honeybee (Apis dorsata) across 4 developmental stages: A comparative study. Insect Sci. 2017, 24, 81–92. [Google Scholar] [CrossRef]
- Russell, J.A.; Funaro, C.F.; Giraldo, Y.M.; Goldman-Huertas, B.; Suh, D.; Kronauer, D.J.C.; Moreau, C.S.; Pierce, N.E. A Veritable Menagerie of Heritable Bacteria from Ants, Butterflies, and Beyond: Broad Molecular Surveys and a Systematic Review. PLoS ONE 2012, 7, e51027. [Google Scholar] [CrossRef]
- Hilgenboecker, K.; Hammerstein, P.; Schlattmann, P.; Telschow, A.; Werren, J.H. How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol. Lett. 2008, 281, 215–220. [Google Scholar] [CrossRef]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008, 6, 741–751. [Google Scholar] [CrossRef]
- Russell, J.A.; Goldman-Huertas, B.; Moreau, C.S.; Baldo, L.; Stahlhut, J.K.; Werren, J.H.; Pierce, N.E. Specialization and geographic isolation among Wolbachia symbionts from ants and lycaenid butterflies. Evolution 2009, 63, 624–640. [Google Scholar] [CrossRef]
- Lefoulon, E.; Clark, T.; Borveto, F.; Perriat-Sanguinet, M.; Moulia, C.; Slatko, B.E.; Gavotte, L. Pseudoscorpion Wolbachia symbionts: Diversity and evidence for a new supergroup S. BMC Microbiol. 2020, 20, 188. [Google Scholar] [CrossRef]
- Baldo, L.; Werren, J.H. Revisiting Wolbachia supergroup typing based on WSP: Spurious lineages and discordance with MLST. Curr. Microbiol. 2007, 55, 81–87. [Google Scholar] [CrossRef]
- Gerth, M. Classification of Wolbachia (Alphaproteobacteria, Rickettsiales): No evidence for a distinct supergroup in cave spiders. Infect. Genet. Evol. 2016, 43, 378–380. [Google Scholar] [CrossRef]
- Bordenstein, S.; Rosengaus, R.B. Discovery of a novel Wolbachia supergroup in isoptera. Curr. Microbiol. 2005, 51, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Glowska, E.; Dragun-Damian, A.; Dabert, M.; Gerth, M. New Wolbachia supergroups detected in quill mites (Acari: Syringophilidae). Infect. Genet. Evol. 2015, 30, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Lo, N.; Casiraghi, M.; Salati, E.; Bazzocchi, C.; Bandi, C. How Many Wolbachia Supergroups Exist? Mol. Biol. Evol. 2002, 19, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Lo, N.; Paraskevopoulos, C.; Bourtzis, K.; O’Neill, S.L.; Werren, J.H.; Bordenstein, S.R.; Bandi, C. Taxonomic status of the intracellular bacterium Wolbachia pipientis. Int. J. Syst. Evol. Microbiol. 2007, 57, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Ros, V.I.D.; Fleming, V.M.; Feil, E.J.; Breeuwer, J.A.J. How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae). Appl. Environ. Microbiol. 2009, 75, 1036–1043. [Google Scholar] [CrossRef]
- Ferri, E.; Bain, O.; Barbuto, M.; Martin, C.; Lo, N.; Uni, S.; Landmann, F.; Baccei, S.G.; Guerrero, R.; de Souza Lima, S.; et al. New Insights into the Evolution of Wolbachia Infections in Filarial Nematodes Inferred from a Large Range of Screened Species. PLoS ONE 2011, 6, e20843. [Google Scholar] [CrossRef]
- Lefoulon, E.; Gavotte, L.; Junker, K.; Barbuto, M.; Uni, S.; Landmann, F.; Laaksonen, S.; Saari, S.; Nikander, S.; de Souza Lima, S.; et al. A new type F Wolbachia from Splendidofilariinae (Onchocercidae) supports the recent emergence of this supergroup. Int. J. Parasitol. 2012, 42, 1025–1036. [Google Scholar] [CrossRef]
- Bolton, B. An Online Catalog of the Ants of the World. Available online: http://www.antcat.org/ (accessed on 20 October 2016).
- Singh, R.; Linksvayer, T.A. Wolbachia-infected ant colonies have increased reproductive investment and an accelerated life cycle. J. Exp. Biol. 2020, 223. [Google Scholar] [CrossRef]
- Cheng, D.; Chen, S.; Huang, Y.; Pierce, N.E.; Riegler, M.; Yang, F.; Zeng, L.; Lu, Y.; Liang, G.; Xu, Y. Symbiotic microbiota may reflect host adaptation by resident to invasive ant species. PLoS Pathog. 2019, 15, e1007942. [Google Scholar] [CrossRef]
- Fernando de Souza, R.; Daivison Silva Ramalho, J.; Santina de Castro Morini, M.; Wolff, J.L.C.; Araújo, R.C.; Mascara, D. Identification and Characterization of Wolbachia in Solenopsis saevissima Fire Ants (Hymenoptera: Formicidae) in Southeastern Brazil. Curr. Microbiol. 2009, 58, 189–194. [Google Scholar] [CrossRef]
- Frost, C.L.; Fernandez-Marin, H.; Smith, J.E.; Hughes, W.O.H. Multiple gains and losses of Wolbachia symbionts across a tribe of fungus-growing ants. Mol. Ecol. 2010, 19, 4077–4085. [Google Scholar] [CrossRef]
- Frost, C.L.; Pollock, S.W.; Smith, J.E.; Hughes, W.O.H. Wolbachia in the flesh: Symbiont intensities in germ-line and somatic tissues challenge the conventional view of Wolbachia transmission routes. PLoS ONE 2014, 9, e95122. [Google Scholar] [CrossRef][Green Version]
- Martins, C.; Souza, R.F.; Bueno, O.C. Presence and distribution of the endosymbiont Wolbachia among Solenopsis spp. (Hymenoptera: Formicidae) from Brazil and its evolutionary history. J. Invertebr. Pathol. 2012, 109, 287–296. [Google Scholar] [CrossRef]
- Kautz, S.; Rubin, B.E.R.; Moreau, C.S. Bacterial Infections across the Ants: Frequency and Prevalence of Wolbachia, Spiroplasma, and Asaia. Psyche A J. Entomol. 2013, 2013, 1–11. [Google Scholar] [CrossRef]
- Ramalho, M.O.; Martins, C.; Silva, L.M.R.; Martins, V.G.; Bueno, O.C. Intracellular symbiotic bacteria of Camponotus textor, Forel (Hymenoptera, Formicidae). Curr. Microbiol. 2017, 74, 589–597. [Google Scholar] [CrossRef]
- Ramalho, M.O.; Vieira, A.S.; Pereira, M.C.; Moreau, C.S.; Bueno, O.C. Transovarian Transmission of Blochmannia and Wolbachia Endosymbionts in the Neotropical Weaver Ant Camponotus textor (Hymenoptera, Formicidae). Curr. Microbiol. 2018, 75, 866–873. [Google Scholar] [CrossRef]
- Ramalho, M.O.; Bueno, O.C.; Moreau, C.S. Species-specific signatures of the microbiome from Camponotus and Colobopsis ants across developmental stages. PLoS ONE 2017, 12, e0187461. [Google Scholar] [CrossRef]
- Ramalho, M.O.; Bueno, O.C.; Moreau, C.S. Microbial composition of spiny ants (Hymenoptera: Formicidae: Polyrhachis) across their geographic range. BMC Evol. Biol. 2017, 17, 96. [Google Scholar] [CrossRef]
- Tseng, S.P.; Wetterer, J.K.; Suarez, A.V.; Lee, C.Y.; Yoshimura, T.; Shoemaker, D.W.; Yang, C.C.S. Genetic Diversity and Wolbachia Infection Patterns in a Globally Distributed Invasive Ant. Front. Genet. 2019, 10, 838. [Google Scholar] [CrossRef]
- Kelly, M.; Price, S.L.; de Oliveira Ramalho, M.; Moreau, C.S. Diversity of Wolbachia Associated with the Giant Turtle Ant, Cephalotes atratus. Curr. Microbiol. 2019, 76, 1330–1337. [Google Scholar] [CrossRef]
- Ramalho, M.O.; Moreau, C.S.; Bueno, O.C. The Potential Role of Environment in Structuring the Microbiota of Camponotus across Parts of the Body. Adv. Entomol. 2019, 7, 47–70. [Google Scholar] [CrossRef][Green Version]
- Reeves, D.D.; Price, S.L.; Ramalho, M.O.; Moreau, C.S. The Diversity and Distribution of Wolbachia, Rhizobiales, and Ophiocordyceps Within the Widespread Neotropical Turtle Ant, Cephalotes atratus (Hymenoptera: Formicidae). Neotrop. Entomol. 2020, 49, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Baldo, L.; Lo, N.; Werren, J.H. Mosaic nature of the wolbachia surface protein. J. Bacteriol. 2005, 187, 5406–5418. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulos, C.; Bordenstein, S.; Wernegreen, J.; Werren, J.; Bourtzis, K. Toward a Wolbachia multilocus sequence typing system: Discrimination of Wolbachia strains present in Drosophila species. Curr. Microbiol. 2006, 53, 388–395. [Google Scholar] [CrossRef]
- Baldo, L.; Dunning Hotopp, J.C.; Jolley, K.A.; Bordenstein, S.R.; Biber, S.A.; Choudhury, R.R.; Hayashi, C.; Maiden, M.C.J.; Tettelin, H.; Werren, J.H. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 2006, 72, 7098–7110. [Google Scholar] [CrossRef]
- Ali, H.; Muhammad, A.; Hou, Y. Infection Density Dynamics and Phylogeny of Wolbachia Associated with Coconut Hispine Beetle, Brontispa longissima (Gestro) (Coleoptera: Chrysomelidae), by Multilocus Sequence Type. Artic. J. Microbiol. Biotechnol. 2018, 28, 796–808. [Google Scholar] [CrossRef]
- Ali, H.; Muhammad, A.; Sanda Bala, N.; Hou, Y. The Endosymbiotic Wolbachia and Host COI Gene Enables to Distinguish Between Two Invasive Palm Pests; Coconut Leaf Beetle, Brontispa longissima and Hispid Leaf Beetle, Octodonta nipae. J. Econ. Entomol. 2018, 111, 2894–2902. [Google Scholar] [CrossRef]
- Betelman, K.; Caspi-Fluger, A.; Shamir, M.; Chiel, E. Identification and characterization of bacterial symbionts in three species of filth fly parasitoids. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Prezotto, L.F.; Perondini, A.L.P.; Hernández-Ortiz, V.; Marino, C.L.; Selivon, D. Wolbachia strains in cryptic species of the Anastrepha fraterculus complex (Diptera, Tephritidae) along the Neotropical Region. Syst. Appl. Microbiol. 2017, 40, 59–67. [Google Scholar] [CrossRef]
- Higgins, D.; Bleasby, A.; Fuchs, R. CLUSTAL V: Improved software for multiple sequence alignment. Comput. Appl. 1992, 8, 189–191. [Google Scholar]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2016, 34, msw260. [Google Scholar] [CrossRef]
- Huelsenbeck, J.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES Portals. Available online: http://www.phylo.org/ (accessed on 20 September 2020).
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: http://www.R-project.org/ (accessed on 20 September 2020).
- Kim, J.; Sanderson, M.J. Penalized Likelihood Phylogenetic Inference: Bridging the Parsimony-Likelihood Gap. Syst. Biol. 2008, 57, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E. Molecular dating of phylogenies by likelihood methods: A comparison of models and a new information criterion. Mol. Phylogenet. Evol. 2013, 67, 436–444. [Google Scholar] [CrossRef]
- Sanderson, M.J. Estimating Absolute Rates of Molecular Evolution and Divergence Times: A Penalized Likelihood Approach. Mol. Biol. Evol. 2002, 19, 101–109. [Google Scholar] [CrossRef]
- Pagel, M. Detecting correlated evolution on phylogenies: A general method for the comparative analysis of discrete characters. Proc. R. Soc. London. Ser. B Biol. Sci. 1994, 255, 37–45. [Google Scholar] [CrossRef]
- Schluter, D.; Price, T.; Mooers, A.Ø.; Ludwig, D. Likelihood of ancestor states in adaptive radiation. Evolution 1997, 51, 1699–1711. [Google Scholar] [CrossRef]
- Maintainer, L.J.R.; Revell, L.J. Package “Phytools” Title Phylogenetic Tools for Comparative Biology (and Other Things). R Package. 2020. Available online: https://besjournals.onlinelibrary.wiley.com/doi/full/10.1111/j.2041-210X.2011.00169.x (accessed on 12 November 2020).
- Huelsenbeck, J.P.; Nielsen, R.; Bollback, J.P. Stochastic Mapping of Morphological Characters. Syst. Biol. 2003, 52, 131–158. [Google Scholar] [CrossRef]
- Bollback, J.P. SIMMAP: Stochastic character mapping of discrete traits on phylogenies. BMC Bioinf. 2006, 7, 88. [Google Scholar] [CrossRef] [PubMed]
- Matzke, N.J. BioGeoBEARS: An R package for model testing and ancestral state estimation in historical biogeography. Methods Ecol. Evol. 2014. [Google Scholar]
- Matzke, N.J. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst. Biol. 2014, 63, 951–970. [Google Scholar] [CrossRef] [PubMed]
- Ree, R.H.; Smith, S.A. Maximum Likelihood Inference of Geographic Range Evolution by Dispersal, Local Extinction, and Cladogenesis. Syst. Biol. 2008, 57, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Castillo, T.; Hernández, H.J.; Pliscoff, P. Hotspots and ecoregion vulnerability driven by climate change velocity in Southern South America. Reg. Environ. Chang. 2020, 20, 1–15. [Google Scholar] [CrossRef]
- Cantidio, L.S.; Souza, A.F. Aridity, soil and biome stability influence plant ecoregions in the Atlantic Forest, a biodiversity hotspot in South America. Ecography 2019, 42, 1887–1898. [Google Scholar] [CrossRef]
- Martins, C.; Correa Bueno, O. Determinação de Cepas de Wolbachia em Populações Naturais de Solenopsis spp. (Hymenoptera: Formicidae) Analisadas via Multilocus Sequence Typing (MLST): Diversidade Genética, Coevolução e Recombinação; Universidade Estadual Paulista (UNESP): Rio Claro, Brazil, 2014. [Google Scholar]
- Comandatore, F.; Sassera, D.; Montagna, M.; Kumar, S.; Koutsovoulos, G.; Thomas, G.; Repton, C.; Babayan, S.A.; Gray, N.; Cordaux, R.; et al. Phylogenomics and Analysis of Shared Genes Suggest a Single Transition to Mutualism in Wolbachia of Nematodes. Genome Biol. Evol. 2013, 5, 1668–1674. [Google Scholar] [CrossRef][Green Version]
- Narita, S.; Shimajiri, Y.; Nomura, M. Strong cytoplasmic incompatibility and high vertical transmission rate can explain the high frequencies of Wolbachia infection in Japanese populations of Colias erate poliographus (Lepidoptera: Pieridae). Bull. Entomol. Res. 2009, 99, 385–391. [Google Scholar] [CrossRef]
- Gerth, M.; Röthe, J.; Bleidorn, C. Tracing horizontal Wolbachia movements among bees (Anthophila): A combined approach using multilocus sequence typing data and host phylogeny. Mol. Ecol. 2013, 22, 6149–6162. [Google Scholar] [CrossRef]
- Duplouy, A.; Couchoux, C.; Hanski, I.; van Nouhuys, S. Wolbachia Infection in a Natural Parasitoid Wasp Population. PLoS ONE 2015, 10, e0134843. [Google Scholar] [CrossRef]
- Bouwma, A.M.; Shoemaker, D. Wolbachia wSinvictaA Infections in Natural Populations of the Fire Ant Solenopsis invicta: Testing for Phenotypic Effects. J. Insect Sci. 2011, 11, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Tolley, S.J.A.; Nonacs, P.; Sapountzis, P. Wolbachia Horizontal Transmission Events in Ants: What Do We Know and What Can We Learn? Front. Microbiol. 2019, 10, 296. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.Z.; Araujo-Jnr, E.V.; Welch, J.J.; Kawahara, A.Y. Wolbachia in butterflies and moths: Geographic structure in infection frequency. Front. Zool. 2015, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.Z.; Breinholt, J.W.; Kawahara, A.Y. Evidence for common horizontal transmission of Wolbachia among butterflies and moths. BMC Evol. Biol. 2016, 16, 1–16. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).