Abstract
The vertebrate toll-like receptor (TLRs) supergene family is a first-line immune defense against viral and non-viral pathogens. Here, comparative evolutionary-genomics of 79 vertebrate species (8 mammals, 48 birds, 11 reptiles, 1 amphibian, and 11 fishes) revealed differential gain/loss of 26 TLRs, including 6 (TLR3, TLR7, TLR8, TLR14, TLR21, and TLR22) that originated early in vertebrate evolution before the diversification of Agnatha and Gnathostomata. Subsequent dynamic gene gain/loss led to lineage-specific diversification with TLR repertoires ranging from 8 subfamilies in birds to 20 in fishes. Lineage-specific loss of TLR8-9 and TLR13 in birds and gains of TLR6 and TLR10-12 in mammals and TLR19-20 and TLR23-27 in fishes. Among avian species, 5–10% of the sites were under positive selection (PS) (omega 1.5–2.5) with radical amino-acid changes likely affecting TLR structure/functionality. In non-viral TLR4 the 20 PS sites (posterior probability PP > 0.99) likely increased ability to cope with diversified ligands (e.g., lipopolysaccharide and lipoteichoic). For viral TLR7, 23 PS sites (PP > 0.99) possibly improved recognition of highly variable viral ssRNAs. Rapid evolution of the TLR supergene family reflects the host–pathogen arms race and the coevolution of ligands/receptors, which follows the premise that birds have been important vectors of zoonotic pathogens and reservoirs for viruses.
1. Introduction
The toll-like receptor (TLRs) supergene family members are type-I transmembrane glycoproteins belonging to the pathogen recognition receptor (PRRs) class of proteins expressed in the cell membrane and intracellular vesicles, including the endoplasmic reticulum, endosomes, lysosomes, and endolysosomes [1,2]. TLRs constitute one of the first lines of the immune defense system and recognize a variety of pathogen-associated-molecular patterns (PAMPs) during pathogen invasion, triggering the cascade of signaling pathways leading to the adaptive immune response [3,4,5]. The vertebrate TLRs studied to date are involved solely in immune response, in contrast with invertebrate TLR–like proteins, which are also associated with developmental functions [6].
The diversity of TLRs has facilitated the detection of a diverse group of pathogenic ligands [7]. The diverse mechanisms used by TLRs paralogs for TLR-ligand recognition and the formation of a m-shaped homo or heterodimeric complex, lead to activation of downstream signaling cascades by Toll/interleukin-1 receptor (TIR) domains [1,8,9,10,11,12,13,14,15]. The TIR dimer is recognized by the TIR domain present in different signaling adaptor proteins—including MyD88, MAL, TRIF, and TRAM [16]—and resulting in the activation of NFkB, the expression of various inflammatory and anti-pathogenic proteins [17], the initiation of the adaptive immune response, and the elimination of the invading pathogens [5,18,19,20].
Each TLR gene consists of a highly conserved intracellular (cytoplasmic) TIR domain, which is responsible for signal transduction [21], a conserved single transmembrane region and a variable extracellular domain (ECD), involved in the ligand recognition and dimerization. ECD consists of variable numbers (~16 to 28) of leucine-rich repeats (LRRs) motifs [22].
TLRs identified to date are grouped phylogenetically into six major families (TLR1, TLR3, TLR4, TLR5, TLR7, and TLR11) [23]. Gene gain and loss have been prominent features in the evolution of this supergene family. The large and variable number of TLR genes facilitate recognition of a wide variety of ligands from diverse pathogens (bacterial, fungal, protozoan, and virus). For example, recent studies have revealed important compensatory mechanisms of the TLR supergene family in both adaptive and innate response in the absence of major histocompatibility complex (MHC) II, CD4 and invariant chain (Ii) in cod fish [24], where TLRs act as an alternative to MHCII, a well-known conserved feature of the adaptive immune system of jawed vertebrates [24,25,26]. The innate immune receptors of the coelacanth are a mixture of mammalian- and teleost-specific TLRs, affirming the transitional position of coelacanth [27], evolutionarily connecting fish and tetrapods, along with its unique immune system lacking IgM [28].
We performed comparative evolutionary genomic analyses of vertebrate TLRs using whole genome sequencing data from 79 species (mammals, birds, reptiles, amphibians, and fishes) to understand the dynamics of gene gain and loss in shaping the TLR gene family repertoire in vertebrate genomes. The most important immunological function of TLRs comprehends protection from pathogens that requires the genes to evolve rapidly in response to selective pressure exerted by constantly evolving pathogens. Because avian species are often important vectors of zoonotic pathogens and reservoirs for viruses we analyzed avian TLRs (viral and non-viral TLRs) to determine how positive diversifying selection affecting the functional and structural variation of TLRs has improved pathogen recognition in birds. We discuss our findings in the context of host–pathogen interactions and the adaptive evolution of avian TLRs to diverse ecological conditions and adaptive requirements.
2. Materials and Methods
2.1. Genome Scan and Synteny Analysis of TLR Supergene Family
We used representative sequences of the vertebrate TLR1-27 as the query to exhaustively perform blast [29] searches to retrieve all available TLRs from 79 species, including the genomes of 48 birds [30,31] plus 31 other vertebrates which included 11 reptiles, a frog, and 11 fish genomes (Supplementary File 1), providing updated TLR information across vertebrate. TLRs, coding sequences retrieved from the Avian genome consortium [30,31] were used in subsequent adaptive evolution analysis. For the synteny analysis of TLRs we scanned vertebrate genomes (Supplementary File 2) for each TLR gene using extensive Blast searches [29] complemented with the Genomicus tool [32,33]. Gene loss can be hard to ascertain due to constraints imposed by levels of genome coverage [34]. Here we define gene loss as genes that are not found in a given assembly after extensive blast searches.
2.2. Phylogenetic Analysis
For the vertebrate TLR supergene family tree, sequences were aligned using ClustalW as implemented in MEGA5 [35] and were corrected manually. The tree was estimated with MEGA5 [35] software using the neighbor joining method with 1000 bootstrap replications. For the phylogenetic and adaptive analyses of the avian TLRs, one to one orthologs were aligned using Muscle [36], implemented in Seaview [37], and manually corrected. We chose the best substitution model based on the Akaike information criteria (AIC) in jModelTest 2 [38]. The sequences were used for maximum likelihood tree construction using PhyML [39] with 500 bootstrap replicates to check the robustness and reliability of the tree [40]. Sequences were tested for nucleotide substitution saturation using DAMBE 5 [41] by plotting the number of transition-transversion against the genetic distance using the F84 model [42], which allows for different equilibrium base frequency and transition-transversion rate bias for nucleotide substitution. The Xia test [43] implemented in DAMBE5 [41] was used to compare the index score (ISS) with critical score (ISS.C) at third and other codon positions to obtain estimates of saturation.
2.3. Gene Conversion and Recombination
The sequence alignments were tested for recombination using the online version of GARD (Genetic Algorithm for Recombination Detection) [44] available at http://www.datamonkey.org. The GENE-CONV [45] software was used for the detection of gene conversion events with 1000 permutation and Bonferroni corrected p-value cutoff of p < 0.01 and mismatch allowed (/g1 = 1).
2.4. Positive Selection in Avian TLRs
The important immunological function of TLRs, i.e., providing protection of host from pathogens, requires them to evolve rapidly in response to selective pressure exerted by rapidly and constantly evolving pathogens. In proteins, different functional sites experience distinct selection pressures with advantageous changes being positively selected due to adaptive benefits. The Avian TLRs are assumed to have similar ligands as those reported in mammalian TLRs [46,47]. In this study, we used multiple approaches to find signals of positive selection (i.e., ω > 1, assessed by comparing the dN-number of non-synonymous substitutions per non-synonymous sites with that of dS-synonymous substitutions per synonymous sites) in avian TLRs (TLR1A, TLR1B, TLR2A, TLR2B, TLR3, TLR4, TLR5, TLR7, TLR15). We excluded TLR21 from positive selection analysis as insufficient sequences were found. The avian TLR7 is reported to have been duplicated in a few passerine birds [48,49,50] while TLR5 is pseudogenized in others [46]. Therefore, the TLR5 pseudogene and duplicated TLR7 sequences were excluded from the positive selection analysis.
We employed codon models implemented in PAML [51,52,53] and Datamonkey [54] together with amino acid models in TreeSAAP [55]. CODEML in PAML [51] implements likelihood ratio tests (LRT) for comparisons of sophisticated nested site-specific models calculated as twice the difference of the log likelihood between the two models following Chi square distribution with the degrees of freedom corresponding to the difference in the number of parameters between the nested model, i.e., null model and alternate model (positive selection). A significant LRT implies that the null model is rejected and sites are under positive selection. We compared M1a (nearly neutral) vs. M2a (positive selection), and M7 (beta: assumes a beta distribution for 0 ≤ ω ≤ 1; therefore this model does not allow for positively selected sites) vs. M8 (beta&ω: same as the M7 plus an extra class of ω > 1, allowing positive selection) to find sites under positive selection. Bayes empirical Bayes (BEB) inferred the posterior probabilities of positive selected sites where higher PP mean high confidence. We also used the Hyphy package (http://www.hyphy.org) [56] (http://www.datamonkey.org/) [54,57], which provides several approaches for detection of positive selected sites, including single-likelihood ancestral counting (SLAC), fixed effects likelihood (FEL), random effects likelihood (REL) [58], mixed effects model of evolution (MEME) [59], fast unconstrained Bayesian approximation (FUBAR) [60], and an integrative approach. The SLAC model uses ancestral sequences reconstruction, FEL calculates site-by-site dN/dS without assuming a prior distribution. REL assumes a prior distribution across site, FUBAR ensures robustness against model misspecification, and MEME is most appropriate to detect episodic diversifying selection affecting individual codon sites. In addition, the integrative approach incorporates all sites detected by SLAC, FEL, REL, FUBAR, and MEME. Sites detected by two different methods are interpreted as having more support of positive selection [61,62,63].
Further support for our results was gained by the complementary protein level approach implemented in TreeSAAP [55]. It uses ancestral sequence reconstruction to detect the physiochemical properties changes of the amino acid replacements considering 31 amino acid properties. The amino acid replacement can lead to conservative or radical change in physiochemical properties. The positive radical changes can modify the protein structure and/or function and the number of radical changes at a site can be used as an indicator of the positive selection strength. To facilitate interpretation of the level of changes at a site we compared sites having six or more radical changes (defined as type I) with sites with less than six properties (defined as type II).
2.5. Comparison of Domain Architecture, Homology Modeling, and Structure Analysis across Vertebrate TLRs
The LRRfinder was used to predict the domain architecture and define locations of specific amino acid residues in the TLR protein domain. This was also verified, whenever possible using the Uniprot protein database (http://www.uniprot.org). The structure of each TLR was predicted using the CPHmodels 3.2 protein homology modeling server, which resulted in significant model structure for the complete regions of TLR5 and the ECD region of TLR1A, TLR2A, TLR2B, TLR3, TLR4, and TLR7. No significant structure was predicted for TLR1B and TLR15. All the highly significant positive selected sites were displayed on the respective predicted structures to show the potential functional or structural significance of specific amino acid residues. The residues were mapped onto the predicted structure using PyMOL (http://www.pymol.org).
3. Results
3.1. Dynamic Gene Gain and Loss Shapes Vertebrate TLR Supergene Family Repertoire
The dynamics of gene gain and loss have an important role in the evolution and diversification of gene families [64,65,66,67,68,69,70,71]. These lineage and species-specific changes are important evolutionary mechanisms of adaptation [72,73,74]. Comprehensive TLR data from varied vertebrate species/lineages is crucial for assessing the major evolutionary events influencing the diversification of the vertebrate TLR supergene family. To describe and interpret TLR gene family evolution across vertebrates, we assessed representatives from major vertebrate groups by comparing closely and distantly related species (e.g., in mammals we covered monotremes-egg laying platypus, marsupial-short-tailed opossum, to placental mammals). TLR data was collected from diverse vertebrates. These included birds [30,31] and eight divergent reptiles from diverse environments (aquatic, semi-aquatic, and terrestrial) and under unique adaptive pressures (pathogens), including three lizards (Anolis carolinensis, Pogona vitticeps, and Gekko Japonicus), two snakes (Python molurus bivittatus and Protobothrops mucrosquamatus), three turtles (Pelodiscus sinensis, Chrysemys picta bellii, and Chelonia mydas), and three crocodilians (Alligator mississippiensis, Gavialis gangeticus, and Crocodylus porosus). Together with mammals, amphibian, and fishes (see Table 1), our analyses revealed a comprehensive picture of the TLR supergene family evolution (Figure 1 and Figure 2).

Table 1.
The TLR supergene family repertoire, showing gene gain and loss in different vertebrate species.

Figure 1.
Phylogeny of vertebrate TLR supergene family from representative species (see Table 1). The NJ tree was made in MEGA5 with 1000 bootstrap replication.
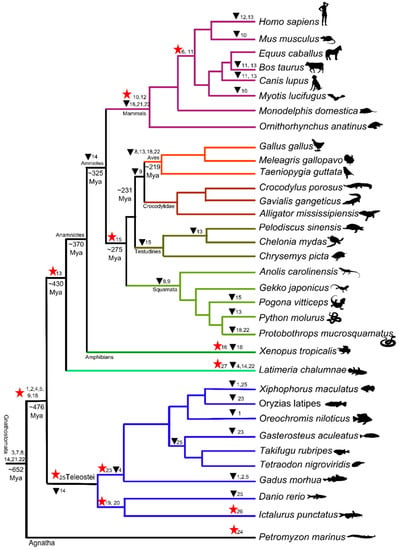
Figure 2.
Molecular evolution of vertebrate TLR gene family with events of TLR subfamily gains and losses shown on the consensus phylogeny made based on published literature. The dates were inferred using http://www.timetree.org/. The gains are represented by a star and the triangle shows loss of TLR subfamily.
The fishes had the highest number of TLR subfamilies (20 subfamilies) with only six subfamilies (TLR6, TLR10-TLR12, TLR15, and TLR16) missing in fishes. This showed that most of the TLR subfamilies originated early in the fish lineage. Fishes also had the highest number of genes derived from duplication events (Table 1 and Figure 2), with cod having 41 TLR gene copies. These patterns highlight the importance of differential gene gain, and its prominence among fish lineages (Table 1 and Figure 2).
The comparative genomics of the TLR supergene family showed lineage and species-specific distribution—e.g., TLR16 is frog-specific, TLR24 lamprey-specific, TLR26 catfish-specific, and TLR27 coelacanth-specific—whereas TLR11-12 are present only in mammals and TLR19-20 and TLR26 are only found in teleost fishes. The TLR18 and TLR22 were lost in both the avian and mammalian lineages, suggesting similar adaptive pressure (Table 1 and Figure 2). Loss of TLR15 and TLR21 in the mammalian lineage possibly occurred after the divergence of sauropsida from the synapsida lineage (Table 1 and Figure 2). Likewise, the finding of TLR13, TLR18, and TLR22, together with TLR15 and TLR21 in reptiles (Table 1) suggests that these TLRs are not limited to particular vertebrate species and lineages, as previously suggested [23,45]. Our study reveals that dynamic gene gain and loss of TLRs in vertebrate genomes facilitated vertebrate TLR supergene family evolution under diverse ecological requirements.
3.2. Synteny of TLR Supergene Family in Vertebrates
Synteny analysis has been fundamental in identifying conserved genomic arrangements and orthologous relationships. Synteny analyses highlighted the genomic organization of TLRs (Figure 3, Supplementary File 2 A-P) which helped evaluate and interpret TLR gene family evolution and levels of homology of TLR genes in different genomes. Lineage-specific TLRs, e.g., fish-specific and tetrapod-specific TLRs, showed highly conserved synteny. In contrast, the coelacanth showed shared features with both fish and tetrapods. Only TLR3, TLR5M, TLR7, and TLR8 genes showed conserved synteny across vertebrates (from fishes to mammals) (Figure 3).

Figure 3.
Genomic organization of TLRs clusters present in two or more classes of vertebrates. (a) Syntenic organization of TLR6, TLR1, and TLR10 genes whereas only TLR1 is widespread across all vertebrates; (b) Syntenic organization of TLR2 genes; (c) Syntenic organization of TLR3; (d) Syntenic organization of TLR4 only present in mammals, avians, and reptiles; (e) Syntenic organization of TLR5; (f) Syntenic organization of TLR7 and TLR8; (g) Syntenic organization of TLR9 which is absent in avian and reptilian; (h) Syntenic organization of TLR13 present in tetrapods (with exception of avian); (i) Syntenic organization of TLR15 exclusive of reptiles and avian; (j) Syntenic organization of TLR21.
3.3. Phylogenetic Analysis of Vertebrate TLR Supergene Family
The TLR data from diverse vertebrates (Figure 1 and Table 1) used for the phylogenetic reconstruction of the TLR supergene family strongly supports the classification of TLRs into six families [23], along with new subfamilies that shape the dynamic distribution of the TLR supergene family (Table 1 and Figure 1). Most of the new subfamilies belong to TLR family 1 and 11, as 10 subfamilies TLR1, TLR2, TLR6, TLR10, TLR14, TLR15, TLR18, TLR24, TLR25, and TLR27 grouped in TLR1, and 10 subfamilies TLR11-TLR13, TLR16, TLR19–TLR23, and TLR26 grouped in TLR11. In contrast, no new subfamilies gain in the remaining TLR families TLR3, TLR4, TLR5, and family TLR7 were recognized (subfamilies TLR7, TLR8, and TLR9).
We found that the TLR13 clade consists of two diverse groups, one with coelacanth and mammals (labeled as 13M) and another with amphibian and reptiles (labeled as 13X) (Figure 1), which were supported by high bootstrap values and our synteny analysis (Supplementary File 2 H1-H2). We clarified the dubious naming of TLR14, TLR18, and TLR25 and address this ambiguity by assessing synteny (genomic arrangement of gene and flanking genes) and relative phylogenetic relationships. Our phylogeny and syntenic analyses suggest that TLR14 [75], which was first described in Fugu, should also be placed within TLR18 with zebrafish and catfish and be renamed TLR18 (Supplementary File 2-I2). The genes that have been previously referred to as TLR18 in medaka, tilapia, and cod are syntenic and form a monophyletic clade with TLR25 from catfish [76] and should thus be classified as TLR25 (Figure 1 and Supplementary File 2P).
Since the frog TLR12 is more similar to the zebrafish TLR19 than the mammalian TLR12, and does not retain a conserved syntenic relationship with either TLR12 and TLR19, we retained it as TLR16 [23]. Overall, this TLR dataset, derived from an extended evaluation of additional newly sequenced genomes, provided an improved evolutionary assessment and a more accurate classification of the TLR supergene family phylogeny across vertebrates (Table 1, Figure 2).
Gene Conversion and Recombination
Gene conversion is common in tandem-duplicated gene paralogs located in close proximity. Gene conversion and recombination can bias phylogenetic inference and therefore care should be taken to identify signals of those events. The duplicated avian TLR gene paralogs of TLR1 and TLR2 are closely related to their orthologous counterpart from other species (i.e., within species paralogs TLR1A is closely related to TLR1B and TLR2A with TLR2B). This is caused by concerted evolution, possibly through gene conversion, leading to homogenization of the paralogs and reduced phylogenetic signals. Gene conversions have been reported in avian TLR1 and TLR2 [77]. TLR1 and TLR2 are closely located and are separated by a short physical distance (~12 kb and ~5 Kb between TLR1 and TLR2 duplicates, respectively) [46,64]. The C terminal of TLR1A/B ranging from LRR14 to the TIR domain is under gene conversion, whereas in TLR2A/B, the N terminal region (N terminal to LRR8) and C terminal region (LRR15 till TIR) are homogenized [46,64,77]. These homogenized regions may have important conserved functions [77].
3.4. Vertebrate TLRs Domain Architecture
TLRs consist of three major characteristic domains (extracellular domain (ECD), transmembrane domain (TM), and toll/interleukin-I receptor domain (TIR)). The one exception is TLR5S, which lacks the characteristic TIR domain. The TM domain connects the ECD with the cytoplasmic domain. The ECD is a solenoid shaped structure involved in interaction with PAMPs present in varied pathogens and is also involved in the formation of a M shaped homo and/or hetero dimer, which leads to signaling cascade by TIR activation [78].
The comparison of predicted architecture of ECD from various lineages of vertebrates: stickleback, coelacanth, Chinese soft-shell turtle, lizard, chicken (Figure 4, Supplementary File 3) [78] showed variable numbers of leucine rich repeats (LRRs). We also compared the domain architecture of TLRs from stickleback, coelacanth, Chinese soft shell turtle, Anolis lizard, and chicken and found a single domain in TLR 3, 5, 7, 8, 9, 13, 15, 18, 21, 22, and 27, whereas three domains were present in TLR 1, 2, and 4 (except in the coelacanth TLR1 exhibiting just a single domain) (Figure 4, Supplementary File 3).
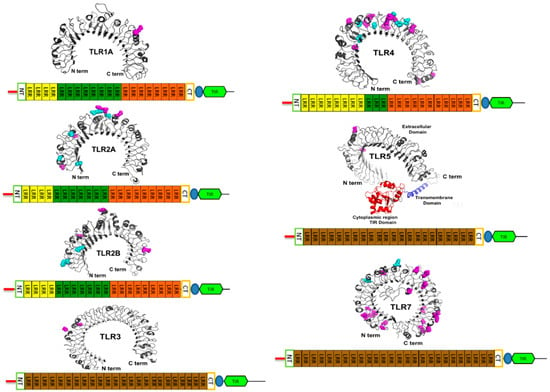
Figure 4.
Scheme of TLR domain architecture for avian TLRs and the positive selected sites with PP > 0.99 in M8 which also had PP > 90 in M2a are shown in the predicted structure of respective TLRs with magenta color. The site with cyan color represents type I changes detected by TreeSAAP. The numbering of positive selected sites in as per sequences given in Supplementary File 4 Tables (For TLR1B and TLR15 structure prediction was not significant).
Each LRR was around 20–30 amino acids long and had a conserved LxxLxLxxN leucine rich motif and a remaining variable region. The hydrophobic leucine residues constitute the conserved concave surface of the parallel beta strands forming the hydrophobic core, where asparagine is involved in the hydrogen bonding providing structural integrity [79]. Leucine can be replaced with other hydrophobic amino acids, whereas asparagine can also be replaced with other hydrogen donor’s like threonine, serine, and cysteine. The variable ‘x’ residues are responsible for the TLR function.
The exposed and convex surface of ECD is formed by the variable part of LRR repeat and this region is involved in PAMPs recognition. The cysteine clusters capping present in the terminal LRRs (LRR-NT and LRR-CT) protect the terminal hydrophobic residues. The human TLR1, 2, 4, 6, and 10 have all three domains, whereas TLR 3, 5, 7, 8, and 9 have a single domain [79]. This categorization is due to the interrupted asparagine in LRRs of the central domain, which results in structural flexibility, whereas uniform LRRs repeats with continuous asparagine results in single domain architecture [78]. The comparison of the domain architecture of representative TLRs from stickleback, coelacanth, Chinese soft shell turtle, Anolis lizard, and chicken was consistent with the human TLRs (Figure 4, Supplementary File 3).
3.5. Rapid Adaptive Evolution of Avian TLRs
We found variable numbers of positive selection sites among different TLR genes. The positive selection model M2a and M8 implemented in PAML4.7 [51] detected significant signals of positive selection in all genes (Table 2). The reliability of sites was well supported by the fact that most positive selected sites with PP > 0.99 under M8 also had PP > 0.90 in M2a. The M8 model found higher number of sites compared to the more conservative model M2a (Table 2). We found significant evidence of positive selection in all avian TLRs studied with different percentage of positively selected sites for different genes ranging from 11% in TLR15 to 5% in TLR7. The highest omega values were found in the TLR4, TLR2A, and TLR7 with 2.6, 2.6, and 2.5, respectively, while TLR15 had the lowest value of 1.5.

Table 2.
PAML results for nested site model comparisons for test of positive selection.
The number of positive selected sites with the highest (BEB) posterior probabilities (PP) also varied and the maximum number of sites with high PP were found in TLR7 (viral) and TLR4 (non-viral), which shows for the first time that both viral and non-viral TLR genes follow a positive selective regime (Table 2). In TLR7, Model M8 detected high numbers of positive selected (PS) sites (4.5% sites, total of 42 positively selected sites, 30 sites PP > 0.90, 25 sites PP > 0.95 and 23 sites PP > 0.99) and a somewhat similar scenario was found in TLR4 (5.5% sites, total of 45 sites, 29 sites PP > 0.90, 27 sites PP > 0.95 and 20 sites PP > 0.99). TLR3 had fewer sites with high PP (e.g., of 5% positive selected sites in TLR3 only five sites had PP > 0.90). Overall these results show that the number and strength of positive selection sites varies in the TLR supergene family and that both viral (TLR7) and non-viral (TLR4) TLRs evolved under strong positive selection.
The avian TLR1/TLR2 forms heterodimers that are activated by both diacyl (Malp-2) and triacyl (Pam3) lipopeptides [13,79,80], with the exception of TLR2a/ TLR1b, which are activated by Pam3 but not by Malp-2. In addition, TLR2a/TLRL1b is activated by peptidoglycan [77]. We found 12 positively-selected (PS) sites in TLR1A, 26 PS sites in TLR1B, 34 PS sites in TLR2A, and 25 PS sites in TLR2B. The previously reported PS site 304 in TLR2A [46,77] and PS sites 293, 295, and 296 found in TLR2B [46], corresponding to the chicken sequence, were also found in our study (Table 3).

Table 3.
Positive selected sites identified by various methods (PAML-M8, SLAC, FEL, REL, MEME, FUBAR, and Integrated).
Overall, of 12 sites in TLR1A (Table 2, Table 3 and Table 4, Figure 4, Supplementary File 4), 10 PS sites were present in LRRs, of which five were in the variable region and possibly involved in the PAMP binding. Only one site each was present in LRR-CT and TM. All 26 sites in TLR1B (Table 2, Table 3 and Table 4, Figure 4, Supplementary File 4) were located in ECD with 7 and 13 sites in conserved and variable regions, respectively, and 4 sites in LRR-NT and 2 in LRR-CT. In TLR2A, 2 of 34 sites (Table 2, Table 3 and Table 4, Supplementary File 4) were in the signal peptide, 4 in LRR-NT, and 17 of the remaining 28 sites present in LRRwere in the variable region. All of the 34 sites found in TLR2A were present in ECD with 2 sites in the signal peptide region, 4 sites in LRR-NT, and the other 28 in LRRs (21 in variable regions and remaining 7 in conserved regions). We found 25 sites in TLR2B (Table 2, Table 3 and Table 4, Figure 4, Supplementary File 4), all of them in LRRs with 14 in the variable region and 11 in the conserved region. The PS sites found in our study could be related with the a wide range of ligands as TLR2 recognizes a variety of compounds other than triacyl lipoproteins, including lipoteichoic acids, lipoarabinomannan, and zymosan [18]. Also, the formation of combinatorial binding sites by selection of TLR1 or TLR6 as the dimerization partner can explain, at least in part, the broad ligand specificity. A similar mechanism might explain the extent that PS sites found in TLR1 and TLR2 are shared with their counterparts in birds. The comparison of the chicken TLR2A and TLR2B genes with respect to the human TLR2s reveals that PS sites 280, 292, 304, 308, 309, 311, 312, 315, 335, 344, 356, 372, 392, 393, and 413 of TLR2A and PS sites 260, 274, 295, 297, 298, 299, 302, 317, 328, 329, 343, 390, and 401 of TLR2B map at or near to the ligand-binding domain and the dimerization surface, as identified from the crystal structure of the complex TLR2–TLR1 with the tri-acylated Pam3CSK4 lipoprotein in humans and mice [13,81].

Table 4.
TreeSAAP results for positive selected sites detected with PP > 0.99 in M8 model which also had PP > 0.90 in M2a. Type I radical changes detected by TreeSAAP are underlined.
TLR3 recognizes dsRNA and prevents the spread of most viruses. TLR3-ECD binds with dsRNA at two sites located at opposite ends of the TLR3 horseshoe structure. The first dsRNA:TLR3 interaction site is located close to the C-terminus, on LRR19-LRR21 and the second dsRNA:TLR3 interaction site is located on the N-terminal end (LRR-NT-LRR3) [82]. The intermolecular contact between the two C-terminal domain regions of TLR3 coordinates and stabilizes the dimer by a series of protein–protein interactions. Of 22 PS sites in TLR3 (Table 2, Table 3 and Table 4, Figure 4, Supplementary File 4), 2 are in signal peptides, 2 in LRR-NT, 2 in TM domains, 3 in the TIR domain, and 13 in LRRs. Of 13 sites in LRRs, 9 were in the variable region. The PS sites found in TLR3 likely favor the recognition and protection against rapidly evolving viral RNA [82].
TLR4 forms a heterodimer with myeloid differentiation factor 2 (MD-2) and recognizes diverse LPS molecules [83,84] along with components of yeast, trypanosoma, and even viruses [20,81,85]. The ECD of TLR4 consists of three subdomains. N subdomain consists of LRR-NT and LRR 1-6, the central subdomain ranges from LRR7-12 and the C subdomain consists of LRR 13-22 and LRR-CT. LPS causes dimerization of the TLR4-MD-2 complex at the central and/or the C-terminal, and interaction between TLR4 and LPS-MD-2 complex takes place at the concave surface between N and central sub-domain [84]. We found 41 PS sites in TLR4 (Table 2, Table 3 and Table 4, Figure 4, Supplementary File 4), three which were present in TM domain and one in TIR domain. The remaining 37 sites are present in ECD, with 3 in LRR-CT and the others 34 in LRRs with 23 of these in the variable region and the 11 others in the conserved region. The majority of the PS sites are present in regions involved in ligand binding and dimerization.
The TLR4 has a 330-amino-acid-long variable region in ECD which is also known as the middle region. Within the middle region there are 82 amino acids that are hyper-variable across species with numerous species-specific changes [86]. The majority of the PS sites found in TLR4 were concentrated near to the hyper-variable region. The primary contact interface between TLR4 and MD-2 involves two chemically distinct regions, the A and B patches provided by the N-terminal and the central domains of TLR4 and main dimerization interface of TLR4 located in the central C-terminal domain. The presence of positive selected sites in TLR4–MD-2–LPS complex may support the remarkable versatility of the ligand recognition mechanisms employed by the TLR family, which is essential for defense against diverse microbial infections [84,86].
The bacterial flagellin is a virulence factor recognized by TLR5 [87]. The residues 174–401 in ECD of TLR5 are responsible for species-specific flagellin recognition [88]. The alpha and epsilon Proteobacteria are able to evade TLR5 recognition by mutating key residues in the TLR5 recognition site [89] and it is suggested that positive selection in primate TLR5 may be related with coevolution between PRRs and their microbial ligands [81]. Twenty-six PS sites were found in TLR5 (Table 2, Table 3 and Table 4, Figure 4, Supplementary File 4), with two sites each in signal peptide, LRR-NT, TM and TIR domains, one site in LRR-CT and the remaining 17 sites in LRRs, of which seven were in the variable region and 10 in the conserved region. Seven of these PS sites were located in 228 amino acid region identified previously [88] and thus could play an important role in the species-specific flagellin recognition and defense.
TLR7 recognize single-stranded RNAs [90]. In TLR7, we found 4, 1 and 4 PS sites in LRR-NT, TM, and TIR domain, respectively, and 39 PS sites were present in LRRs of which 33 sites were in the variable region (Table 2, Table 3 and Table 4, Figure 4, Supplementary File 4). This variable region directly interacts with the ssRNA. Thus, the high proportion of PS sites in viral TLR7 is probably related with the host pathogen arms race. TLR7 is also known to be evolving under strong selection pressure in mammals [79,85]. Recent studies found TLR7 to be under positive selection in bats, which are natural reservoirs and carriers of numerous deadly viruses [91]. The long-term coexistence of bats and viruses imposed strong selective pressures on the bat genome, with strong positive selection occurring in genes like TLR7 involved in the innate immune system, the first line of anti-viral defense [92]. The rapid evolution of viral TLR and its associated role in maintaining and disseminating viruses (e.g., in bats), suggest that it might have a similar role in the rapid evolution of viral TLR7 in birds. The avian TLR7 is a candidate for the detection of influenza [93] and has evolved to recognize ssRNA (with very high mutation rates due to lack of mismatch repair), pointing towards the long coexistence of viruses and birds. This may help understand the role of birds as natural reservoirs and vectors of zoonotic pathogens [93].
TLR15s recognize yeast-derived agonists [94]. We found 41 PS sites (Table 3 and Supplementary File 4) with 1, 3, 4, 2, and 1 PS sites in the signal peptide, LRR-NT, LRR-CT, TM, and TIR domains, respectively, and 30 sites in LRRs, of which 14 were present in the variable region. None of the PS sites were located in the highly conserved three box regions of the TIR domain [95,96]. The exact mechanism of action of TLR15 is unknown but the presence of PS sites is in consistent with other TLRs involved in immune defense.
Evidence of positive-selected sites was further assessed using by multiple complementary approaches (SLAC, FEL, REL, MEME, and FUBAR) implemented in the HyPhy package (Table 3) (http://www.hyphy.org) [56] (http://www.datamonkey.org/) [54,57] in viral and non-viral avian TLRs (Table 3). Sites detected by more than one method (concordant of two or more methods) were thus considered to be robust candidates for positive selection (Table 3) [79]. We further implemented TreeSAAP [55] to detect positive radical changes in physiochemical properties of amino acids, which in turn can affect the structure and function of proteins. The TreeSAAP results also complemented our findings of positive selection (Table 4 and Figure 4), with most sites under selection belonging to type I (sites with six or more positive radical changes) or type II radical changes (sites with less than six positive radical changes).
Finally, homology modeling elucidated the role of the highly significant positive selected sites (Table 4) on the structural and functional diversification of TLRs (Figure 4). Since the majority of positive selected sites restricted to the variable extracellular domain (ECD) are involved in the ligand recognition and dimerization, this highlights their importance in the structure and function of these proteins (Figure 4). This strongly supports the hypothesis that the host–pathogen arms race drives the rapid evolution of TLRs.
4. Discussion
The vertebrate toll-like receptors (TLRs) supergene family constitutes the first line of immune defense against diverse pathogens and provides a fascinating example of the host–pathogen evolutionary arms race. Here, employing state-of-the-art DNA and protein level analyses, we provided a comprehensive comparative evolutionary genomics characterization of the vertebrate TLR supergene family using whole genome sequencing data from 79 species (mammals, birds, reptiles, amphibians, and fishes) together with strong positive selection of avian TLRs (viral and non-viral TLRs). Our results elucidated the dynamic evolution of TLR super gene family with different level of gene gain and loss leading to variable species and lineages specific TLR repertoire across vertebrates and positive selection analysis of avian TLRs showed extensive adaptive evolution shaping host pathogen arms race in the avian TLRs.
Our results show that the TLR supergene family is broadly divided into six major families, including TLR1, TLR3, TLR4, TLR5, TLR7, and TLR11, with each having different numbers of subfamilies—e.g., 10 subfamilies in TLR1, 10 subfamilies in TLR11, 3 subfamilies in TLR7, and a single subfamily in families TLR3, TLR4, and TLR5. We conclude that most of the TLRs originated early during vertebrate evolution and that genome duplication together with differential rate of gene gain and loss shaped the TLR gene family evolution in vertebrates, leading to species and lineage-specific TLR variations. We found that gene loss was common in the avian lineage, resulting in only eight extant subfamilies, the least observed among vertebrates. In contrast, 20 subfamilies were detected in fishes, reflecting the strong impact of gene duplication. The genomic scan of diverse reptilian genomes confirmed the presence of TLR13, TLR15, TLR18, TLR21, and TLR22 in reptiles (Table 1), supporting other evidence that these TLRs are widely distributed and not limited to a particular vertebrate species or group [23,45]. For example, TLR15 was not bird-specific and is also found in most reptilian genomes. Similarly, TLR21 was also found in reptiles, as well as fishes, frogs, and birds. The subfamilies TLR11-TLR12 are mammal-specific and subfamilies TLR19-20 and TLR23-27 are specific to fishes. More duplication events occurred in fishes (TLR2, TLR4, TLR5, TLR7, TLR8, TLR9, TLR14, TLR24, and TLR19-TLR23) compared with tetrapods (TLR1, TLR2, TLR5, TLR8, TLR13, and TLR14), clearly showing a reduction of the TLR supergene family repertoire of fishes relative to other vertebrates (Table 1), e.g., maximum of 41 TLR copies in cod genome among fishes compared to maximum 14 copies in mammals suggest the greater role of TLRs in fishes. The importance of TLRs in fishes is also supported by the loss of the MHCII, CD4, and invariant chain (Ii) in cod [24] and IgM in coelacanth [28], which suggests that TLRs have an important alternative and compensatory role in the fish lineage.
The synteny of TLR supports fish- and tetrapod-specific genomic arrangements, with the exception of subfamilies TLR3, TLR5M, TLR7, and TLR8 having conserved syntenic organization across all vertebrates. The coelacanth was an exception to this trend as its TLRs had features shared with both tetrapods (TLR1, TLR2, and TLR13) and fishes (TLR21), supporting its evolutionary proximity with both groups. This pattern also probably reflects its unique immune system, which lacks IgM as part of its adaptive immune system [27,28].
The important immunological function of TLRs—i.e., protection of host from pathogens—requires them to evolve rapidly in response to selective pressure exerted by rapidly and constantly evolving pathogens. Birds have been important vectors of zoonotic pathogens and reservoirs for viruses, thus the availability of avian genomes allowed us to test the adaptive evolution of avian TLRs responsible for both viral and non-viral pathogens.
Positive selection is one of the hallmarks of immune-defense-related genes [97,98] and especially those encoding recognition proteins, evolve under positive selection [99]. There is growing evidence of positive selected sites in TLR loci [46,50,65,76,86]. These positive-selected sites can provide increased number of advantageous variations, which is important for pathogen recognition and the host–pathogen arms-race that facilitate successful adaptation against changing environments and pathogens [99]. The use of different codon and protein-level approaches revealed extensive positive selected sites in birds TLRs and the homology modeling of these important changes in TLR proteins (Table 2, Table 3 and Table 4, Figure 4 and Supplementary File 4) confirms their possible effect on structural and functional diversification of the respective TLRs, and is discussed below.
Our results were further supported by protein level approaches. Most PS sites found under high PP > 0.99 (Table 4 and Figure 4), had the highest number of positive radical physiochemical changes. Of the 71 sites with high PP > 0.99 (Table 4), 59 PS sites showed positive radical changes in physiochemical properties (TreeSAAP categories 6, 7, and 8). We also found 15 PS sites with type-I radical changes (sites with six or more positive radical changes). TLR4 (non-viral) and TLR7 (viral) had the most sites with PP > 0.99 (Table 4). In TLR4 we found 20 such sites (Table 4 and Figure 4), among which 17 showed type II changes (sites with less than six positive radical changes) and 6 PS sites showed type I changes. Most of the type I changes were restricted to TLR2A (5 sites) and TLR4 (6 sites) (Table 4 and Figure 4). In TLR7 we found 23 sites with PP > 0.99, among which 19 showed type II changes and two PS sites showed type I changes (Table 4). The homology modeling showed that the majority of sites having PP > 0.99 were found in the ECD with only few (4 sites) found outside the ECD (Figure 4)—e.g., site 16 from TLR2A, sites 640 and 655 from TLR4, and site 920 from TLR7—were found in the signal peptide, TM domain, and TIR domain of the respective gene.
Overall positive selection in the avian TLRs suggests that the host pathogen arms race has played an important role in the rapid evolution of the avian TLRs immune genes to adapt against pathogens. Our finding suggests rapid positive selection in both viral and non-viral TLRs (Table 3) with non-viral TLR4 having the highest PS sites (20 sites with highest PP > 0.99) and among viral TLRs, TLR7 had the highest number of PS sites (23 sites with PP > 0.99). The majority of PS sites with type I and II positive radical changes were located in the LRRs of ECD, which is mainly involved in PAMPs recognition. The finding of non-viral TLR4 and viral TLR7 genes with highest number of PS cites coincides with similar finding for TLR7 [87] and non-viral TLR4 [92] in mammals. The role of non-viral TLR4 in recognizing diverse ligands (e.g., LPS-lipopolysaccharide and LTA-lipoteichoic) and TLR7 in recognizing ssRNA and possibly influenza [94] may be linked with the mechanism of how birds maintain and disseminate numerous deadly viruses [95].
These findings support the premise that the host–pathogen arms race led to co-evolution and possibly explains the strong selective pressure imposed by the long-term coexistence of viruses and birds and is consistent with the proposed role of birds as natural reservoirs and vectors of zoonotic pathogens [100]. The rapid evolution of the TLR supergene family that we have documented using newly-available bird genomes fits well with other evidence that these immune genes experienced strong adaptive selection due to the rapid evolution of pathogens [101] and that the host–pathogen arms race increases the chance of developing novel protection against diverse species of pathogens.
5. Conclusions
This study revealed lineage and species-specific differences in the distribution of the TLR supergene family among vertebrates (n = 79 species), with varied rates of gene gain/loss resulting in different repertoires of TLRs that would have facilitated recognition and protection from diverse pathogens. Our results provide strong support for the rapid evolution of both viral and non-viral avian TLRs and strengthens the hypothesis that the long-term coexistence of birds and viruses contributed to the strong selective pressure observed in the viral TLR immune genes. The patterns of gene gain, gene loss, and positive selection in the TLR gene superfamily provides compelling support of the co-evolutionary host–pathogen arms race.
Supplementary Materials
The following are available online at https://www.mdpi.com/1424-2818/11/8/131/s1, Supplementary file 1: Table S1—Information of gene location used from vertebrates genomes as in Table 1, Sequences used for phylogeny of vertebrates TLRs, Avian sequences used in this study; Supplementary file 2: Synteny information of vertebrates TLRs; Supplementary file 3: Comparative domain architecture of vertebrates TLRs; Supplementary file 4: Table S2—Domain characterization of TLR1A, Table S3—Domain characterization of TLR1B, Table S4—Domain characterization of TLR2A, Table S5—Domain characterization of TLR2B, Table S6—Domain characterization of TLR3, Table S7—Domain characterization of TLR4, Table S8—Domain characterization of TLR5, Table S9—Domain characterization of TLR7, Table S10—Domain characterization of TLR15.
Author Contributions
Conceptualization, I.K. and A.A.; Methodology, I.K., E.M., and L.S.; Software, I.K. and E.M.; Validation, I.K., E.M., L.S., D.A., and A.A.; Formal analysis, I.K., E.M., L.S., and D.A.; Resources, A.A.; Data curation, I.K. and E.M.; Writing—original draft preparation, I.K.; Writing—review and editing, I.K., E.M., L.S., D.A., W.E.J., S.J.O., G.Z., E.D.J., M.T.P.G., and A.A.; Supervision and design, A.A.; Funding acquisition, A.A.
Funding
I.K., D.A. and L.S. were funded with a PhD grant from “Fundação para a Ciência e a Tecnologia” (FCT) (IK: SFRH/BD/48518/2008, DA: SFRH/BD/79766/2011, LS: SFRH/BD/134565/2017). W.E.J. prepared portions of this manuscript while holding a National Research Council Research Associateship Award at the Walter Reed Army Institute of Research and the published material reflects the views of the authors and should not be construed to represent those of the Department of the Army or the Department of Defense. S.J.O. was supported, in part, by the Russian Science Foundation grant (project no. 17-14-01138) and by St. Petersburg State University (Genome Russia Grant no. 1.52.1647.2016). E.D.J. was supported by the Howard Hughes Medical Institute. A.A. was partially supported by the Strategic Funding UID/Multi/04423/2019 through national funds provided by FCT and the European Regional Development Fund (ERDF) in the framework of the program PT2020, by the European Structural and Investment Funds (ESIF) through the Competitiveness and Internationalization Operational Program—COMPETE 2020 and by National Funds through the FCT under the project PTDC/CTA-AMB/31774/2017 (POCI-01-0145-FEDER/031774/2017).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen Recognition and Innate Immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001, 1, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Schnare, M.; Barton, G.M.; Holt, A.C.; Takeda, K.; Akira, S.; Medzhitov, R. Toll-like receptors control activation of adaptive immune responses. Nat. Immunol. 2001, 2, 947–950. [Google Scholar] [CrossRef]
- Iwasaki, A.; Medzhitov, R. Regulation of adaptive immunity by the innate immune system. Science 2010, 327, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Imler, J.L.; Hoffmann, J.A. Toll Receptors in Drosophila: A Family of Molecules Regulating Development and Immunity. Curr. Top. Microbiol. Immunol. 2002, 270, 63–79. [Google Scholar]
- O’Neill, L.A.J.; Golenbock, D.; Bowie, A.G. The history of Toll-like receptors—Redefining innate immunity. Nat. Rev. Immunol. 2013, 13, 453–460. [Google Scholar] [CrossRef]
- Janeway, C.A.J.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef]
- Takeuchi, O.; Sato, S.; Horiuchi, T.; Hoshino, K.; Takeda, K.; Dong, Z.; Modlin, R.L.; Akira, S. Cutting edge: Role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. 2002, 169, 10–14. [Google Scholar] [CrossRef]
- Gantner, B.N.; Simmons, R.M.; Canavera, S.J.; Akira, S.; Underhill, D.M. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J. Exp. Med. 2003, 197, 1107–1117. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. TLR signaling. Cell Death Differ. 2006, 13, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, I.; Medzhitov, R. Two Modes of Ligand Recognition by TLRs. Cell 2007, 130, 979–981. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jin, M.; Kim, S.; Heo, J. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 2007, 130, 1071–1082. [Google Scholar]
- Jin, M.S.; Lee, J.O. Structures of TLR-ligand complexes. Curr. Opin. Immunol. 2008, 20, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Botos, I.; Segal, D.M.; Davies, D.R. The Structural Biology of Toll-Like Receptors. Structure 2011, 19, 447–459. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.J.; Bowie, A.G. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 2007, 7, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Doyle, S.L.; O’Neill, L.A.J. Toll-like receptors: From the discovery of NFkappaB to new insights into transcriptional regulations in innate immunity. Biochem. Pharmacol. 2006, 72, 1102–1113. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Akira, S. Microbial recognition by Toll-like receptors. J. Dermatol. Sci. 2004, 34, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R. Recognition of microorganisms and activation of the immune response. Nature 2007, 449, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Kawai, T.; Akira, S. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009, 388, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Preston-Hurlburt, P.; Janeway, C.A. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 1997, 388, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, N.; Tanaka, T.; Enkhbayar, P.; Mikami, T.; Taga, M.; Yamada, K.; Kuroki, Y. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC Genom. 2007, 8, 124. [Google Scholar] [CrossRef] [PubMed]
- Roach, J.C.; Glusman, G.; Rowen, L.; Kaur, A.; Purcell, M.K.; Smith, K.D.; Hood, L.E.; Aderem, A. The evolution of vertebrate Toll-like receptors. Proc. Natl. Acad. Sci. USA 2005, 102, 9577–9582. [Google Scholar] [CrossRef] [PubMed]
- Star, B.; Nederbragt, A.J.; Jentoft, S.; Grimholt, U.; Malmstrøm, M.; Gregers, T.F.; Rounge, T.B.; Paulsen, J.; Solbakken, M.H.; Sharma, A.; et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature 2011, 477, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Flajnik, M.F.; Kasahara, M. Origin and evolution of the adaptive immune system: Genetic events and selective pressures. Nat. Rev. Genet. 2010, 11, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Litman, G.W.; Rast, J.P.; Fugmann, S.D. The origins of vertebrate adaptive immunity. Nat. Rev. Immunol. 2010, 10, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Boudinot, P.; Zou, J.; Ota, T.; Buonocore, F.; Scapigliati, G.; Canapa, A.; Cannon, J.; Litman, G.; Hansen, J.D. A tetrapod-like repertoire of innate immune receptors and effectors for coelacanths. J. Exp. Zool. Part B Mol. Dev. Evol. 2014, 322, 415–437. [Google Scholar] [CrossRef] [PubMed]
- Amemiya, C.T.; Alföldi, J.; Lee, A.P.; Fan, S.; Philippe, H.; MacCallum, I.; Braasch, I.; Manousaki, T.; Schneider, I.; Rohner, N.; et al. The African coelacanth genome provides insights into tetrapod evolution. Nature 2013, 496, 311–316. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Jarvis, E.D.; Mirarab, S.; Aberer, A.J.; Li, B.; Houde, P.; Li, C.; Ho, S.Y.W.; Faircloth, B.C.; Nabholz, B.; Howard, J.T.; et al. Whole Genome Analyses Resolve Early Branches in the Tree of Life of Modern Birds. Science 2014, 346, 1320–1331. [Google Scholar] [CrossRef]
- Zhang, G.; Li, C.; Li, Q.; Li, B.; Larkin, D.M.; Lee, C.; Storz, J.F.; Antunes, A.; Greenwold, M.J.; Meredith, R.W.; et al. Comparative Genomics Reveals Insights into Avian Genome Evolution and Adaptation. Science 2014, 346, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Muffato, M.; Louis, A.; Poisnel, C.E.; Crollius, H.R. Genomicus: A database and a browser to study gene synteny in modern and ancestral genomes. Bioinformatics 2010, 26, 1119–1121. [Google Scholar] [CrossRef]
- Louis, A.; Muffato, M.; Roest Crollius, H. Genomicus: Five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res. 2012, 41, D700–D705. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.; Vasconcelos, V.M.; Antunes, A. The phosphoprotein phosphatase family of Ser/Thr phosphatases as principal targets of naturally occurring toxins. Crit. Rev. Toxicol. 2011, 41, 83–110. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Xia, X. DAMBE5: A Comprehensive Software Package for Data Analysis in Molecular Biology and Evolution. Mol. Biol. Evol. 2013, 30, 1720–1728. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Rannala, B. Phylogenetic Methods Come of Age: Testing Hypotheses in an Evolutionary Context. Science 1997, 276, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Xie, Z.; Salemi, M.; Chen, L.; Wang, Y. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 2003, 26, 1–7. [Google Scholar] [CrossRef]
- Pond, S.L.K.; Posada, D.; Gravenor, M.B.; Woelk, C.H.; Frost, S.D.W. GARD: A genetic algorithm for recombination detection. Bioinformatics 2006, 22, 3096–3098. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, S. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 1989, 6, 526–538. [Google Scholar]
- Alcaide, M.; Edwards, S.V. Molecular Evolution of the Toll-Like Receptor Multigene Family in Birds. Mol. Biol. Evol. 2011, 28, 1703–1715. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.R.; Vasconcelos, V.; Antunes, A. Computational Study of the Covalent Bonding of Microcystins to Cysteine Residues—A Reaction Involved in the Inhibition of the PPP Family of Protein Phosphatases. FEBS J. 2013, 280, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Cormican, P.; Lloyd, A.T.; Downing, T.; Connell, S.J.; Bradley, D.; O’Farrelly, C. The avian Toll-Like receptor pathway—Subtle differences amidst general conformity. Dev. Comp. Immunol. 2009, 33, 967–973. [Google Scholar] [CrossRef]
- Grueber, C.E.; Wallis, G.P.; King, T.M.; Jamieson, I.G. Variation at Innate Immunity Toll-Like Receptor Genes in a Bottlenecked Population of a New Zealand Robin. PLoS ONE 2012, 7, e45011. [Google Scholar] [CrossRef]
- Velová, H.; Gutowska-Ding, M.W.; Burt, D.W.; Vinkler, M. Toll-like receptor evolution in birds: Gene duplication, pseudogenisation and diversifying selection. Mol. Biol. Evol. 2018, 35, 2170–2184. [Google Scholar] [CrossRef]
- Yang, Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Bioinformatics 1997, 13, 555–556. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, E.; Almeida, D.; Escalona, T.; Khan, I.; Vasconcelos, V.; Antunes, A. LMAP: Lightweight Multigene Analyses in PAML. BMC Bioinform. 2016, 17, 354. [Google Scholar] [CrossRef] [PubMed]
- Pond, S.L.K.; Frost, S.D.W.; Pond, S.L.K. Datamonkey: Rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 2005, 21, 2531–2533. [Google Scholar] [CrossRef] [PubMed]
- Woolley, S.; Johnson, J.; Smith, M.J.; Crandall, K.A.; McClellan, D.A. TreeSAAP: Selection on Amino Acid Properties using phylogenetic trees. Bioinformatics 2003, 19, 671–672. [Google Scholar] [CrossRef] [PubMed]
- Pond, S.L.K.; Muse, S.V. HyPhy: Hypothesis testing using phylogenies. Bioinformatics 2005, 21, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Delport, W.; Poon, A.F.Y.; Frost, S.D.W.; Kosakovsky Pond, S.L. Datamonkey 2010: A suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 2010, 26, 2455–2457. [Google Scholar] [CrossRef] [PubMed]
- Pond, S.L.K.; Frost, S.D.W. Not So Different After All: A Comparison of Methods for Detecting Amino Acid Sites Under Selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Pond, S.L.K. Detecting Individual Sites Subject to Episodic Diversifying Selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Pond, S.L.K.; Scheffler, K. FUBAR: A Fast, Unconstrained Bayesian AppRoximation for Inferring Selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef]
- Fry, B.G.; Undheim, E.A.B.; Ali, S.A.; Debono, J.; Scheib, H.; Ruder, T.; Jackson, T.N.W.; Morgenstern, D.; Cadwallader, L.; Whitehead, D.; et al. Squeezers and leaf-cutters: Differential diversification and degeneration of the venom system in toxicoferan reptiles. Mol. Cell. Proteom. 2013, 12, 1881–1899. [Google Scholar] [CrossRef]
- Undheim, E.A.B.; Sunagar, K.; Herzig, V.; Kely, L.; Low, D.H.W.; Jackson, T.N.W.; Jones, A.; Kurniawan, N.; King, G.F.; Ali, S.A.; et al. A Proteomics and Transcriptomics Investigation of the Venom from the Barychelid Spider Trittame loki (Brush-Foot Trapdoor). Toxins 2013, 5, 2488–2503. [Google Scholar] [CrossRef] [PubMed]
- Sunagar, K.; Fry, B.; Jackson, T.N.W.; Casewell, N.R.; Undheim, E.A.B.; Vidal, N.; Ali, S.A.; King, G.F.; Vasudevan, K.; Vasconcelos, V.; et al. Molecular evolution of vertebrate neurotrophins: Co-option of the highly conserved nerve growth factor gene into the advanced snake venom arsenal. PLoS ONE 2013, 8, e81827. [Google Scholar] [CrossRef]
- Temperley, N.D.; Berlin, S.; Paton, I.R.; Griffin, D.K.; Burt, D.W. Evolution of the chicken Toll-like receptor gene family: A story of gene gain and gene loss. BMC Genom. 2008, 9, 62. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, A.Y.; Kiron, V.; Dopazo, J.; Fernandes, J.M. Diversification of the expanded teleost-specific toll-like receptor family in Atlantic cod, Gadus morhua. BMC Evol. Biol. 2012, 12, 256. [Google Scholar] [CrossRef] [PubMed]
- Blomme, T.; Vandepoele, K.; De Bodt, S.; Simillion, C.; Maere, S.; Van De Peer, Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006, 7, R43. [Google Scholar] [CrossRef] [PubMed]
- DeMuth, J.P.; De Bie, T.; Stajich, J.E.; Cristianini, N.; Hahn, M.W. The Evolution of Mammalian Gene Families. PLoS ONE 2006, 1, e85. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.; Maldonado, E.; Khan, I.; Silva, L.; Gilbert, M.T.P.; Zhang, G.; Jarvis, E.D.; O’Brien, S.J.; Johnson, W.E.; Antunes, A. Whole-Genome Identification, Phylogeny, and Evolution of the Cytochrome P450 Family 2 (CYP2) Subfamilies in Birds. Genome Biol. Evol. 2016, 8, 1115–1131. [Google Scholar] [CrossRef] [PubMed]
- da Fonseca, R.; Antunes, A.; Melo, A.; Ramos, M.J. Structural divergence and adaptive evolution in mammalian cytochromes P450 2C. Gene 2007, 387, 58–66. [Google Scholar] [CrossRef]
- Machado, J.P.; Johnson, E.W.; O’Brien, S.J.; Vasconcelos, V.; Antunes, A. Adaptive evolution of the matrix extracellular phosphoglycoprotein in mammals. BMC Evol. Biol. 2011, 11, 342. [Google Scholar] [CrossRef]
- Khan, I.; Maldonado, E.; Vasconcelos, V.; O’Brien, S.J.; Johnson, W.E.; Antunes, A. Mammalian keratin associated proteins (KRTAPs) subgenomes: Disentangling hair diversity and adaptation to terrestrial and aquatic environments. BMC Genom. 2014, 15, 779. [Google Scholar] [CrossRef]
- Khan, I.; Yang, Z.; Maldonado, E.; Li, C.; Zhang, G.; Gilbert, M.T.P.; Jarvis, E.D.; O’Brien, S.J.; Johnson, W.E.; Antunes, A. Olfactory Receptor Subgenomes Linked with Broad Ecological Adaptations in Sauropsida. Mol. Biol. Evol. 2015, 32, 2832–2843. [Google Scholar] [CrossRef]
- Silva, L.; Antunes, A. Vomeronasal Receptors in Vertebrates and the Evolution of Pheromone Detection. Annu. Rev. Anim. Biosci. 2017, 5, 353–370. [Google Scholar] [CrossRef]
- Luo, S.J.; Johnson, W.E.; Martenson, J.; Antunes, A.; Martelli, P.; Uphyrkina, O.; Traylor-Holzer, K.; Smith, J.L.; O’Brien, S.J. Subspecies Genetic Assignments of Worldwide Captive Tigers Increase Conservation Value of Captive Populations. Curr. Biol. 2008, 18, 592–596. [Google Scholar] [CrossRef]
- Kasamatsu, J.; Oshiumi, H.; Matsumoto, M.; Kasahara, M.; Seya, T. Phylogenetic and expression analysis of lamprey toll-like receptors. Dev. Comp. Immunol. 2010, 34, 855–865. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, S.; Rajendran, K.; Sun, L.; Zhang, Y.; Sun, F.; Kucuktas, H.; Liu, H.; Liu, Z. Pathogen recognition receptors in channel catfish: III Phylogeny and expression analysis of Toll-like receptors. Dev. Comp. Immunol. 2013, 40, 185–194. [Google Scholar] [CrossRef]
- Huang, Y.; Temperley, N.D.; Ren, L.; Smith, J.; Li, N.; Burt, D.W. Molecular evolution of the vertebrate TLR1 gene family—A complex history of gene duplication, gene conversion, positive selection and co-evolution. BMC Evol. Biol. 2011, 11, 149. [Google Scholar] [CrossRef]
- Kang, J.Y.; Lee, J.O. Structural Biology of the Toll-Like Receptor Family. Annu. Rev. Biochem. 2011, 80, 917–941. [Google Scholar] [CrossRef]
- Kang, J.Y.; Nan, X.; Jin, M.S.; Youn, S.J.; Ryu, Y.H.; Mah, S.; Han, S.H.; Lee, H.; Paik, S.G.; Lee, J.O. Recognition of Lipopeptide Patterns by Toll-like Receptor 2-Toll-like Receptor 6 Heterodimer. Immunity 2009, 31, 873–884. [Google Scholar] [CrossRef]
- Guan, Y.; Ranoa, D.R.E.; Jiang, S.; Mutha, S.K.; Li, X.; Baudry, J.; Tapping, R.I. Human TLRs 10 and 1 share common mechanisms of innate immune sensing but not signaling. J. Immunol. 2010, 184, 5094–5103. [Google Scholar] [CrossRef]
- Wlasiuk, G.; Nachman, M.W. Adaptation and Constraint at Toll-Like Receptors in Primates. Mol. Biol. Evol. 2010, 27, 2172–2186. [Google Scholar] [CrossRef]
- Liu, L.; Botos, I.; Wang, Y.; Leonard, J.N.; Shiloach, J.; Segal, D.M.; Davies, D.R. Structural basis of Toll-Like Receptor 3 Signaling with double-stranded RNA. Science 2008, 320, 379–381. [Google Scholar] [CrossRef]
- Kim, H.M.; Park, B.S.; Kim, J.I.; Kim, S.E.; Lee, J.; Oh, S.C.; Enkhbayar, P.; Matsushima, N.; Lee, H.; Yoo, O.J.; et al. Crystal Structure of the TLR4-MD-2 Complex with Bound Endotoxin Antagonist Eritoran. Cell 2007, 130, 906–917. [Google Scholar] [CrossRef]
- Park, B.S.; Song, D.H.; Kim, H.M.; Choi, B.S.; Lee, H.; Lee, J.O. The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature 2009, 458, 1191–1195. [Google Scholar] [CrossRef]
- Kumar, H.; Kawai, T.; Akira, S. Pathogen recognition in the innate immune response. Biochem. J. 2009, 420, 1–16. [Google Scholar] [CrossRef]
- Hajjar, A.M.; Ernst, R.K.; Tsai, J.H.; Wilson, C.B.; Miller, S.I. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat. Immunol. 2002, 3, 354–359. [Google Scholar] [CrossRef]
- Hayashi, F.; Smith, K.D.; Ozinsky, A.; Hawn, T.R.; Yi, E.C.; Goodlett, D.R.; Eng, J.K.; Akira, S.; Underhill, D.M.; Aderem, A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor. Nature 2001, 410, 1099–1103. [Google Scholar] [CrossRef]
- Andersen-Nissen, E.; Smith, K.D.; Bonneau, R.; Strong, R.K.; Aderem, A. A conserved surface on Toll-like receptor 5 recognizes bacterial flagellin. J. Exp. Med. 2007, 204, 393–403. [Google Scholar] [CrossRef]
- Andersen-Nissen, E.; Smith, K.D.; Strobe, K.L.; Barrett, S.L.R.; Cookson, B.T.; Logan, S.M.; Aderem, A. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc. Natl. Acad. Sci. USA 2005, 102, 9247–9252. [Google Scholar] [CrossRef]
- Gantier, M.P.; Tong, S.; Behlke, M.A.; Xu, D.; Phipps, S.; Foster, P.S.; Williams, B.R.G. TLR7 is involved in sequence-specific sensing of single-stranded RNAs in human macrophages. J. Immunol. 2008, 180, 2117–2124. [Google Scholar] [CrossRef]
- Wang, L.F.; Walker, P.J.; Poon, L.L. Mass extinctions, biodiversity and mitochondrial function: Are bats ‘special’ as reservoirs for emerging viruses? Curr. Opin. Virol. 2011, 1, 649–657. [Google Scholar] [CrossRef]
- Zhang, G.; Cowled, C.; Shi, Z.; Huang, Z.; Bishop-Lilly, K.A.; Fang, X.; Wynne, J.W.; Xiong, Z.; Baker, M.L.; Zhao, W.; et al. Comparative Analysis of Bat Genomes Provides Insight into the Evolution of Flight and Immunity. Science 2013, 339, 456–460. [Google Scholar] [CrossRef]
- Lund, J.M.; Alexopoulou, L.; Sato, A.; Karow, M.; Adams, N.C.; Gale, N.W.; Iwasaki, A.; Flavell, R.A. Recognition of single-stranded RNA viruses by Toll-like receptor. Proc. Natl. Acad. Sci. USA 2004, 101, 5598–5603. [Google Scholar] [CrossRef]
- Boyd, A.C.; Peroval, M.Y.; Hammond, J.A.; Prickett, M.D.; Young, J.R.; Smith, A.L. TLR15 Is Unique to Avian and Reptilian Lineages and Recognizes a Yeast-Derived Agonist. J. Immunol. 2012, 189, 4930–4938. [Google Scholar] [CrossRef]
- Areal, H.; Abrantes, J.; Esteves, P.J. Signatures of positive selection in Toll-like receptor (TLR) genes in mammals. BMC Evol. Biol. 2011, 11, 368. [Google Scholar] [CrossRef]
- Slack, J.L. Identification of Two Major Sites in the Type I Interleukin-1 Receptor Cytoplasmic Region Responsible for Coupling to Pro-inflammatory Signaling Pathways. J. Biol. Chem. 2000, 275, 4670–4678. [Google Scholar] [CrossRef]
- Lahn, B.T.; Vallender, E.J. Positive selection on the human genome. Hum. Mol. Genet. 2004, 13 (Suppl. S2), R245–R254. [Google Scholar]
- Sackton, T.B.; Lazzaro, B.P.; A Schlenke, T.; Evans, J.D.; Hultmark, D.; Clark, A.G. Dynamic evolution of the innate immune system in Drosophila. Nat. Genet. 2007, 39, 1461–1468. [Google Scholar] [CrossRef]
- Kosiol, C.; Vinar, T.; Da Fonseca, R.R.; Hubisz, M.J.; Bustamante, C.D.; Nielsen, R.; Siepel, A. Patterns of Positive Selection in Six Mammalian Genomes. PLoS Genet. 2008, 4, e1000144. [Google Scholar] [CrossRef]
- Reed, K.D.; Meece, J.K.; Henkel, J.S.; Shukla, S.K. Birds, Migration and Emerging Zoonoses: West Nile Virus, Lyme Disease, Influenza A and Enteropathogens. Clin. Med. Res. 2003, 1, 5–12. [Google Scholar] [CrossRef]
- Nielsen, R.; Bustamante, C.; Clark, A.G.; Glanowski, S.; Sackton, T.B.; Hubisz, M.J.; Fledel-Alon, A.; Tanenbaum, D.M.; Civello, D.; White, T.J.; et al. A Scan for Positively Selected Genes in the Genomes of Humans and Chimpanzees. PLoS Biol. 2005, 3, e170. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).