Marine Invertebrates: Underexplored Sources of Bacteria Producing Biologically Active Molecules
Abstract
1. Introduction
2. Relationships between Microorganisms and Marine Invertebrates
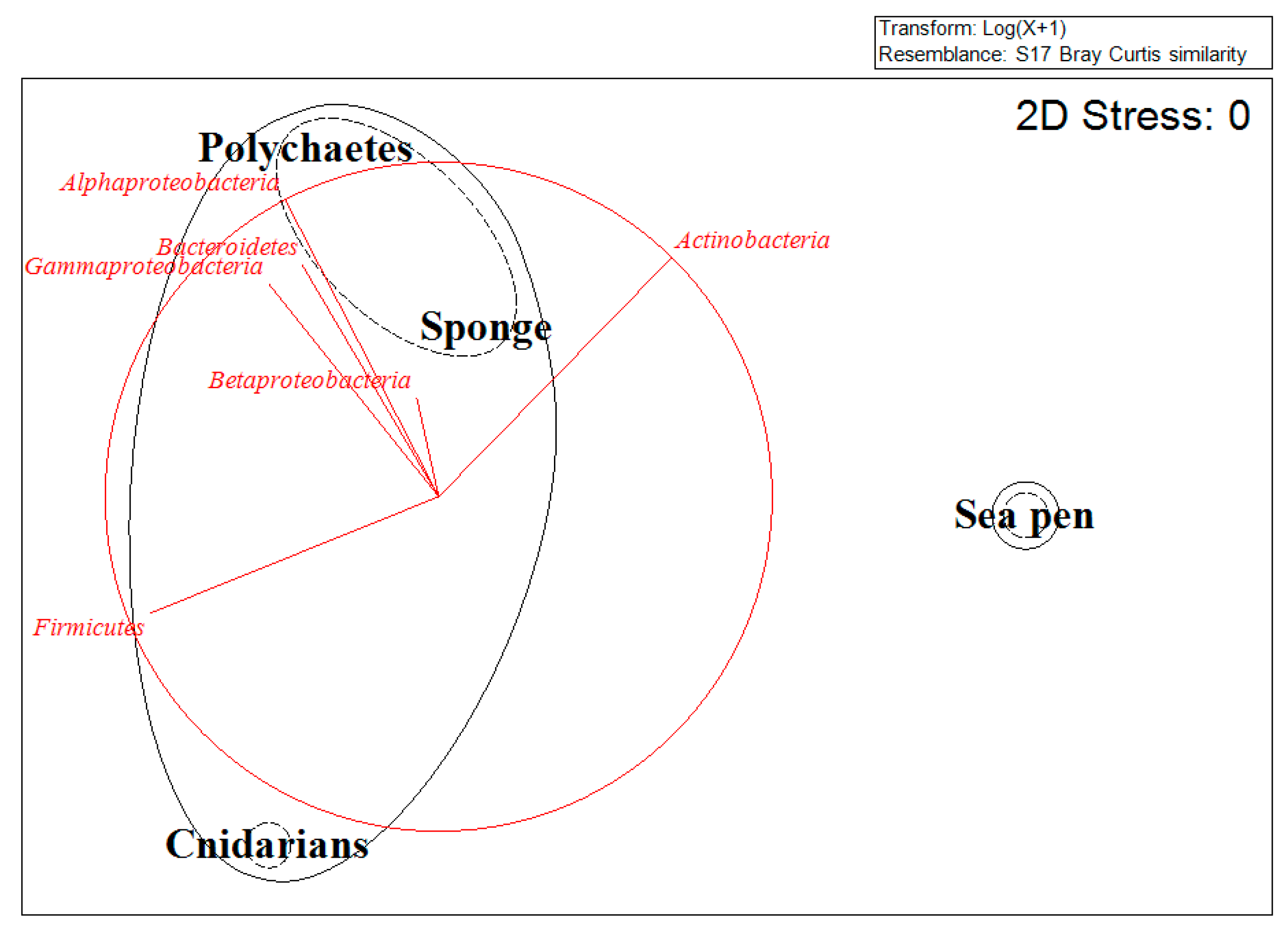
2.1. Mutualistic Symbiosis
2.2. Commensalistic Symbiosis
2.3. Parasitic Symbiosis
2.4. Synergism and Sintrofia
3. Marine Invertebrates Involved in Microbial Association Relationships
3.1. Porifera
3.2. Annellids
3.3. Cnidaria
4. Production of Bioactive Molecules by Bacteria Associated with Marine Invertebrates
4.1. Biosurfactants
4.1.1. Main Screening Methods Applied for the Search of Biosurfactants from Marine Invertebrates
4.1.2. Phylogenetic Affiliation of BS-Producing Bacteria from Marine Invertebrates
4.1.3. Applications of BSs Produced by Bacteria Associated with Marine Invertebrates
4.2. Extracellular Polymeric Substances (EPSs)
Applications of EPSs Produced by Bacteria Associated with Marine Invertebrates
4.3. Biomedicals
5. Concluding Remarks
Conflicts of Interest
References
- Blunt, J.W.; Copp, B.R.; Munro, M.H.G.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2011, 28, 196–268. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, C.M.; Kearns, P.S.; Evans-Illidge, E.; Kurtböke, D.I. Diversity and bioactivity of marine bacteria associated with the pponges Candidaspongia flabellata and Rhopaloeides odorabile from the Great Barrier Reef in Australia. Diversity 2017, 9, 39. [Google Scholar] [CrossRef]
- Jensen, P.R.; Fenical, W. Marine microorganisms and drug discovery: Current status and future potential. In Drugs from the Sea; Fusetani, N., Ed.; Karger: Basel, Switzerland, 2000; pp. 6–29. [Google Scholar]
- Skariyachan, S.; Rao, A.G.; Patil, M.R.; Saikia, B.; Bharadwaj, K.N.; Rao, G.S. Antimicrobial potential of metabolites extracted from bacterial symbionts associated with marine sponges in coastal area of gulf of mannar biosphere, India. Lett. Appl. Microbiol. 2013, 58, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Hoffmeister, M.; Martin, W. Interspecific evolution: Microbial symbiosis, endosymbiosis and gene transfer. Environ. Microbiol. 2003, 5, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Webster, N.S. Communication, cooperation and coevolution: Grand challenges in microbial symbiosis research. Front. Microbiol. 2014, 5, 164. [Google Scholar] [CrossRef] [PubMed]
- Horn, M.; Collingro, A.; Schmitz-Esser, S.; Beier, C.L.; Purkhold, U.; Fartmann, B.; Brandt, P.; Nyakatura, G.J.; Droege, M.; Frishman, D.; et al. Illuminating the evolutionary history of Chlamydiae. Science 2004, 304, 728–730. [Google Scholar] [CrossRef] [PubMed]
- Kojima, A.; Hirose, E. Transmission of cyanobacterial symbionts during embryogenesis in the coral reef ascidians Trididemnum nubilum and T. clinides (Didemnidae, Ascidiacea, Chordata). Biol. Bull. 2012, 222, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Usher, K.M.; Kuo, J.; Fromont, J.; Sutton, D.C. Vertical transmission of cyanobacterial symbionts in the marine sponge Chondrilla australiensis (Demospongiae). Hydrobiologia 2001, 461, 9–13. [Google Scholar] [CrossRef]
- Webster, N.; Taylor, M.W.; Behnam, F.; Lücker, S.; Rattei, T.; Whalan, S.; Horn, M.; Wagner, M. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. 2010, 12, 2070–2082. [Google Scholar] [CrossRef] [PubMed]
- Bright, M.; Bulgheresi, S. A complex journey: Transmission of microbial symbionts. Nat. Rev. Microbiol. 2010, 8, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Cordes, E.E.; Arthur, M.A.; Shea, K.; Arvidson, R.S.; Fisher, C.R. Modeling the mutualistic interactions between tubeworms and microbial consortia. PLoS Biol. 2005, 3, e77. [Google Scholar] [CrossRef] [PubMed]
- Bull, A.T.; Stach, J.E.M. Marine actinobacteria: New opportunities for natural product search and discovery. Trends Microbiol. 2007, 15, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, L.M.; Blanco, J.A.; Baz, J.P.; Puentes, J.L.; Millan, F.R.; Vazquez, F.E.; Chimeno, R.I.; Grávalos, D.G. 4′-N-methyl-5′-hydroxystaurosporine and 50hydroxystaurosporine, new indolocarbazole alkaloids from a marine Micromonospora sp. strain. J. Antibiot. (Tokyo) 2000, 53, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Schneemann, I.; Nagel, K.; Kajahn, I.; Labes, A.; Wiese, J.; Imhoff, J.F. Comprehensive investigation of marine Actinobacteria associated with the sponge Halichondriapanicea. J. Appl. Microbiol. 2010, 76, 3702–3714. [Google Scholar] [CrossRef] [PubMed]
- Dubilier, N.; Bergin, C.; Lott, C. Symbiotic diversity in marine animals: The art of harnessing chemosynthesis. Nat. Rev. Microbiol. 2008, 6, 725–740. [Google Scholar] [CrossRef] [PubMed]
- McFall-Ngai, M. Host-microbe symbiosis: The squid-Vibrio association—A naturally occurring, experimental model of animal/bacterial partnerships. Adv. Exp. Med. Biol. 2008, 635, 102–112. [Google Scholar] [PubMed]
- Anoop, A.; Antunes, A. Pyrosequencing characterization of the microbiota from Atlantic intertidal marine sponges reveals high microbial diversity and the lack of co-occurrence patterns. PLoS ONE 2015, 10, e0127455. [Google Scholar]
- Bourne, D.G.; Dennis, P.G.; Uthicke, S.; Soo, R.M.; Tyson, G.W.; Webster, N. Coral reef invertebrate microbiomes correlate with the presence of photosymbionts. ISME J. 2013, 7, 1452–1458. [Google Scholar] [CrossRef] [PubMed]
- Glasl, B.; Herndl, G.J.; Frade, P.R. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 2016, 10, 2280–2292. [Google Scholar] [CrossRef] [PubMed]
- Lokmer, A.; Mathias Wegner, K. Hemolymph microbiome of Pacific oysters in response to temperature, temperature stress and infection. ISME J. 2015, 9, 670–682. [Google Scholar] [CrossRef] [PubMed]
- Hogan, C. Commensalism, the Encyclopedia of Earth. 2012. Available online: http://www.eoearth.org/view/article/171918 (accessed on 16 June 2014).
- McCoy, W.M.; Holdredge, C.; Silliman, B.R.; Altieri, A.H.; Thomsen, M.S. Facilitation. In Encyclopedia of Theoretical Ecology; University of California Press: Berkeley, CA, USA, 2012; pp. 276–280. [Google Scholar]
- Vannier-Santos, M.A.; Lenzi, H.L. Parasites or Cohabitants: Cruel Omnipresent Usurpers or Creative “Eminences Grises”? J. Parasitol. Res. 2011, 214174. [Google Scholar] [CrossRef] [PubMed]
- Grossart, H.P.; Riemann, L.; Tang, K.W. Molecular and functional ecology of aquatic microbial symbionts. Front. Microbiol. 2013, 4, 59. [Google Scholar] [CrossRef] [PubMed]
- Borchiellini, C.; Manuel, M.; Alivon, E.; Boury-Esnault, N.; Vacelet, J.; Le Parco, Y. Sponge paraphyly and the origin of metazoa. J. Evol. Biol. 2001, 14, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Hooper, J.N.A.; van Soest, R.W.M. Systema Porifera: A Guide to the Classification of Sponges; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2002. [Google Scholar]
- Pile, A.J.; Patterson, M.R.; Witman, J.D. In situ grazing on plankton <10μm by the boreal sponge Mycale lingua. Mar. Ecol. Prog. Ser. 1996, 141, 95–102. [Google Scholar]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Boil. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [PubMed]
- Bavestrello, G.; Arillo, A.; Calcinai, B.; Cattaneo-Vietti, R.; Cerrano, C.; Gaino, E.; Penna, A.; Sara, M. Parasitic diatoms inside Antarctic sponges. Biol. Bull. 2000, 198, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Webster, N.S.; Negri, A.P.; Webb, R.I.; Hill, R.T. A sponginboring alpha-proteobacterium is the etiological agent of disease in the Great Barrier Reef sponge Rhopaloeides odorabile. Mar. Ecol. Prog. Ser. 2002, 232, 305–309. [Google Scholar] [CrossRef]
- Giles, E.C.; Kamke, J.; Moitinho-Silva, L.; Taylor, M.W.; Hentschel, U.; Ravasi, T.; Schmitt, S. Bacterial community profiles in low microbial abundance sponges. FEMS Microbiol. Ecol. 2013, 83, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Fieseler, L.; Horn, M.; Wagner, M.; Hentschel, U. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl. Environ. Microbiol. 2004, 70, 3724–3732. [Google Scholar] [CrossRef] [PubMed]
- Webster, N.S.; Hill, R.T. The culturable microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile is dominated by an α-Proteobacterium. Mar. Biol. 2001, 138, 843–851. [Google Scholar] [CrossRef]
- Hentschel, U.; Hopke, J.; Horn, M.; Friedrich, A.B.; Wagner, M.; Hacker, J.; Moore, B.S. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 2002, 68, 4431–4440. [Google Scholar] [CrossRef] [PubMed]
- Alex, A.; Silva, V.; Vasconcelos, V.; Antunes, A. Evidence of unique and generalist microbes in distantly related sympatric intertidal marine sponges (Porifera: Demospongiae). PLoS ONE 2013, 8, e80653. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.; Flemer, B.; Jackson, S.A.; Morrissey, J.P.; O’Gara, F.; Dobson, A.D. Evidence of a putative deep sea specific microbiome in marine sponges. PLoS ONE 2014, 9, e91092. [Google Scholar] [CrossRef] [PubMed]
- Simister, R.L.; Deines, P.; Botté, E.S.; Webster, N.S.; Taylor, M.W. Sponge-specific clusters revisited: A comprehensive phylogeny of sponge-associated microorganisms. Environ. Microbiol. 2012, 14, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Marconi, S.; De la Iglesia, R.; Díez, B.; Fonseca, C.A.; Hajdu, E.; Trefault, N. Characterization of Bacterial, Archaeal and Eukaryote Symbionts from Antarctic sponges reveals a high diversity at a three-domain level and a particular signature for this ecosystem. PLoS ONE 2015, 10, e0138837. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, S.; Tsai, P.; Bell, J.; Fromont, J.; Ilan, M.; Lindquist, N.; Perez, T.; Rodrigo, A.; Schupp, P.J.; Vacelet, J.; et al. Assessing the complex sponge microbiota: Core, variable and species-specific bacterial communities in marine sponges. ISME J. 2012, 6, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Reynolds, D.; Liua, M.; Stark, M.; Kjelleberg, S.; Webster, N.S.; Torsten, T. Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc. Natl. Acad. Sci. USA 2012, 109, 1878–1887. [Google Scholar] [CrossRef] [PubMed]
- Lee, O.O.; Wang, Y.; Yang, J.; Lafi, F.F.; Al-Suwailem, A.; Qian, P.Y. Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the Red Sea. ISME J. 2011, 5, 650–664. [Google Scholar] [CrossRef] [PubMed]
- Struck, T.H.; Schult, N.; Kusen, T.; Hickman, E.; Bleidorn, C.; McHugh, D.; Halanych, K.M. Annelid phylogeny and the status of Sipuncula and Echiura. BMC Evol. Biol. 2007, 7, 57. [Google Scholar] [CrossRef] [PubMed]
- Stabili, L.; Licciano, M.; Giangrande, A.; Fanelli, G.; Cavallo, R.A. Sabella spallanzanii filter-feeding on bacterial community: Ecological implications and applications. Mar. Environ. Res. 2006, 61, 74–92. [Google Scholar] [CrossRef] [PubMed]
- Licciano, M.; Stabili, L.; Giangrande, A. Clearance rates of Sabella spallanzanii and Branchiommaluctuosum (Annelida: Polychaeta) on a pure culture of Vibrio alginolyticus. Water Res. 2005, 39, 4375–4384. [Google Scholar] [CrossRef] [PubMed]
- Licciano, M.; Stabili, L.; Giangrande, A.; Cavallo, R.A. Bacterial accumulation by Branchiommaluctuosum (Annelida: Polychaeta): A tool for biomonitoring marine systems and restoring polluted waters. Mar. Environ. Res. 2007, 63, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Neave, M.J.; Streten-Joyce, C.; Glasby, C.J.; McGuinness, K.A.; Parry, D.L.; Gibb, K.S. The bacterial community associated with the marine polychaete Ophelina sp.1 (Annelida: Opheliidae) is altered by copper and zinc contamination in sediments. Microb. Ecol. 2012, 63, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Shankar, C.V.S.; Satheesh, S.; Viju, N.; Punitha, S.M.J. Antibacterial and biofilm inhibitory activities of bacteria associated with polychaetes. J. Coast Life Med. 2015, 3, 495–502. [Google Scholar]
- Haddad, A.; Camacho, F.; Durand, P.; Cary, S.C. Phylogenetic characterization of the epibiotic bacteria associated with the hydrothermal vent Polychaete Alvinella pompejana. Appl. Environ. Microbiol. 1995, 61, 1679–1687. [Google Scholar] [PubMed]
- Cary, S.C.; Cottrell, M.T.; Stein, J.L.; Camacho, F.; Desbruyères, D. Molecular identification and localization of a filamentous symbiotic bacteria associated with the hydrothermal vent annelid, Alvinella pompejana. Appl. Environ. Microbiol. 1997, 63, 1124–1130. [Google Scholar] [PubMed]
- Campbell, B.J.; Cary, S.C. Characterization of a novel spirochete associated with the hydrothermal vent polychaete annelid, Alvinella pompejana. Appl. Environ. Microbiol. 2001, 67, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, M.T.; Cary, S.C. Diversity of dissimilatory bisulfite reductase genes of bacteria associated with the deep-sea hydrothermal vent polychaete annelid Alvinella pompejana. Appl. Environ. Microbiol. 1999, 65, 1127–1132. [Google Scholar] [PubMed]
- Jeanthon, C.; Prieur, D. Susceptibility to heavy metals and characterization of heterotrophic bacteria isolated from two hydrothermal vent polychaete annelids, Alvinella pompejana and Alvinella caudata. Appl. Environ. Microbiol. 1990, 56, 3308–3314. [Google Scholar] [PubMed]
- Alain, K.; Olagnon, M.; Desbruyères, D.; Pagé, A.; Barbier, G.; Juniper, S.K.; Quérellou, J.; Cambon-Bonavita, M.-A. Phylogenetic characterization of the bacterial assemblage accociated with mucous secretions of the hydrothermal vent polychaete Paralvinella palmiformis. FEMS Microbiol. Ecol. 2002, 42, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Takai, K. Deep-sea vent chemoautotrophs: Diversity, biochemistry and ecological significance. FEMS Microbiol. Ecol. 2008, 65, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Desbruyères, D.; Chevaldonné, P.; Alayse, A.M.; Jollivet, D.; Lallier, F.H.; Jouin-Toulmond, C.; Zal, F.; Sarradin, P.M.; Cosson, R.; Caprais, J.C.; et al. Biology and ecology of the “Pompeii worm” (Alvinella pompejana Desbruyères and Laubier), a normal dweller of an extreme deep-sea environment: A synthesis of current knowledge and recent developments. Deep Sea Res. II Top Stud. Oceanogr. 1998, 45, 383–422. [Google Scholar] [CrossRef]
- Csotonyi, J.T.; Stackebrandt, E.; Yurkov, V. Anaerobic respiration on tellurate and other metalloids in bacteria from hydrothermal vent fields in the eastern Pacific Ocean. Appl. Environ. Microbiol. 2006, 72, 4950–4956. [Google Scholar] [CrossRef] [PubMed]
- Vincent, P.; Pignet, P.; Talmont, F.; Bozzi, L.; Fournet, B.; Guezennec, J.; Jeanthon, C.; Prieuri, D. Production and characterization of an exopolysaccharide excreted by a deep-sea hydrothermal vent bacterium isolated from the Polychaete Annelid Alvinella pompejana. Appl. Environ. Microbiol. 1994, 60, 4134–4141. [Google Scholar] [PubMed]
- Robidart, J.C.; Bench, S.R.; Feldman, R.A.; Novoradovsky, A.; Podell, S.B.; Gaasterland, T.; Allen, E.E.; Felbeck, H. Metabolic versatility of the Riftiapachyptila endosymbiont revealed through metagenomics. Environ. Microbiol. 2008, 10, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, H.; Gu, J.-D. Phylogenetic diversity and axial distribution of microbes in the intestinal tract of the polychaete Neanthes glandicincta. Microb. Ecol. 2009, 58, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Knowlton, N.; Rohwer, F. Multispecies microbial mutualisms on coral reefs: The host as a habitat. Am. Nat. 2003, 162, S51–S62. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, T.D.; Krause, L.; Bridge, T.; Torda, G.; Raina, J.-B.; Zakrzewski, M.; Gates, R.D.; Padilla-Gamino, J.L.; Spalding, H.L.; Smith, C.; et al. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 2015, 9, 2261–2274. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Agreda, A.; Leggat, W.; Bongaerts, P.; Ainsworth, T.D. The microbial signature provides insight into the mechanistic basis of coral success across reef habitats. mBio 2016, 7, e00560-16. [Google Scholar] [CrossRef] [PubMed]
- van de Water, J.A.J.M.; Melkonian, R.; Voolstra, C.R.; Junca, H.; Beraud, E.; Allemand, D.; Ferrier-Pagès, C. Comparative assessment of Mediterranean gorgonian associated microbial communities reveals conserved core and locally variant bacteria. Microb. Ecol. 2017, 73, 466–478. [Google Scholar] [CrossRef] [PubMed]
- van de Water, J.A.J.M.; Voolstra, C.R.; Rottier, C.; Cocito, S.; Peirano, A.; Allemand, D.; Ferrier-Pagès, C. Seasonal stability in the microbiomes of temperate gorgonians and the red coral Corallium rubrum across the Mediterranean Sea. Microb. Ecol. 2018, 75, 1–15. [Google Scholar] [CrossRef] [PubMed]
- McFall-Ngai, M.; Hadfield, M.G.; Bosch, T.C.G.; Carey, H.V.; Domazet-Lošo, T.; Douglas, A.E.; Dubilier, N.; Eberl, G.; Fukami, T.; Gilbert, S.F.; et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA 2013, 110, 3229–3236. [Google Scholar] [CrossRef] [PubMed]
- Raina, J.-B.; Tapiolas, D.; Willis, B.L.; Bourne, D.G. Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 2009, 75, 3492–3501. [Google Scholar] [CrossRef] [PubMed]
- Kvennefors, E.C.; Sampayo, E.; Kerr, C.; Vieira, G.; Roff, G.; Barnes, A.C. Regulation of bacterial communities through antimicrobial activity by the coral holobiont. Microb. Ecol. 2012, 63, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Bednarz, V.N.; Grover, R.; Maguer, J.-F.; Fine, M.; Ferrier-Pagès, C. The assimilation of diazotroph-derived nitrogen by Scleractinian corals depends on their metabolic status. mBio 2017, 8, e02058-16. [Google Scholar] [CrossRef] [PubMed]
- Bourne, D.G.; Garren, M.; Work, T.M.; Rosenberg, E.; Smith, G.W.; Harvell, C.D. Microbial disease and the coral holobiont. Trends Microbiol. 2009, 17, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, T.D.; Thurber, R.V.; Gates, R.D. The future of coral reefs: A microbial perspective. Trends Ecol. Evol. 2010, 25, 233. [Google Scholar] [CrossRef] [PubMed]
- Mouchka, M.E.; Hewson, I.; Harvell, C.D. Coral-associated bacterial assemblages: Current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 2010, 50, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Bayer, T.; Arif, C.; Ferrier-Pagès, C.; Zoccola, D.; Aranda, M.; Voolstra, C. Bacteria of the genus Endozoicomonas dominate the microbiome of the Mediterranean gorgonian coral Eunicella cavolini. Mar. Ecol. Prog. Ser. 2013, 479, 75–84. [Google Scholar] [CrossRef]
- Vezzulli, L.; Pezzati, E.; Huete-Stauffer, C.; Pruzzo, C.; Cerrano, C. 16SrDNA pyrosequencing of the Mediterranean gorgonian Paramuricea clavata reveals a link among alterations in bacterial holobiont members, anthropogenic influence and disease outbreaks. PLoS ONE 2013, 8, e67745. [Google Scholar] [CrossRef] [PubMed]
- La Rivière, M.; Roumagnac, M.; Garrabou, J.; Bally, M. Transient shifts in bacterial communities associated with the temperate gorgonian Paramuricea clavata in the northwestern Mediterranean Sea. PLoS ONE 2013, 8, e57385. [Google Scholar] [CrossRef] [PubMed]
- La Rivière, M.; Garrabou, J.; Bally, M. Evidence for host specificity among dominant bacterial symbionts in temperate gorgonian corals. Coral Reefs 2015, 34, 1087–1098. [Google Scholar] [CrossRef]
- Ransome, E.; Rowley, S.J.; Thomas, S.; Tait, K.; Munn, C.B. Disturbance to conserved bacterial communities in the cold-water gorgonian coral Eunicella verrucosa. FEMS Microbiol. Ecol. 2014, 90, 404–416. [Google Scholar] [PubMed]
- Van de Water, J.A.J.M.; Melkonian, R.; Junca, H.; Voolstra, C.R.; Reynaud, S.; Allemand, D.; Ferrier-Pagès, C. Spirochaetes dominate the microbial community associated with the red coral Corallium rubrum on a broad geographic scale. Sci. Rep. 2016, 6, 27277. [Google Scholar] [CrossRef] [PubMed]
- Penn, K.; Wu, D.; Eisen, J.A.; Ward, N. Characterization of bacterial communities associated with deep-sea corals on Gulf of Alaska seamounts. Appl. Environ. Microbiol. 2006, 72, 1680–1683. [Google Scholar] [CrossRef] [PubMed]
- Webster, N.S.; Bourne, D. Bacterial community structure associated with the antarctic soft coral, Alcyonium antarcticum. FEMS Microbiol. Ecol. 2007, 59, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Sunagawa, S.; Woodley, C.M.; Medina, M. Threatened corals provide underexplored microbial habitats. PLoS ONE 2010, 5, e9554. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.A.; Stone, R.P.; McLaughlin, M.R.; Kellogg, C.A. Microbial consortia of gorgonian corals from the Aleutian Islands. FEMS Microbiol. Ecol. 2011, 76, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Robertson, V.; Haltli, B.; McCauley, E.P.; Overy, D.P.; Kerr, R.G. Highly variable bacterial communities associated with the Octocoral Antillogorgiaelisabethae. Microorganisms 2016, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Correa, H.; Haltli, B.; Duque, C.; Kerr, R. Bacterial communities of the gorgonian octocoral Pseudopterogorgia elisabethae. Microb. Ecol. 2013, 66, 972–985. [Google Scholar] [CrossRef] [PubMed]
- Van de Water, J.A.J.M.; Allemand, D.; Ferrier-Pagès, C. Host-microbe interactions in octocoral holobionts-recent advances and perspectives. Microbiome 2018, 6, 64. [Google Scholar] [CrossRef] [PubMed]
- Rohwer, F.; Breitbart, M.; Jara, J.; Azam, F.; Knowlton, N. Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 2001, 20, 86–91. [Google Scholar]
- Rohwer, F.; Knowlton, N. Diversity and distribution of coral-associated bacteria. Mar Ecol. Prog. Ser. 2002, 243, 1–10. [Google Scholar] [CrossRef]
- Rosenberg, E.; Koren, O.; Reshef, L.; Efrony, R.; Zilber-Rosenberg, I. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 2007, 5, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Neulinger, S.C.; Jarnegren, J.; Ludvigsen, M.; Lochte, K.; Dullo, W.C. Phenotype-specific bacterial communities in the cold-water coral Lophelia pertusa (Scleractinia) and their implications for the coral’s nutrition, health, and distribution. Appl. Environ. Microbiol. 2008, 74, 7272–7285. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, C.A.; Lisle, J.T.; Galkiewicz, J.P. Culture-independent characterization of bacterial communities associated with the cold-water coral Lophelia purtusa in the north eastern Gulf of Mexico. Appl. Environ. Microbiol. 2009, 75, 2294–2303. [Google Scholar] [CrossRef] [PubMed]
- Littman, R.A.; Willis, B.L.; Bourne, D.G. Bacterial communities of juvenile corals infected with different Symbiodinium (dinoflagellate) clades. Mar. Ecol. Prog. Ser. 2009, 389, 45–59. [Google Scholar] [CrossRef]
- Kvennefors, E.C.; Sampayo, E.; Ridgway, T.; Barnes, A.C.; Hoegh-Guldberg, O. Bacterial communities of two ubiquitous Great Barrier Reef corals reveals both site- and species-specificity of common bacterial associates. PLoS ONE 2010, 5, e10401. [Google Scholar] [CrossRef] [PubMed]
- Sweet, M.J.; Croquer, A.; Bythell, J.C. Dynamics of bacterial community development in the reef coral Acropora muricata following experimental antibiotic treatment. Coral Reefs 2011, 30, 1121–1133. [Google Scholar] [CrossRef]
- Roder, C.; Bayer, T.; Aranda, M.; Kruse, M.; Voolstra, C.R. Microbiome structure of the fungid coral Ctenactis echinata aligns with environmental differences. Mol. Ecol. 2015, 24, 3501–3511. [Google Scholar] [CrossRef] [PubMed]
- Brück, T.B.; Brück, W.M.; Santiago-Vázquez, L.Z.; McCarthy, P.J.; Kerr, R.G. Diversity of the bacterial communities associated with the azooxanthellate deep water octocorals Leptogorgia minimata, Iciligorgia schrammi, and Swiftiaexertia. Mar. Biotechnol. (N. Y.) 2007, 9, 561–576. [Google Scholar] [CrossRef] [PubMed]
- Gil-Agudelo, D.L.; Myers, C.; Smith, G.W.; Kim, K. Changes in the microbial communities associated with Gorgonia ventalina during aspergillosis infection. Dis. Aquat. Organ. 2006, 69, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Neave, M.J.; Rachmawati, R.; Xun, L.; Michell, C.T.; Bourne, D.G.; Apprill, A.; Voolstra, C.R. Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J. 2017, 11, 186–200. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.; Duperron, S.; Birkeland, N.K.; Hovland, M. Intracellular Oceanospirillales bacteria inhabit gills of Acesta bivalves. FEMS Microbiol. Ecol. 2010, 74, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Morrow, K.M.; Moss, A.G.; Chadwick, N.E.; Liles, M.R. Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability. Appl. Environ. Microbiol. 2012, 78, 6438–6449. [Google Scholar] [CrossRef] [PubMed]
- Forget, N.L.; Juniper, K.S. Free-living bacterial communities associated with tubeworm (Ridgeia piscesae) aggregations in contrasting diffuse flow hydrothermal vent habitats at the Main Endeavour Field, Juan de Fuca Ridge. Microbiologyopen 2013, 2, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Fiore, C.L.; Labrie, M.; Jarett, J.K.; Lesser, M.P. Transcriptional activity of the giant barrel sponge, Xestospongia muta holobiont: Molecular evidence for metabolic interchange. Front. Microbiol. 2015, 6, 364. [Google Scholar] [CrossRef] [PubMed]
- Katharios, P.; Seth-Smith, H.M.B.; Fehr, A.; Mateos, J.M.; Qi, W.; Richter, D.; Nufer, L.; Ruetten, M.; Guevara Soto, M.; Ziegler, U.; et al. Environmental marine pathogen isolation using mesocosm culture of sharpsnout seabream: Striking genomic and morphological features of novel Endozoicomonas sp. Sci. Rep. 2015, 5, 17609. [Google Scholar] [CrossRef] [PubMed]
- Neave, M.J.; Apprill, A.; Ferrier-Pagès, C.; Voolstra, C.R. Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 2016, 100, 8315–8324. [Google Scholar] [CrossRef] [PubMed]
- Bourne, D.G.; Munn, C.B. Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ. Microbiol. 2005, 7, 1162–1174. [Google Scholar] [CrossRef] [PubMed]
- Reshef, L.; Koren, O.; Loya, Y.; Zilber-Rosenberg, I.; Rosenberg, E. The coral probiotic hypothesis. Environ. Microbiol. 2006, 8, 2068–2073. [Google Scholar] [CrossRef] [PubMed]
- Lema, K.A.; Bourne, D.G.; Willis, B.L. Onset and establishment of diazotrophs and other bacterial associates in the early life history stages of the coral Acropora millepora. Mol. Ecol. 2014, 23, 4682–4695. [Google Scholar] [CrossRef] [PubMed]
- Kimes, N.E.; Van Nostrand, J.D.; Weil, E.; Zhou, J.; Morris, P.J. Microbial functional structure of Montastraea faveolata, an important Caribbean reef-building coral, differs between healthy and yellow-band diseased colonies. Environ. Microbiol. 2010, 12, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.T. Microbes from marine sponges: A treasure trove of biodiversity for natural products discovery. In Microbial Diversity and Bioprospecting; Bull, A.T., Ed.; ASM Press: Washington, DC, USA, 2004; pp. 177–190. [Google Scholar]
- Wakimoto, T.; Egami, Y.; Nakashima, Y.; Wakimoto, Y.; Mori, T.; Awakawa, T.; Ito, T.; Kenmoku, H.; Asakawa, Y.; Piel, J.; et al. Calyculin biogenesis from a pyrophosphate protoxin produced by a sponge symbiont. Nat. Chem. Biol. 2014, 10, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Salomon, C.; Deerinck, T.; Ellisman, M.; Faulkner, D. The cellular localization of dercitamide in the Palauan sponge Oceanapia sagittaria. Mar. Biol. 2001, 139, 313–319. [Google Scholar]
- Gandhimathi, R.; Kiran, S.G.; Hema, T.A.; Selvin, J. Production and characterization of lipopeptide biosurfactant by a sponge associated marine actinomycetes Nocardiopsis alba MSA10. Bioprocess Biosyst. Eng. 2009, 32, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Dhasayan, A.; Selvin, J.; Kiran, S. Biosurfactant production from marine bacteria associated with sponge Callyspongia diffusa. Biotech 2015, 5, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, C.; Syldatk, C.; Hausmann, R.; Gerçe, B.; Longo, C.; Papale, M.; Conte, A.; De Domenico, E.; Michaud, L.; Lo Giudice, A. The demosponge Halichondria (Halichondria) panicea (Pallas, 1766) as a novel source of biosurfactant-producing bacteria. J. Basic Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Kiran, G.S.; Sabarathnam, B.; Thajuddin, N.; Selvin, J. Production of Glycolipid Biosurfactant from Sponge-Associated Marine actinobacterium Brachybacteriumparaconglomeratum MSA21. Surfact. Deterg. 2014, 17, 531–542. [Google Scholar] [CrossRef]
- Kiran, G.S.; Anto, T.T.; Selvin, J.; Sabarathnam, B. Optimization and characterization of a new lipopeptide biosurfactant produced by marine Brevibacterium aureum MSA13 in solid state culture. Biores. Technol. 2010, 101, 2389–2396. [Google Scholar] [CrossRef] [PubMed]
- Selvin, J.; Lipton, A.P. Biopotentials of secondary metabolites isolated from marine sponges. Hydrobiologia 2004, 513, 231–238. [Google Scholar] [CrossRef]
- Caruso, C.; Rizzo, C.; Mangano, S.; Poli, A.; Di Donato, P.; Finore, I.; Nicolaus, B.; Di Marco, G.; Michaud, L.; Lo Giudice, A. Production and biotechnological potential of extracellular polymeric substances from sponge-associated Antarctic bacteria. Appl. Environ. Microbiol. 2018, 84, e01624-17. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, M.C.; Fondi, M.; Maida, I.; Perrin, E.; Lo Giudice, A.; Michaud, L.; Mangano, S.; Bartolucci, G.; Romoli, R.; Fani, R. Sponge-associated microbial Antarctic communities exhibiting antimicrobial activity against Burkholderia cepacia complex bacteria. Biotechnol. Adv. 2012, 30, 272–293. [Google Scholar] [CrossRef] [PubMed]
- Mabrouk, M.E.M.; Youssif, E.M.; Sabry, S.A. Biosurfactant production by a newly isolated soft coral-associated marine Bacillus sp. E34: Statistical optimization and characterization. Life Sci. J. 2014, 11, 10. [Google Scholar]
- Nithyanand, P.; Pandian, S.K. Phylogenetic characterization of culturable bacterial diversity associated with the mucus and tissue of the coral Acropora digitifera from Gulf of Mannar. FEMS Microbiol. Ecol. 2009, 69, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Nithyanand, P.; Thenmozhi, R.; Rathna, J.; Pandian, S.K. Inhibition of biofilm formation in Streptococcus pyogenes by coral associated Actinomycetes. Curr. Microbiol. 2010, 60, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Padmavathi, A.R.; Pandian, S.K. Antibiofilm Activity of Biosurfactant Producing Coral Associated Bacteria Isolated from Gulf of Mannar. Indian J. Microbiol. 2014, 54, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Thenmozhi, R.; Nithyanand, P.; Rathna, J.; Pandian, S.K. Antibiofilm activity of coral associated bacteria against different clinical M serotypes of Streptococcuspyogenes. FEMS Immunol. Med. Microbiol. 2009, 57, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, C.; Michaud, L.; Hörmann, B.; Gerçe, B.; Syldatk, C.; Hausmann, R.; De Domenico, E.; Lo Giudice, A. Bacteria associated with sabellids (Polychaeta: Annelida) as a novel source of surface active compounds. Mar. Poll. Bull. 2013, 70, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, C.; Michaud, L.; Syldatk, C.; Hausmann, R.; De Domenico, E.; Lo Giudice, A. Influence of salinity and temperature on the activity of biosurfactants by polychaeteassociated isolates. Environ. Sci. Pollut. Res. 2014, 21, 2988–3004. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, C.; Michaud, L.; Graziano, M.; de Domenico, E. Biosurfactant activity, heavy metal tolerance and characterization of Joostella strain A8 from the Mediterranean polychaete Megalomma claparedei (Gravier, 1906). Ecotoxicology 2015, 24, 1294–1304. [Google Scholar] [CrossRef] [PubMed]
- Graziano, M.; Rizzo, C.; Michaud, L.; Porporato, E.M.D. Biosurfactant production by hydrocarbon-degrading Brevibacterium and Vibrio isolates from the sea pen Pteroeides spinosum (Ellis, 1764). J. Basic Microbiol. 2016, 56, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Banat, I.M.; Makkar, R.S.; Cameotra, S.S. Potential commercial applications of microbial surfactants. Appl. Microbiol. Biotechnol. 2000, 53, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Pacwa-Płociniczak, M.; Płaza, G.A.; Piotrowska-Seget, Z.; Cameotra, S.S. Environmental applications of biosurfactants: Recent advances. Int. J. Mol. Sci. 2011, 12, 633–654. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Mukherjee, A.K. Crude petroleum-oil biodegradation efficiency of Bacillus subtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. Bioresour. Technol. 2007, 98, 1339–1345. [Google Scholar] [CrossRef] [PubMed]
- Franzetti, A.; Bestetti, G.; Caredda, P.; Colla La, P.; Tamburini, E. Surface-active compounds and their role in the access to hydrocarbons in Gordonia strains. FEMS Microbiol. Ecol. 2008, 63, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.W.; Kim, Y.B.; Shin, J.D.; Kim, E.K. Enhanced biodegradation of hydrocarbons in soil by microbial biosurfactant, sophorolipid. Appl. Biochem. Biotechnol. 2010, 160, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.C.; Huang, Y.C.; Wei, Y.H.; Chang, J.S. Biosurfactant-enhanced removal of total petroleum hydrocarbons from contaminated soil. J. Hazard. Mater. 2009, 167, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Urum, K.; Grigson, S.; Pekdemir, T.; McMenamy, S. A Comparison of the efficiency of different surfactants for removal of crude oil from contaminated soils. Chemosphere 2006, 62, 1403–1410. [Google Scholar] [CrossRef] [PubMed]
- Aşçı, Y.; Nurbaş, M.; Açıkel, Y.S. Investigation of sorption/desorption equilibria of heavy metal ions on/from quartz using rhamnolipid biosurfactant. J. Environ. Manag. 2010, 91, 724–731. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Mukherjee, S.; Sen, R. Biosurfactant of marine origin exhibiting heavy metal remediation properties. Bioresour. Technol. 2009, 100, 4887–4890. [Google Scholar] [CrossRef] [PubMed]
- Juwarkar, A.A.; Nair, A.; Dubey, K.V.; Singh, S.K.; Devotta, S. Biosurfactant technology for remediation of cadmium and lead contaminated soils. Chemosphere 2007, 68, 1996–2002. [Google Scholar] [CrossRef] [PubMed]
- Gnanamani, A.; Kavitha, V.; Radhakrishnan, N.; Rajakumar, G.S.; Sekaran, G.; Mandal, A.B. Microbial products (biosurfactant and extracellular chromate reductase) of marine microorganism are the potential agents reduce the oxidative stress induced by toxic heavy metals. Colloid Surf. B 2010, 79, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.H.; Liao, Z.Y.; Wang, C.L.; Cai, P.; Yang, W.Y.; Lu, M.F.; Huang, G.W. Purification and antitumour activity of a lipopeptide biosurfactant produced by Bacillus natto TK-1. Biotechnol. Appl. Biochem. 2009, 52, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Remichkova, M.; Galabova, D.; Roeva, I.; Karpenko, E.; Shulga, A.; Galabov, A.S. Anti-herpes virus activities of Pseudomonas sp. S-17 rhamnolipid and its complex with alginate. Z. Naturforsch. C 2008, 63, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Satpute, S.K.; Bhawsar, B.D.; Dhakephalkar, P.K.; Chopade, B.A. Assessment of different screening methods for selecting biosurfactant producing marine bacteria. Ind. J. Mar. Sci. 2008, 37, 243–250. [Google Scholar]
- Tonkova, E.; Galabova, D.; Stoimenova, E. Characterization of bacterial isolates from industrial wastewater according to probable modes of hexadecane uptake. Microbiol. Res. 2008, 4, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Sivapathasekaran, C.; Mukherjee, S.; Sen, R. Optimization of a marine medium for augmented biosurfactant production. Int. J. Chem. React. Eng. 2010, 8, 47–54. [Google Scholar] [CrossRef]
- Salihu, A.; Abdulkadir, I.; Almustapha, M.N. An investigation for potential development on biosurfactants. Biotechnol. Mol. Biol. Rev. 2009, 3, 111–117. [Google Scholar]
- Walter, V.; Syldatk, C.; Hausmann, R. Screening concepts for the isolation of biosurfactant producing microorganisms. Biosurfactants 2010, 672, 1–13. [Google Scholar]
- Montalvo, N.F.; Mohamed, N.M.; Enticknap, J.J.; Hill, R.T. Novel Actinobacteria from marine sponges. Antonie Van Leeuwenhoek 2005, 87, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, C.; Rappazzo, A.C.; Michaud, L.; De Domenico, E.; Rochera, C.; Camacho, A.; Lo Giudice, A. Efficiency in hydrocarbon degradation and biosurfactant production by Joostella sp. A8 when grown in pure culture and consortia. J. Environ. Sci. 2018, 67, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Tsuge, K.; Ano, T.; Shado, M. Isolation of a gene essential for biosynthesis of the lipopeptide antibiotics lipastatin B and surfactin in Bacillus subtilis YB8. Arch. Microbiol. 1996, 165, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J.; Griebe, T.; Mayer, C. Physico-Chemical properties of Biofilms. In Biofilms: Recent Advances in their study and Control; Evans, L.V., Ed.; CRC Press: Boca Raton, FL, USA, 2000; pp. 19–34. ISBN 978-9058230935. [Google Scholar]
- Kumar, A.K.; Mody, K.; Jha, B. Bacterial exopolysaccharides—A perception. J. Basic Microbiol. 2007, 47, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Delbarre-Ladrat, C.; Sinquin, C.; Lebellenger, L.; Zykwinska, A.; Colliec-Jouault, S. Exopolysaccharides produced by marine bacteria and their applications as glycosaminoglycan-like molecules. Front. Chem. 2014, 2, 85. [Google Scholar] [CrossRef] [PubMed]
- Rehm, B.H.A. Bacterial polymers: Biosynthesis, modifications and applications. Nat. Rev. Microbiol. 2010, 8, 578–592. [Google Scholar] [CrossRef] [PubMed]
- Ruas-Madiedo, P.; Hugenholdtz, J.; Zoon, P. An overview of the functionality of exopolysaccharides produced by lactic acid bacteria. Int. Diary J. 2002, 12, 163–171. [Google Scholar] [CrossRef]
- More, T.T.; Yadav, J.S.S.; Yan, S.; Tyagi, R.D.; Surampalli, R.Y. Extracellular polymeric substances of bacteria and their potential environmental applications. J. Environ. Manag. 2014, 144, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Jannasch, H.W.; Taylor, C.D. Deep-sea microbiology. Ann. Rev. Microbiol. 1984, 38, 487. [Google Scholar] [CrossRef] [PubMed]
- Decho, A. Microbial exopolymer secretions in ocean enviroments: Their role(s) in food webs and marine processes. Oceanogr. Mar. Biol. Ann. Rev. 1990, 28, 73–153. [Google Scholar]
- Finore, I.; Di Donato, P.; Mastascusa, V.; Nicolaus, B.; Poli, A. Fermentation technologies for the optimization of marine microbial exopolysaccharide production. Mar. Drugs 2014, 12, 3005–3024. [Google Scholar] [CrossRef] [PubMed]
- Nicolaus, B.; Kambourova, M.; Oner, E.T. Exopolysaccharides from extremophiles: From fundamentals to biotechnology. Enz. Technol. 2010, 31, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Poli, A.; Anzelmo, G.; Nicolaus, B. Bacterial exopolysaccharides from extreme marine habitats: Production, characterization and biological activities. Mar. Drugs 2010, 8, 1779–1802. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, T.L.; Gugliandolo, C.; Caccamo, D.; Panico, A.; Lama, L.; Gambacorta, A.; Nicolaus, B. A halophilic thermotolerant Bacillus isolated from a marine hot spring able to produce a new exopolysaccharide. Biotechnol. Lett. 2002, 24, 515–519. [Google Scholar] [CrossRef]
- Vanfossen, A.L.; Lewis, D.L.; Nichols, J.D.; Kelly, R.M. Polysaccharide degradation and synthesis by extremely thermophilic anaerobes. Ann. N. Y. Acad. Sci. 2008, 1125, 322–337. [Google Scholar] [CrossRef] [PubMed]
- Rinker, K.D.; Kelly, R.M. Effect of carbon and nitrogen sources on growth dynamics and exopolysaccharide production for the hyperthermophilic archaeon Thermococcuslitoralis and bacterium Thermotogamaritima. Biotechnol. Bioeng. 2000, 69, 537–547. [Google Scholar] [CrossRef]
- Nicolaus, B.; Manca, M.C.; Ramano, I.; Lama, L. Production of an exopolysaccharide from two thermophilic archaea belonging to the genus Sulfolobus. FEMS Microbiol. Lett. 1993, 109, 203–206. [Google Scholar] [CrossRef]
- Carrion, O.; Delgado, L.; Mercade, E. New emulsifying and cryoprotective exopolysaccharides from Antarctic Pseudomonas sp. ID1. Carbohydr. Polym. 2015, 117, 1028–1034. [Google Scholar] [CrossRef] [PubMed]
- Corsaro, M.M.; Lanzetta, R.; Parrilli, E.; Parrilli, M.; Tutino, M.L.; Ammarino, S. Influence of growth temperature on lipid and phosphate contents of surface polysaccharides from the Antarctic bacterium Pseudoalteromonas haloplanktis TAC 125. J. Bacteriol. 2004, 186, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Yim, J.H. Cryoprotective properties of exopolysaccharide (P-21653) produced by the Antarctic bacterium, Pseudoalteromonas arctica KOPRI 21653. J. Microbiol. 2007, 45, 510–514. [Google Scholar] [PubMed]
- Nichols, C.M.; Bowman, J.P.; Guézennec, J. Effects of incubation temperature on growth and production of exopolysaccharides by an Antarctic sea ice bacterium grown in batch culture. Appl. Environ. Microbiol. 2005, 71, 3519–3523. [Google Scholar] [CrossRef] [PubMed]
- Nichols, C.M.; Bowman, J.P.; Guézennec, J. Olleya marilimosa gen nov, sp nov, an exopolysaccharide-producing marine bacterium from the family Flavobacteriaceae, isolated from the Southern Ocean. Int. J. Syst. Evol. Microbiol. 2005, 55, 1557–1561. [Google Scholar] [CrossRef] [PubMed]
- Nichols, C.A.; Guézennec, J.; Bowman, J.P. Bacterial exopolysaccharides from extreme environments with special consideration of the Southern Ocean, sea ice, and deep-sea hydrothermal vents: A review. Mar. Biotechnol. 2005, 7, 253–271. [Google Scholar] [CrossRef] [PubMed]
- Nichols, C.M.; Lardiere, S.G.; Bowman, J.P.; Nichols, P.D.; Gibson, J.A.E.; Guézennec, J. Chemical characterization of exopolysaccharides from Antarctic marine bacteria. Microb. Ecol. 2005, 49, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Mancuso Nichols, C.A.; Garron, S.; Bowman, J.P.; Raguénès, G.; Guèzennec, J. Production of exopolysaccharides by Antarctic marine bacterial isolates. J. Appl. Microbiol. 2004, 96, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Krembs, C.; Eicken, H.; Junge, K.; Deming, J.W. High concentrations of exopolymeric substance in Arctic winter sea ice: Implications for the polar ocean carbon cycle and cryoprotection of diatoms. Deep Sea Res. 2002, 49, 2163–2181. [Google Scholar] [CrossRef]
- Manca, M.C.; Lama, L.; Improta, R.; Esposito, E.; Gambacorta, A.; Nicolaus, B. Chemical composition of two exopolysaccharides from Bacillus thermoantarcticus. Appl. Environ. Microbiol. 1996, 62, 3265–3269. [Google Scholar] [PubMed]
- Arias, S.; Moral, A.D.; Ferrer, M.R.; Tallon, R.; Quesada, E.; Bejar, V. Mauran, an exopolysaccharide produced by the halophilic bacterium Halomonas maura, with a novel composition and interesting properties for biotechnology. Extremophiles 2003, 7, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Mata, J.A.; Bejar, V.; Llamas, I.; Arias, S.; Bressollier, P.; Tallon, R.; Urdaci, M.C.; Quesada, E. Exopolysaccharides produced by the recently described bacteria Halomonas ventosae and Halomonas anticariensis. Res. Microbiol. 2006, 157, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Poli, H.; Kazak, B.; Gürleyendag, B.; Tommonaro, G.; Pieretti, G.; Öner, E.T.; Nicolaus, B. High level synthesis of levan by a novel Halomonas species growing on defined media. Carbohydr. Polym. 2009, 78, 651–657. [Google Scholar] [CrossRef]
- Corsaro, M.M.; Grant, W.D.; Grant, S.; Marciano, C.E.; Parrilli, M. Structure determination of an exopolysaccharide from an alkaliphilic bacterium closely related to Bacillus spp. Eur. J. Biochem. 1999, 264, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Romano, I.; Giordano, A.; Lama, L.; Nicolaus, B.; Gambacorta, A. Halomonas campaniensis sp. nov., a haloalkaliphilic bacterium isolated from a mineral pool of Campania Region, Italy. Syst. Appl. Microbiol. 2005, 28, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Park, S.J.; Lee, Y.; Lee, S.H. Economic aspects of biopolymer production. Biopolymers 2003, 10, 307–337. [Google Scholar]
- Rohwerder, T.; Gehrke, T.; Kinzler, K.; Sand, W. Bioleaching review, part A: Progress in bioleaching: Fundamentals and mechanisms of bacterial metal sulphide oxidation. Appl. Microbiol. Biotechnol. 2003, 63, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Mangold, S.; Harneit, K.; Rohwerder, T.; Claus, G.; Sand, W. Novel combination of atomic force microscopy and epifluorescence microscopy for visualization of leaching bacteria on pyrite. Appl. Environ. Microbiol. 2008, 74, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Michel, C.; Bény, C.; Delorme, F.; Poirier, L.; Spolaore, P.; Morin, D.; d’Hugues, P. New protocol for the rapid quantification of exopolysaccharides in continuous culture systems of acidophilic bioleaching bacteria. Appl. Microbiol. Biotechnol. 2009, 82, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Raguenes, G.; Christen, R.; Guezennec, J.; Pignet, P.; Barbier, G. Vibrio diabolicus sp. nov., a new polysaccharide-secreting organism isolated from a deep-sea hydrothermal vent polychaete annelid, Alvinella pompejana. Int. J. Syst. Bacteriol. 1997, 47, 989–995. [Google Scholar] [CrossRef] [PubMed]
- Priyankaa, P.; Arun, A.B.; Young, C.C.; Rekha, P.D. Prospecting exopolysaccharides produced by selected bacteria associated with marine organisms for biotechnological applications. Chin. J. Polym. Sci. 2015, 33, 236–244. [Google Scholar] [CrossRef]
- Sayem, S.M.A.; Manzo, E.; Ciavatta, L.; Tramice, A.; Cordone, A.; Zanfardino, A.; De Felice, M.; Varcamonti, M. Anti-biofilm activity of an exopolysaccharide from a sponge-associated strain of Bacilluslicheniformis. Microb. Cell Fact. 2011, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Lahaye, M.; Robic, A. Structure and functional properties of Ulvan, a polysaccharide from green seaweeds. Biomacromolecules 2007, 8, 1765–1774. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, C.; Genovese, G.; Morabito, M.; Faggio, C.; Pagano, M.; Spanò, A.; Zammuto, V.; Armeli Minicante, S.; Manghisi, A.; Cigala, R.M.; et al. Potential antibacterial activity of marine macroalgae against pathogens relevant for aquaculture and human health. J. Pure. Appl. Microbiol. 2017, 11, 1695–1706. [Google Scholar] [CrossRef]
- Caruso, C.; Rizzo, C.; Mangano, S.; Poli, A.; Di Donato, P.; Nicolaus, B.; Di Marco, G.; Michaud, L.; Lo Giudice, A. Extracellular polymeric substances with metal adsorption capacity produced by Pseudoalteromonas sp. MER144 from Antarctic seawater. Environ. Sci. Pollut. Res. 2018, 25, 4667. [Google Scholar] [CrossRef] [PubMed]
- Dubreucq, G.; Domon, B.; Fournet, B. Structure determination of a novel uronic acid residue isolated from the exopolysaccharide produced by a bacterium originating from deep sea hydrothermal vents. Carbohydr. Res. 1996, 290, 175–181. [Google Scholar] [CrossRef]
- Rougeaux, H.; Guezennec, J.; Carlson, R.W.; Kervarec, N.; Pichon, R.; Talaga, P. Structural determination of the exopolysaccharide of Pseudoalteromonas strain HYD721 isolated from a deep-sea hydrothermal vent. Carbohydr. Res. 1999, 315, 273–285. [Google Scholar] [CrossRef]
- Cambon-Bonavita, M.A.; Raguenes, G.; Jean, J.; Vincent, P.; Guezennec, J. A novel polymer produced by a bacterium isolated from a deep-sea hydrothermal vent polychaete annelid. J. Appl. Microbiol. 2002, 93, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Rougeaux, H.; Kervarec, N.; Pichon, R.; Guezennec, J. Structureofthe exopolysaccharide of Vibrio diabolicus isolated from a deep-sea hydrothermal vent. Carbohydr. Res. 1999, 322, 40–45. [Google Scholar] [CrossRef]
- Sinquin, C.; Colliec-Jouault, S. Les Polysaccharides Marin Set Leurs Applications Dansledomaine Dela Santé. Bioprocédés Danslesdomaines de Lasanté, Del’agroalimentaire et Del Achimie. Techniques Del’ingénieur BIO6250 1-5. 2014. Available online: http://www.techniques-ingenieur.fr/base-documentaire/biomedical-pharma-th15/chimie-pharmaceutique-42609210/les-polysaccharides-marins-et-leurs-applications-dans-le-domaine-de-la-sante-bio6250/ (accessed on 27 June 2018).
- Raguenes, G.; Cambon-Bonavita, M.A.; Lohier, J.F.; Boisset, C.; Guezennec, J. A novel, highly viscous polysaccharide excreted by an Alteromonas isolated from a deep-sea hydrothermal vent shrimp. Curr. Microbiol. 2003, 46, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Raguenes, G.; Pignet, P.; Gauthier, G.; Peres, A.; Christen, R.; Rougeaux, H.; Barbier, G.; Guezennec, G.J. Description of a new polymer-secreting bacterium from a deepsea hydrothermal vent, Alteromonas macleodii subsp. fijiensis, and preliminary characterization of the polymer. Appl. Environ. Microbiol. 1996, 6, 67–73. [Google Scholar]
- Rougeaux, H.; Talaga, P.; Carlson, R.W.; Guezennec, J. Structural studies of an exopolysaccharide produced by Alteromonas macleodii subsp. fijiensis originating from a deep-sea hydrothermal vent. Carbohydr. Res. 1998, 312, 53–59. [Google Scholar] [CrossRef]
- Raguenes, G.H.; Peres, A.; Ruimy, R.; Pignet, P.; Christen, R.; Loaec, M.; Rougeaux, H.; Barbier, G.; Guezennec, G.J. Alteromonas infernus sp. nov., a new polysaccharide-producing bacterium isolated from a deep-sea hydrothermal vent. J. Appl. Microbiol. 1997, 82, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Roger, O.; Kervarec, N.; Ratiskol, J.; Colliec-Jouault, S.; Chevolot, L. Structural studies of the main exopolysaccharide produced by the deep-sea bacterium Alteromonas infernus. Carbohydr. Res. 2004, 339, 2371–2380. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, S.Y.; Anzelmo, G.; Ozer, T.; Radchenkova, N.; Genc, S.; Di Donato, P.; Nicolaus, B.; Oner, E.T.; Kambourova, M. Brevibacillus themoruber: A promising microbial cell factory for exopolysaccharide production. J. Appl. Microbiol. 2014, 116, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Shukla, M.K.; Mishra, A.; Kumari, P.; Reddy, C.R.K.; Jha, B. Isolation and characterization of exopolysaccharides from seaweed associated bacteria Bacillus licheniformis. Carbohydr. Pol. 2011, 84, 1019–1026. [Google Scholar] [CrossRef]
- Manzoni, M.; Rollini, M. Isolation and characterization of the exopolysaccharide produced by Daedalea quercina. Biotechnol. Lett. 2001, 23, 1491–1497. [Google Scholar] [CrossRef]
- Liu, S.B.; Chen, X.L.; He, H.L.; Zhang, X.Y.; Xie, B.B.; Yu, Y.; Chen, B.; Zhou, B.C.; Zhang, Y.Z. Structure and ecological roles of a novel exopolysaccharide from the arctic sea ice bacterium Pseudoalteromonas sp. strain SM20310. Appl. Environ. Microbiol. 2013, 79, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Marx, J.G.; Carpenter, S.D.; Deming, J.W. Production of cryoprotectant extracellular polysaccharide substance (EPS) by the marine psychrophilic bacterium Colwellia psychrerythraea strain 34H under extreme conditions. Can. J. Microbiol. 2009, 55, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Zhu, L.; Chen, X.; Wang, P.G.; Zhang, Y. Structural characterization and ecological roles of a novel exopolysaccharide from deep-sea psychrotolerant bacterium Pseudoalteromonas sp. SM9913. Microbiology 2007, 153, 1566–1572. [Google Scholar] [CrossRef] [PubMed]
- Selvin, J.; Ninawe, A.S.; Kiran, G.S.; Lipton, A.P. Sponge-microbial interactions: Ecological implications and bioprospecting avenues. Crit. Rev. Microbiol. 2010, 36, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Moskovitz, J.; Yim, M.B.; Chock, P.B. Free radicals and disease. Arch. Biochem. Biophys. 2002, 397, 354. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, I. A sticky business. Microbial polysaccharides: Current products and future trends. Microbiol. Today 2002, 29, 70. [Google Scholar]
- Radjasa, O.K. Bioprospecting of Marine Microbial Symbionts: Exploitation of Underexplored Marine Microorganisms. In Marine Microbiology. Bioactive Compounds and Biotechnological Applications; Kim, S.K., Ed.; Wiley-VCH Verlag GmbH & Co., KGaA: Weinheim, Germany, 2013; pp. 369–377. [Google Scholar]
- Satheesh, S.; Soniyamby, A.R.; Shankar, C.V.S.; Punitha, S.M.J. Antifouling activities of marine bacteria associated with the sponge (Sigmodocia sp.). J. Ocean. Univ. China 2012, 11, 354–360. [Google Scholar] [CrossRef]
- Radjasa, O.K.; Salasia, S.I.O.; Sabdono, A.; Weise, J.; Imhoff, J.F.; Lammler, C.; Risk, M.J. Antibacterial activity of marine bacterium Pseudomonas sp. associated with soft coral Sinulariapolydactyla against Streptococcus equi Subsp. zooepidemicus. Int. J. Pharmacol. 2007, 3, 170–174. [Google Scholar]
- Kadiri, S.K.; Yarla, N.S.; Vidavalur, S. Screening and isolation of antagonistic actinobacteria associated with marine sponges from Indian coast. J. Microb. Biochem. Technol. 2014. [Google Scholar] [CrossRef]
- Devi, P.; Wahidullah, S.; Rodrigues, C.; Souza, L.D. The sponge-associated bacterium Bacillus licheniformis SAB1: A source of antimicrobial compounds. Mar. Drugs 2010, 8, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Wahl, M.; Goecke, F.; Labes, A.; Dobretsov, S.; Weinberger, F. The second skin: Ecological role of epibiotic biofilms on marine organisms. Front. Microbiol. 2012, 3, 292. [Google Scholar] [CrossRef] [PubMed]
- Barresi, G.; Di Carlo, E.; Trapani, M.R.; Parisi, M.G.; Chille, C.; Mule, M.F.; Cammarata, M.; Palla, F. Marine organisms as source of bioactive molecules applied in restoration projects. Herritage Sci. 2015, 3, 17. [Google Scholar] [CrossRef]
- Ivanova, E.P.; Vysotskii, M.V.; Svetashev, V.I.; Nedashkovskayal, O.I.; Gorshkoval, N.M.; Mikhailovl, V.V.; Yumoto, N.; Shigeri, Y.; Taguchi, T.; Yoshikawa, S. Characterization of Bacillus strains of marine origin. Int. Microbiol. 1999, 2, 267–271. [Google Scholar] [PubMed]
- Aishwarya, M.S.; Lipton, A.P.; Sarika, A.R. Phylogenetic appraisal of the drug bearing marine sponge Callyspongia subarmigera (Ridley, 1884) from South India. Indian J. Geo-Mar. Sci. 2013, 42, 139–145. [Google Scholar]
- Aboul-Ela, H.M.; Shreadah, M.A.; Abdel-Monem, N.M.; Yakout, G.A.; van Soest, R.W.M. Isolation, cytotoxic activity and phylogenetic analysis of Bacillus spp. bacteria associated with the red sea sponge Amphimedonochracea. Adv. Biosci. Biotechnol. 2012, 3, 815–823. [Google Scholar] [CrossRef]
- Costa Leal, M.; Sheridan, C.; Osinga, R.; Dionísio, G.; Rocha, R.J.M.; Silva, B.; Rosa, R.; Calado, R. Marine Microorganism-Invertebrate Assemblages: Perspectives to Solve the “Supply Problem” in the Initial Steps of Drug Discovery. Mar. Drugs 2014, 12, 3929–3952. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, K.; Baringhaus, K.-H.; Schneider, G. Scaffold diversity of natural products: Inspiration for combinatorial library design. Nat. Prod. Rep. 2008, 25, 892–904. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F.; Labes, A.; Wieses, J. Bio-mining the microbial treasures of the ocean: New natural products. Biotechnol. Adv. 2011, 29, 468–482. [Google Scholar] [CrossRef] [PubMed]
- Otero-Gonzàlez, A.J.; Magalhanes, B.S.; Garcia-Villarino, M.; Lòpez-Abarrategui, C.; Sousa, D.A.; Dias, S.C.; Franco, O.L. Antimicrobial peptides from marine invertebrates is a new frontier for microbial infection control. FASEB J. 2010, 24, 1320–1334. [Google Scholar] [CrossRef] [PubMed]
- Smith, V.J.; Desbois, A.P.; Dyrunda, E.A. Conventional and unconventional antimicrobials from fish, marine invertebrates and micro-algae. Mar. Drugs 2010, 8, 1213–1262. [Google Scholar] [CrossRef] [PubMed]
- Waters, A.L.; Hill, R.T.; Place, A.R.; Hamann, M.T. The expanding role of marine microbes in pharmaceutical development. Curr. Opin. Biotechnol. 2010, 21, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Radjasa, O.K.; Vaske, Y.M.; Navarro, G.; Vervoort, H.C.; Tenney, K.; Linington, R.G.; Crews, P. Highlights of marine invertebrate-derived biosynthetic products: Their biomedical potential and possible production by microbial associants. Bioorg. Med. Chem. 2011, 19, 6658–6674. [Google Scholar] [CrossRef] [PubMed]

| Organism | Species | Bioactive Compound | Reference |
|---|---|---|---|
| Sponges | Callyspongia diffusa | Biosurfactant | [111] |
| Callyspongia diffusa | Biosurfactant | [112] | |
| Halicondria panicea | Biosurfactant | [113] | |
| Dendrilla nigra | Biosurfactant | [114] | |
| Dendrilla nigra | Biosurfactant | [115] | |
| Dendrilla nigra | Antibacterial | [116] | |
| Haliclonissa verrucosa | EPS | [117] | |
| Hemigellius pilosus | EPS | [117] | |
| Tedania charcoti | EPS | [117] | |
| Callyspongia | Antibacterial | [4] | |
| Haliclona sp. | Antibacterial | [4] | |
| Haliclonissa verrucosa, Anoxycalyx joubini and Lissodendoryx nobilis | Antibacterial | [118] | |
| Cnidarians | Sarchophyton glaucum | Biosurfactant | [119] |
| Acropora digitifera | Antibacterial | [120] | |
| Acropora digitifera | Antibacterial | [121] | |
| Acropora digitifera | Antibiofilm | [122] | |
| Acropora digitifera | Antibiofilm | [123] | |
| Polichaetes | Megalomma claparedei | Biosurfactant | [124,125] |
| Branchiomma luctuosum | Biosurfactant | [124,126] | |
| Sabella spallanzanii | Biosurfactant | [124,125] | |
| Sea pens | Pteroeides spinosum | Biosurfactant | [127] |
| Phylum or Class | Strain (Accession Number) | Isolation Source | Reference |
|---|---|---|---|
| Actinobacteria | Brachybacterium paraconglomeratum MSA21 (GQ153943) | Dendrilla nigra | [114] |
| Actinobacteria | Brevibacterium sp. PBE178 (KR185336) | Pteroides spinousm | [127] |
| Actinobacteria | Brevibacterium sp. PBE181 (KR185337) | Pteroides spinousm | [127] |
| Actinobacteria | Brevibacterium sp. PBE190 (KR185338) | Pteroides spinousm | [127] |
| Actinobacteria | Brevibacterium sp. PBE209 (KR1853312) | Pteroides spinousm | [127] |
| Actinobacteria | Nocardiopsis alba MSA10 (EU563352) | Callyspongia diffusa | [111] |
| Actinobacteria | Citricoccus sp. Bl52 (KF032914) | Branchiomma luctuosum | [125] |
| Actinobacteria | Citricoccus sp. Bl54 (KF032915) | Branchiomma luctuosum | [125] |
| Actinobacteria | Citricoccus sp. Bl55 (KF032924) | Branchiomma luctuosum | [125] |
| Actinobacteria | Brevibacteriumaureum MSA13 (GQ153943) | Dendrilla nigra | [115] |
| Alfaproteobacteria | Pseudovibrio sp. SpE85 (KY129823) | Halicondria panicea | [113] |
| Alfaproteobacteria | Pseudovibrio sp. SpE86 (KY129822) | Halicondria panicea | [113] |
| Alfaproteobacteria | Pseudovibrio sp. SpE90 (KY129821) | Halicondria panicea | [113] |
| Alfaproteobacteria | Pseudovibrio sp. SpE93 (KY129820) | Halicondria panicea | [113] |
| Alfaproteobacteria | Thalassospira sp. A46 (JX298566) | Sabella spallanzanii enr. | [124] |
| Alfaproteobacteria | Thalassospira sp. A57 (JX298539) | Branchiomma luctuosum enr. | [124] |
| Alfaproteobacteria | Pseudovibrio sp. A27 (JX298538) | Megalomma claparedei enr. | [124] |
| Alfaproteobacteria | Cohaesibacter sp. A25 (JX298537) | Megalomma claparedei enr. | [124] |
| Alfaproteobacteria | Cohaesibacter sp. A55 (JX298551) | Branchiomma luctuosum enr. | [124] |
| Alfaproteobacteria | Cohaesibacter sp. A60 (JX298552) | Branchiomma luctuosum enr. | [124] |
| Alfaproteobacteria | Cohaesibacter sp. A49 (JX298548) | Branchiomma luctuosum enr. | [124] |
| Betaproteobacteria | Alcaligenes sp. MB-19 (KJ540940) | Callyspongia diffusa | [112] |
| Gammaproteobacteria | Vibrio sp. PBN295 (KR185340) | Pteroides spinousm | [127] |
| Gammaproteobacteria | Providencia rettgeri U7 (JN315773) | Acropora digitifera | [122] |
| Gammaproteobacteria | Psychrobacter sp. U9 (JN315776) | Acropora digitifera | [122] |
| Gammaproteobacteria | Psychrobacter sp. U14 (JN315774) | Acropora digitifera | [122] |
| Gammaproteobacteria | Halomonas sp. MB-30 (KJ414418) | Callyspongia diffusa | [112] |
| Chemical Characterization | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Organism | Species | Strain | Monosaccharides | CRB ● | PRT ●● | UA □ | SULF □□ | Properties | Ref. |
| Sponge | Haliclonissa verrucosa | Shewanella sp. CAL606 | glucose, galactose, mannose, galactosamine, glucuronic acid, galacturonic acid (1:1:0.9:0.6:0.3:0.1) | 26 (mg/100 mg EPS) | 3 (mg/100 mg EPS) | 6.07 (mg/100 mg EPS) | 2.4% | Emulsifying activities, cryoprotective effect, heavy metal tolerance | [117] |
| Sponge | Hemigelliuspilosus | Colwellia sp. GW185 | glucose, mannose, galactose, galactosamine, glucuronic acid, galacturonic acid (1:1:0.7:0.7:0.3:0.04) | 28 (mg/100 mg EPS) | 2.08 (mg/100 mg EPS) | 6.09 (mg/100 mg EPS) | 3.8% | Emulsifying activities, cryoprotective effect, heavy metal tolerance | [117] |
| Sponge | Tedaniacharcoti | Winogradskyella sp. CAL396 | mannose, arabinose, galacturonic acid, glucuronic acid, galactose, glucose, glucosamine (1:0.9:0.4:0.3:0.2:0.2:0.01) | 21 (mg/100 mg EPS) | 8.8 (mg/100 mg EPS) | 3.2 (mg/100 mg EPS) | 8.9% | Emulsifying activities, cryoprotective effect, heavy metal tolerance | [117] |
| Sponge | Tedaniacharcoti | Winogradskyella sp. CAL384 | glucose, mannose, galacturonic acid, arabinose, galactose, glucosamine, glucuronic acid (1:0.5:0.3:0.25:0.1:0.1:0.1) | 15 (mg/100 mg EPS) | 2.4 (mg/100 mg EPS) | 11.9 (mg/100 mg EPS) | 7.7% | Emulsifying activities, cryoprotective effect, heavy metal tolerance | [117] |
| Crab | NS * | Nitratireductor sp. PRIM-24 | - | 390.7 (mg/g EPS) | 119.9 (mg/g EPS) | 218.7 (mg/L g EPS) | 22 (mg/g EPS) | AntioxidantActivities, emulsifyingactivities | [184] |
| Sea anemone | NS * | Enterobacter sp. PRIM-26 | - | 625.2 (mg/g EPS) | 31.7 (mg/g EPS) | 253.3 (mg/L g EPS) | ND | AntioxidantActivities, emulsifyingactivities | [184] |
| Polychaete | Alvinellapompejana | Vibriodiabolicus | glucosamine, galactosamine, glucuronic acid, galacturonicacids | - | - | - | - | ND ° | [197] |
| Sponge | Spongiaofficinalis | Bacilluslicheniformis | α-d-galactopyranosyl-(1→2)-glycerol-phosphate monomeric units | - | - | - | - | Antibacterial activity, Anti-biofilm effect | [185] |
| Polychaete | Alvinellapompejana | Alteromonas sp. strain HYD-154 | Rhamnose, glucose, galactose, mannose, glucuronic acid, galacturonic acid | 88.2/927 (g/100 g EPS) | 0.2/3 (g/100 g EPS) | 32.5/39, 33/36 (g/100g EPS) | - | ND ° | [58] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rizzo, C.; Lo Giudice, A. Marine Invertebrates: Underexplored Sources of Bacteria Producing Biologically Active Molecules. Diversity 2018, 10, 52. https://doi.org/10.3390/d10030052
Rizzo C, Lo Giudice A. Marine Invertebrates: Underexplored Sources of Bacteria Producing Biologically Active Molecules. Diversity. 2018; 10(3):52. https://doi.org/10.3390/d10030052
Chicago/Turabian StyleRizzo, Carmen, and Angelina Lo Giudice. 2018. "Marine Invertebrates: Underexplored Sources of Bacteria Producing Biologically Active Molecules" Diversity 10, no. 3: 52. https://doi.org/10.3390/d10030052
APA StyleRizzo, C., & Lo Giudice, A. (2018). Marine Invertebrates: Underexplored Sources of Bacteria Producing Biologically Active Molecules. Diversity, 10(3), 52. https://doi.org/10.3390/d10030052





