Abstract
Kale (Brassica oleracea L. var. acephala) is a widely appreciated vegetable with a century-old history of cultivation in Italy. The present study was addressed to the collection and characterization of kale germplasm traditionally cultivated in Apulia, Southern Italy, nowadays at risk of genetic erosion. In total, nineteen Apulian kale accessions were acquired. Genotyping by means of simple sequence repeat (SSR) DNA markers led to the identification of highly informative primer combinations and highlighted significant patterns of molecular variation among accessions. Consistently, significant differences were observed with respect to morpho-agronomic traits, including yield and harvesting time, and the content of bioactive compounds, namely total phenols, flavonoids, and anthocyanins, associated with antioxidant activity. Overall, this study led to the establishment of an ex situ collection of great importance to preserve endangered Apulian kale germplasm and to provide seed access to potential growers. Meanwhile, it offers a first characterization of Apulian kale, useful to promote its consumption and valorisation through breeding programmes.
1. Introduction
The species Brassica oleracea L. (2n = 2x = 18) encompasses an extraordinary variety of vegetables, each one with a different domestication history: kales (var. acephala); Chinese broccoli (var. alboglabra); cauliflowers (var. botrytis); cabbages (var. capitata); Brussels sprouts (var. gemmifera); kohlrabies (var. gongylodes); broccoli (var. italica); and savoy cabbages (var. sabauda). The non-heading varietal group of kales is the closest to B. oleracea wild relatives, and is thought to originate from a domestication process that occurred either in the Mediterranean area or in the European northwest [1]. Possibly used as a food crop since 2000 years BC [2], kale is nowadays grown throughout the world. There are no statistics for kale cultivation area at the FAOSTAT database, which considers this vegetable together with other Brassica species. From available data, it emerges that kale is extensively grown in some European areas, namely parts of the Black Sea coast, central and northern parts of the Iberian Peninsula, and Tuscany [1,3,4,5], in relation to local traditions and/or its adaptation to extremely low winter temperatures, whereas in other regions it has a minor economic importance. However, in the United States a significant increase in kale production was reported, from 3994 to 6256 harvested acres in the period from 2007 to 2012 [6].
In Italy, kale was present already during Roman times. In Tuscany, a kale type displaying dark and embossed leaves, referred to as ‘cavolo nero’ (black cabbage), is widely cultivated [4]. In contrast, kale is sporadically grown in other Regions, as a last relic of former wider occurrence. In Apulia, Southern Italy, kale displays leaves with peculiar indented margins, and is therefore known as ‘cavolo riccio’ (curly cabbage). According to farmers’ information, it is generally transplanted in September–October (in-row distance of 30–50 cm and between-row distance of 70–100 cm), and axillary buds and small inflorescences are generally harvested from two to four times from November to March, each harvest yielding 50–200 g of edible part per plant. The time spanning from transplanting to the first harvest is mostly depending on the date of transplanting and climatic conditions, influencing the rate of plant growth, and the genotype. Having a century-old history of cultivation, the Apulian kale is one of the main ingredients of a number of traditional recipes, including a well-known dish, consumed at Christmas time, in which it is prepared in combination with smashed broad beans [7]. However, it is today a secondary vegetable, whose presence is limited to a few peri-urban vegetable gardens, most of them located in the metropolitan area of the city of Bari [8]. Recently, kale was mentioned in the documentation accompanying the 2013–2017 Apulian Rural Development Programme as one of the typical vegetables at risk of genetic erosion [8].
The characterization of plant genetic resources (PGRs) is pivotal to guide appropriate conservation strategies and to improve local economies through the identification of valuable genotypes. PGR characterization is often carried out by means of simple sequence repeat (SSR) DNA markers, due to their high reproducibility, multi-allelic nature and co-dominant inheritance [9]. Alternatively, it may encompass morpho-agronomic and/or nutritional features. The nutritional characterization of Brassica species is of great interest, as they are widely appreciated as low-calorie vegetables rich in vitamins and bioactive compounds, including anthocyanin and flavonoids, associated with antioxidant and anti-inflammatory properties [4,10,11,12]. Kale in particular is gaining a great popularity as it is perceived, especially in the US, as a “superfood” due to its health benefits [6]. A number of epidemiological studies reports the negative correlation between consumption of Brassicaceae and risk of chronic diseases, including cardiovascular diseases and cancer [13,14,15,16]. The bioactive molecule content of Brassicaceae mainly relates to the genotype, although environmental conditions may also plays a role [17,18,19].
The present study was promoted by the Apulian Regional Government in the framework of a series of initiatives for the safeguard and valorisation of local agricultural biodiversity. The objective of this research was the establishment and the characterization of a representative collection of Apulian kale, in order to describe patterns of genetic diversity and identify superior populations with respect to agronomic and nutritional traits.
2. Materials and Methods
2.1. Germplasm Collection
Information on Apulian kale collection sites was gathered by setting-up a network among Institutions, farmers, and stakeholders interested in the protection and valorisation of Apulian vegetable biodiversity (further information available at the website www.biodiversitapuglia.it). Collection sites of each accession were mapped through the Geographic Information System (GIS) (Table S1). For each individual accession, seeds were pooled together from the population, and about 10 g were randomly selected to establish an ex situ germplasm collection.
2.2. Phenotypic Evaluation
Apulian kale accessions were grown at the experimental farm “P. Martucci” of the University of Bari Aldo Moro (41°01′22.1′′ N 16°54′21.0′′ E) during the growing season 2016–2017, according to a randomized block design with three replications, each block formed by 20 plants, and adopting traditional agronomic practices. Quantitative traits, namely plant height, flowering time, first harvesting time, edible yield per plant (at the first two harvesting times), and 1000 seed weight, were taken into account together with morphological descriptors for leaf margin (indented or entire), leaf colour (silver-green or green-bluish), and petal colour (yellow or white).
2.3. DNA Extraction and SSR Marker analysis
From each accession, eight individuals also assayed for phenotypic evaluation were randomly sampled. Young leaf tissues were ground in liquid nitrogen and stored at −80 °C until use. DNA was extracted following a CTAB–based protocol [20], and then checked for quality and concentration using 0.8% gel electrophoresis.
A set of 25 SSR primer pairs, previously tested on Brassica species, were retrieved from scientific literature [21,22,23] (Table S2). Each PCR reaction was carried out in a final volume of 20 μL containing about 50 ng of template DNA, 1× of PCR buffer, 0.032 μM of forward primer tagged at the 5′ end with the M13 universal sequence, 0.16 μM of reverse primer, 0.08 μM of a primer complementary to M13 and labelled with one of the FAM, VIC, PET and NED fluorescent dyes (Sigma-Aldrich, St. Louis, MO, USA), 0.2 mM of each dNTP and 0.8 U of DreamTaq polymerase (Thermo Fisher, Waltham, MA, USA). PCR conditions were as following: 94 °C for 5 min, 40 cycles at 94 °C for 30 s, 55–61 °C for 45 s and 72 °C for 45 s and 72 °C for 5 min. PCR products (2 μL) were mixed with 14 μL of formamide and 0.5 μL of the GeneScan 500 LIZ size standard (Life Technologies, Carlsbad, CA, USA), and then used for capillary electrophoresis. This was performed using an ABI PRISM 3100 Genetic Analyzer (Life Technologies). Allele sizes were assigned by using the software GeneMapper 3.7 (Life Technologies).
SSR loci were scored as co-dominant markers, thus individuals displaying one allele were considered to be homozygous. For each marker, the following parameters were calculated using the softwares Genalex 6.5 and Cervus 3.0.7 [24,25]: number of alleles (Na), number of effective alleles (Ne), Shannon’s information index (I), polymorphic information content (PIC), observed heterozygosity (Ho), expected heterozygosity (He), and fixation index (F). The software Genalex 6.5 was also used to perform molecular variance analysis (AMOVA), using 9999 bootstrap iterations, and principal component analysis (PCoA). A dendrogram of genetic distances was constructed using the Unweighted Pair-Group Method with Arithmetic mean (UPGMA) algorithm implemented by POPTREEW, the web version of the POPTREE2 software [26].
2.4. Bioactive Compounds and Antioxidant Activity
For each accession, three lots were created, each one formed by leaf samples of three individual plants. Each lot was then freeze-dried, ground to powder, and stored at −20 °C until use. When required, a methanolic extract was prepared by adding 10 mL of methanol to 0.1 g of powder, and by filtering the supernatant through a 0.45 μM nylon filter.
Total phenolic content (TPC) was determined by mixing 0.1 mL of methanolic extract with Folin-Ciocalteu reagent as reported in [27]. TPC was expressed as mg of gallic acid equivalent (GAE)/g of dry weight, resulting from the mean of three technical replicates.
Total anthocyanin compounds (TAC) were extracted by adding 10 mL of 85:15 (v/v) methanol/1 M HCl to 0.5 g of freeze-dried samples and adjusting the pH to 1.0. The mixture was kept on an orbital shaker at 500 rpm for 30 min in the dark and centrifuged at 12,000× g for 5 min. After recovering the supernatant, the absorbance of the solution was determined at 535 nm using a Cary 60 UV–Vis spectrophotometer (Agilent Technologies, Santa Clara, CA, USA). All the concentrations were estimated using a calibration curve determined by using solutions of the standard cyanidin 3-O-glucoside [28], as the results of three technical replicates.
Total flavonoid content (TFC) was determined using a spectrophotometric method [29]. Freeze-dried sample powder (100 mg) was mixed with 10 mL of 70:29.5:0.5 methanol:water:hydrochloric acid (v/v/v). The UV absorbance was measured at 420 nm, using a calibration curve prepared with quercetin (ranging from 20 to 200 μM, r2 = 0.991), and results were expressed as mg quercetin equivalent/g dry weight (dw).
Leaf antioxidant activity was assessed by two complementary assays based on 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) and 1,1-Diphenyl-2-picrylhydrazyl (DPPH). For the ABTS assay, the ABTS•+ radical cation was generated by the interaction of ABTS (7 mmol/L) with K2S2O8 (2.45 mmol/L). The two solutions were mixed, allowed to stand for 16 h at room temperature, and then diluted with methanol until the absorbance at 734 nm reached 0.7 [30]. The resulting solution (1 mL) was mixed with 10 μL of methanolic extract and, after 6 min of incubation, absorbance was read at 734 nm. Antioxidant capacities of extracts were expressed as μM trolox equivalent/g of dry weight, resulting from the mean of three technical replicates. For the DPPH assay, reaction mixtures containing 0.1 mL of methanolic extracts and 1 mL of 50 μM DPPH• (prepared in methanol) were incubated for 30 min in the dark at room temperature. Then, absorbance at 517 nm was measured for an aliquot of each sample. Antioxidant capacities of extracts were expressed as μM trolox equivalent/g of dry weight, resulting from the mean of three technical replicates.
3. Results
3.1. Establishment of a Kale Germplasm Collection
Seeds from a set of 19 Apulian kale (AK) accessions were collected by missions performed through the province of Bari, Apulia, Italy, which were named sequentially from AK-1 to AK-19 (Table S1). All the accessions are now part of the ex situ germplasm collection of the Department of Plant, Soil, and Food Science of the University of Bari Aldo Moro.
3.2. SSR Polymorphism and Genetic Relationships among Accessions
Out of 25 SSR markers tested, 12 revealed polymorphism in a preliminary assay and were further used to genotype eight individuals for each accession. Information and statistics for each marker are reported in Table 1. In total, 46 SSR alleles were identified within the Apulian kale germplasm collection. The number of alleles per locus ranged from two (BRMS-005, BRMS-019, and BRMS-033) to seven (OL12-A04). Shannon’s Information Index (I) values ranged from 0.123 (BRMS-033) to 1.017 (OL12-A04). Consistently, polymorphic information content (PIC) was lowest for BRMS-033 (0.086) and highest for OL12-A04 (0.743). The inbreeding coefficient (F), calculated from the values of observed (Ho) and expected (He) heterozygosity, ranged from −0.657 (BRMS-036) to 0.481 (BRMS-005).

Table 1.
Genetic diversity indices for individual simple sequence repeat (SSR) markers used in this study. Na: number of alleles; Ne: number of effective alleles; Ho: observed heterozygosity; He: expected heterozygosity; I: Shannon’s information index; F: fixation index; and PIC: polymorphic information content.
AMOVA highlighted a significant level of molecular variance among accessions (p < 0.001), corresponding to 34% of the total variance (Table 2). Pairwise values of the PhiPT parameter, analogous to the Wright’s FST fixation index, ranged from 0.06 (AK-12/AK-13) to 0.603 (AK-2/AK-16). PhiPT pairwise values were always significant (p > 0.05), except for the comparisons AK-5/AK-10, AK-12/AK-13, AK-6/AK-7, AK-6/AK-11, and AK-7/AK-11 (Table S3).

Table 2.
Analysis of molecular variance among and within Apulian kale accessions.
Diversity among accessions was further assessed by the construction of an UPGMA dendrogram of genetic similarity (Figure 1a). Here, strong bootstrap support values separated the accessions AK-9, AK-12, AK-13, and AK-14 from the rest of the germplasm collection, and highlighted high level of genetic similarity within groups formed by AK-6/AK-11/AK-7, AK-12/AK-13, and AK-5/AK-10. In addition, AK-16 appeared as a divergent lineage (Figure 1a). Finally, PCoA analysis substantiated most of the results obtained by AMOVA and hierarchical clustering, including the occurrence of a group formed by the accessions AK-9, AK-12, AK-13, and AK-14, together included in the lower-right panel of the PCoA plot. However, the first two components did not differentiate AK-16, which grouped together with AK-5/AK-10 (Figure 1b).
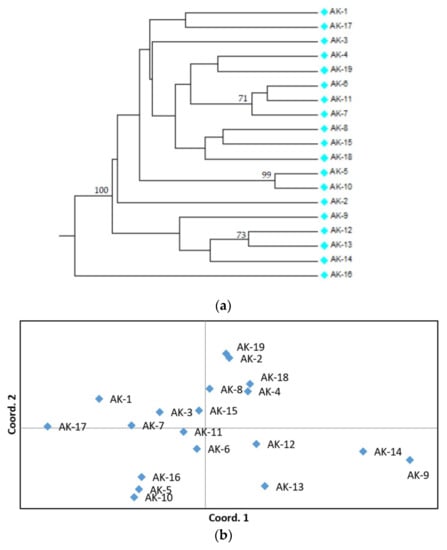
Figure 1.
Genetic diversity among Apulian kale accessions characterized in this study, based on their SSR profile. (a) Unweighted Pair-Group Method with Arithmetic mean (UPGMA) dendrogram of genetic similarity. Bootstrap support values above 70 are indicated above corresponding nodes; (b) principal component analysis (PCoA).
3.3. Phenotypic Variation
All the accessions displayed individuals with indented leaves typical of the Apulian kale (Figure 2). However, accessions from AK-1 to AK-4 also included individuals with entire leaf margin. Leaf and flowers appeared silver-green and yellow, respectively, except for AK-8 and AK-19, displaying green-bluish leaves and white flowers, and AK-3, displaying white flowers. Concerning quantitative traits, analysis of variance (ANOVA) resulted in the detection of significant phenotypic variation with respect to plant height, harvesting time, yield, and flowering time (Table 3). Plant height ranged from 18.6 (AK-14) to 34.2 cm (AK-1), first harvesting time ranged from 120 (AK-9) to 159 (AK-17) days after transplanting and flowering time ranged from 138 (AK-9) to 177 (AK-17) days after transplanting. Concerning the edible yield per plant, this ranged from 34.6 (AK-15) to 188.8 (AK-2) g/plant for the first harvest, and from 38.6 (AK-1) to 176.4 (AK-7) g/plant for the second harvest. Statistical comparisons among means associated with all the traits and accessions are presented in Table 4.

Figure 2.
Apulian kale phenotype with indented leaves.

Table 3.
Analysis of variance (ANOVA) for plant height (PH), first harvesting time (HT), flowering time (FT), edible yield per plant at the first two harvests (Y1 and Y2), and 1000 seed weight (SW) in the Apulian kale germplasm collection under study.

Table 4.
Comparison of individual Apulian kale accessions for plant height (PH), first harvesting time (HT), flowering time (FT), and edible yield per plant at the first two harvests (Y1 and Y2). Values represent means ± standard errors.
3.4. Bioactive Compounds and Correlation with Antioxidant Activity
Results from the characterization of kale germplasm for bioactive compounds and antioxidant activity are presented in Table 5. Low level of variation was observed for total phenols content. Significant differences were only observed between the accession AK-13 (4.5 mg gallic acid/g dw) and the accessions AK-14 (3.0 mg gallic acid/g dw), AK-17 (3.1 mg gallic acid/g dw), and AK-18 (3.0 mg gallic acid/g dw). A greater variability was found with respect to total flavonoid content, with values ranging from 51.1 mg quercetin/g dw (AK-14) to 81.9 mg quercetin/g dw (AK-4). A considerable level of variation was also found for total anthocyanins, with an average value of 15.5 mg 3-O glucoside/100 g dw. The accessions AK-5, AK-8, AK-12, and AK-19 displayed the highest total anthocyanin content (about 20 mg 3-O glucoside/100 g dw), whereas the accessions AK-3 and AK-7 were associated with the lowest anthocyanin content (about 10 mg 3-O glucoside/100 d dw).

Table 5.
Comparison of individual Apulian kale accessions for total phenol content, total flavonoid content, total anthocyanin content, and antioxidant activities. Values represent means ± standard errors.
Antioxidant activity, evaluated through two different assays (ABTS and DPPH tests) was significantly correlated with the total phenolic content (R2 = 0.883 and R2 = 0.684, respectively) and the total flavonoid content (R2 = 0.602 and R2 = 0.454, respectively), while the correlation with the total anthocyanins content did not have statistical significance. Overall, AK-13, characterized by the highest phenols content and a high content of flavonoids, showed the greatest antioxidant activity, while AK-14, AK-17, and AK-18, characterized by the lowest total phenolic content and low total flavonoids content, are the samples with the lowest antioxidant activity. Figure 3 shows the results of PCoA analysis performed on Apulian kale accessions, based on bioactive molecule content and antioxidant activity. PC1, responsible for over 73% of the variability, leads to a distribution of samples in relation to the total phenol and flavonoid contents and the antioxidant activity, while PC2 distributes samples according to the total anthocyanin content.
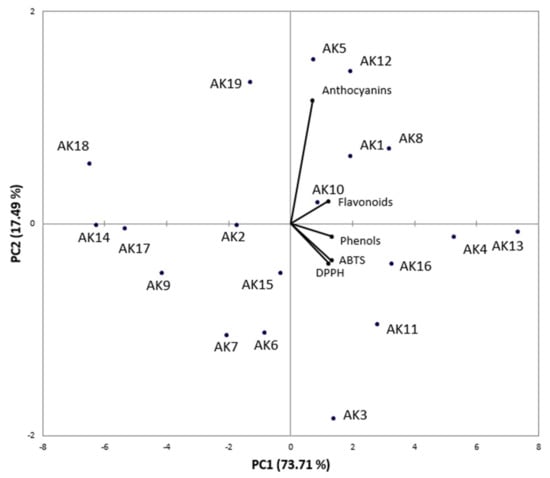
Figure 3.
Genotype principal component (PCoA) analysis based on bioactive molecules content and antioxidant activity.
4. Discussion
This work reports the establishment of a germplasm collection of Apulian kale, a typical vegetable production endangered by genetic erosion [8]. The collection, besides representing a valuable resource for kale ex situ conservation, might facilitate seed access to growers and thus on-farm conservation. Genotyping by means of SSR markers highlighted extremely informative primer combinations (Table 1), which could be conveniently used for future molecular characterizations of kale germplasm. When averaged over all markers and accessions, observed and expected heterozygosity values were close to each other, resulting in a low value of fixation index (−0.039, Table 1). Similar results were obtained when individual accessions were considered (Table S4). Together, these results are in line with the open pollinated nature of kale and a general low level of inbreeding in Apulian populations.
High level of molecular variance within accessions (66% of the total) is also consistent with low level of inbreeding in Apulian germplasm. On the other hand, significant level of variation among accessions (34% of the total) suggests the occurrence of genetic structure and the possible fixation of some traits in individual accessions. Pairwise PhiPT estimates, UPGMA hierarchical clustering and PCoA together provided indication of genetic relationships among individual accessions. All these analysis suggested the occurrence of a distinct group formed by the accessions AK-9, AK-12, AK-13, and AK-14, which might thus be associated with peculiar features. Moreover, they highlight close relationships between the accessions by AK-5/AK-10, AK-12/AK-13, and AK-6/AK-7/AK-11 (Figure 1 and Table S3). The accession AK-16, appearing as a divergent lineage in the dendrogram, does not appear genetically separated by PCoA analysis (Figure 1). It might be thus possible that the patterns of variation differentiating AK-16 from other Apulian kale accessions are caught by other components than the two represented by the PCoA plot. Interestingly, AK-16 is, together with AK-19, one of the two accessions sampled outside the metropolitan area of Bari, thus suggesting the occurrence of geographical stratification.
In line with molecular analyses, bio-agronomic characterization of the Apulian kale germplasm collection also revealed significant differences among accessions, suggesting the opportunity to carry out breeding programs aimed to the selection of superior genotypes and populations. This might ultimately contribute to the commercial valorisation of this vegetable, currently mainly grown for self-consumption [7], in local and international markets. Noteworthy, besides relatively high-yielding accessions, this study also characterized an accession, AK-9, associated with relatively earlier first harvesting time, about one-month before all the other accessions characterized in this study (Table 4). This trait might be of interest for varietal development and for basic studies on flowering time in kale. However, it should be highlighted that quantitative trait data, referring to one single location and one year, are only meant to provide a preliminary evaluation of plant genetic resources collected in this study, thus further trials should be carried out to carefully assess the performance of individual accessions.
Finally, this study also provides a first characterization of Apulian kale germplasm with respect to bioactive molecules and antioxidant proprieties. This is of great interest, as Brassica vegetables, and kale in particular, have gained the attention of the public and the scientific community for their health potential [31]. Notably, high variation with respect to flavonoid and anthocyanin content (Table 5) indicates polymorphism in genes involved in the biosynthesis of these compounds and the possibility to exploit this feature for breeding purposes. Average kale total phenolic and anthocyanin content were similar to those reported for white cabbage [19,32]. As for the flavonoid content, this can be assessed in Brassica species by several quantification methods [33]. The spectrophotometric method used in this study, expressing total flavonoid content as quercetin equivalent, was previously used to estimate flavonoid content in broccoli heads, resulting in a range from 4.52 to 9.27 mg quercetin/g dw [34], and in broccoli extracts, resulting in a range from to values from 12.5 to 17.5 mg quercetin/g dw [35]. Therefore, results of this study suggest that the Apulian kale has, on average, a much higher flavonoid content (65.2 quercetin/g dw, Table 5). However, we used a different extraction process based on low amounts of hydrochloric acid, thus the methods might not be perfectly overlapping. Besides this, it is known that estimates of phenolic compounds, including anthocyanins and flavonoids, are strongly influenced by environmental factors [30], so caution should be taken when comparing results from different experiments. Clearly, future investigations specifically addressing comparisons among different Brassicaceae, which were beyond the scope of this work, might provide additional elements for the nutritional characterization of the Apulian kale.
Supplementary Materials
The following are available online at http://www.mdpi.com/1424-2818/10/2/25/s1, Table S1: GPS coordinates and collection sites of the Apulian kale accessions characterized in this study. Table S2: Polymorphic primer pairs tested on the whole germplasm collection of Apulian kale. Asterisks indicate polymorphic primer pairs used for germplasm genetic characterization. Table S3: Pairwise population PhiPT values among Apulian kale accessions. PhiPT values are shown below diagonal. Probability values based on 999 permutations are shown above diagonal. Table S4: Fixation Index (F) among the 19 Apulian kale (AK) accessions collected in the study.
Acknowledgments
This work was funded by the Regional Apulian project “Biodiversity of Apulian vegetable species”—Programma di Sviluppo Rurale per la Puglia 2014–2020. Misura 10—Sottomisura 10.2; grant CUP H92C15000270002, Italy.
Author Contributions
C.L. and L.R. conceived the study and designed the experiment; P.I. and V.F. performed SSR marker analysis; I.C. performed biochemical analyses; A.R.M. and G.M. performed field trials; P.I., S.P., A.R.M., I.C., C.S., and C.L. were involved in data analysis; S.P., P.I., and I.C. wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Balkaya, A.; Yanmaz, R. Promising kale (Brassica oleracea var. acephala) populations from Black Sea region in Turkey. N. Z. J. Crop Hortic. Sci. 2005, 33, 1–7. [Google Scholar] [CrossRef]
- Dixon, G.R. Vegetable Brassicas and Related Crucifers; Crop Production Science in Horticulture Series, 14; CABI: Wallingford, UK, 2007; Volume 60, pp. 205–211. ISBN 9780851993959. [Google Scholar]
- Cartea, M.E.; Picoaga, A.; Soengas, P.; Ordás, A. Morphological characterization of kale populations from northwestern Spain. Euphytica 2003, 29, 25–32. [Google Scholar] [CrossRef]
- Giorgetti, L.; Giorgi, G.; Cherubini, E.; Gervasi, P.G.; Della Croce, C.M.; Longo, V.; Bellani, L. Screening and identification of major phytochemical compounds in seeds, sprouts and leaves of Tuscan black kale Brassica oleracea (L.) ssp acephala (DC) var. sabellica L. Nat. Prod. Res. 2017, 23, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.; von Bothmer, R.; Poulsen, G.; Maggioni, L.; Phillip, M.; Andersen, B.A. AFLP analysis of genetic diversity in leafy kale (Brassica oleracea L. convar. acephala (DC.) Alef.) landraces, cultivars and wild populations in Europe. Genet. Res. Crop Evol. 2011, 58, 657–666. [Google Scholar] [CrossRef]
- Šamec, D.; Urlić, B.; Salopek-Sondi, B. Kale (Brassica oleracea var. acephala) as a superfood: Review of the scientific evidence behind. Crit. Rev. Food Sci. Nutr. 2018, 20, 1–37. [Google Scholar] [CrossRef]
- Greifenberg, A. Cavolo da foglia (Brassica oleracea L. Var. acephala DC.). In Orticoltura, 1st ed.; Bianco, V.V., Pimpini, F., Eds.; Pàtron: Bologna, Italy, 1990; pp. 1–985. ISBN 9788855520997. [Google Scholar]
- Elia, A.; Santamaria, P. Biodiversity in vegetable crops: A heritage to save. The case of the Puglia region. Ital. J. Agron. 2013, 8, 21–34. [Google Scholar] [CrossRef]
- Tautz, D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989, 17, 6463–6471. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, L.; Branca, F.; Licciardello, F.; Restuccia, C.; Melilli, M.G.; Argento, S.; Raccuia, S.A. Development of modified atmosphere packages on the quality of sicilian kale (Brassica oleracea var. acephala) shoots. Acta Hortic. 2013, 1005, 315–321. [Google Scholar] [CrossRef]
- Sikora, E.; Bodziarczyk, I. Composition and antioxidant activity of kale (Brassica oleracea L. var. acephala) raw and cooked. Acta Sci. Pol. Technol. Aliment. 2012, 11, 239–248. [Google Scholar] [PubMed]
- Velasco, P.; Cartea, M.E.; Gonzalez, C.; Vilar, M.; Ordás, A. Factors affecting the glucosinolate content of kale (Brassica oleracea acephala group). J. Agric. Food Chem. 2007, 55, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Ambrosone, C.B.; McCann, S.E.; Freudenheim, J.L.; Marshall, J.R.; Zhang, Y.; Shields, P.G. Breast cancer risk in premenopausal women is inversely associated with consumption of broccoli, a source of isothiocyanates, but is not modified by GST genotype. J. Nutr. 2004, 134, 1134–1138. [Google Scholar] [CrossRef] [PubMed]
- Cartea, M.; Francisco, M.; Soengas, P.; Velasco, P. Phenolic compounds in Brassica vegetables. Molecules 2011, 16, 251–280. [Google Scholar] [CrossRef] [PubMed]
- Heber, D.; Bowerman, S. Applying science to changing dietary patterns. J. Nutr. 2001, 131, 3078–3081. [Google Scholar] [CrossRef]
- Verhoeven, D.T.; Goldbohm, R.A.; Van Poppel, G.; Verhagen, H.; Van den Brandt, P.A. Epidemiological studies on brassica vegetables and cancer risk. Cancer Epidemiol. Biomark. Prev. 1996, 5, 733–748. [Google Scholar]
- Gawlik-Dziki, U. Effect of hydrothermal treatment on the antioxidant properties of broccoli (Brassica oleracea var. botrytis italica) florets. Food Chem. 2008, 109, 393–401. [Google Scholar] [CrossRef] [PubMed]
- De Pascale, S.; Maggio, A.; Pernice, R.; Fogliano, V.; Barbieri, G. Sulphur fertilization may improve the nutritional value of Brassica rapa L. subsp. Sylvestris. Eur. J. Agron. 2017, 26, 418–424. [Google Scholar] [CrossRef]
- Kusznierewicz, B.; Śmiechowska, A.; Bartoszek, A.; Namieśnik, J. The effect of heating and fermenting on antioxidant properties of white cabbage. Food Chem. 2008, 108, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–14. [Google Scholar]
- Suwabe, K.; Iketani, H.; Nunome, T.; Kage, T.; Hirai, M. Isolation and characterization of microsatellites in Brassica rapa L. Theor. Appl. Genet. 2002, 104, 1092–1098. [Google Scholar] [CrossRef] [PubMed]
- Lowe, A.J.; Moule, C.; Trick, M.; Edwards, K.J. Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theor. Appl. Genet. 2004, 108, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- El-Esawi, M.A.; Germaine, K.; Bourke, P.; Malone, R. Genetic diversity and population structure of Brassica oleracea germplasm in Ireland using SSR markers. C. R. Biol. 2016, 339, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research: An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Takezaki, N.; Nei, M.; Tamura, K. POPTREEW: Web version of POPTREE for constructing population trees from allele frequency data and computing some other quantities. Mol. Biol. Evol. 2014, 31, 1622–1624. [Google Scholar] [CrossRef] [PubMed]
- Cosmai, L.; Caponio, F.; Pasqualone, A.; Paradiso, V.M; Summo, C. Evolution of the oxidative stability, bio-active compounds and color characteristics of non-thermally treated vegetable pâtés during frozen storage. J. Sci. Food Agric. 2017, 9, 4904–4911. [Google Scholar] [CrossRef] [PubMed]
- Pasqualone, A.; Bianco, A.M.; Paradiso, V.M.; Summo, C.; Gambacorta, G.; Caponio, F.; Blanco, A. Production and characterization of functional biscuits obtained from purple wheat. Food Chem. 2015, 180, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Downes, K.; Chope, G.A.; Terry, L.A. Postharvest application of ethylene and methylcyclopropene either before or after curing affects onion (Allium cepa L.) bulb. Postharvest Biol. Technol. 2010, 55, 36–44. [Google Scholar] [CrossRef]
- Huang, D.; Ou, B.; Prior, R.L. The chemistry behind antioxidant capacity assays. J. Agric. Food Res. 2005, 53, 1841–1856. [Google Scholar] [CrossRef] [PubMed]
- Kasprzak, K.; Oniszczuk, T.; Wójtowicz, A.; Waksmundzka-Hajnos, M.; Olech, M.R.; Polak, R.; Oniszczuk, A. Phenolic acid content and antioxidant properties of extruded corn snacks enriched with kale. J. Anal. Methods Chem. 2018, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Leja, M.; Kamińska, I.; Kołton, A. phenolic compounds as the major antioxidants in red cabbage. Folia Hort. 2010, 22, 19–24. [Google Scholar] [CrossRef]
- Mageney, V.; Neugart, S.; Albach, D.C. A guide to the variability of flavonoids in Brassica oleracea. Molecules 2017, 22, 252. [Google Scholar] [CrossRef] [PubMed]
- Naguib, A.E.-M.M.; El-Baz, F.K.; Salama, Z.A.; Hanaa, H.A.E.B; Ali, H.F.; Gaafar, A.A. Enhancement of phenolics, flavonoids and glucosinolates of Broccoli (Brassica olaracea, var. Italica) as antioxidants in response to organic and bio-organic fertilizers. J. Saudi Soc. Agric. Sci. 2012, 11, 135–142. [Google Scholar] [CrossRef]
- Jaiswal, A.K.; Abu-Ghannam, N.; Gupta, S. A comparative study on the polyphenolic content, antibacterial activity and antioxidant capacity of different solvent extracts of Brassica oleracea vegetables. Int. J. Food Sci. Technol. 2012, 47, 223–231. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).