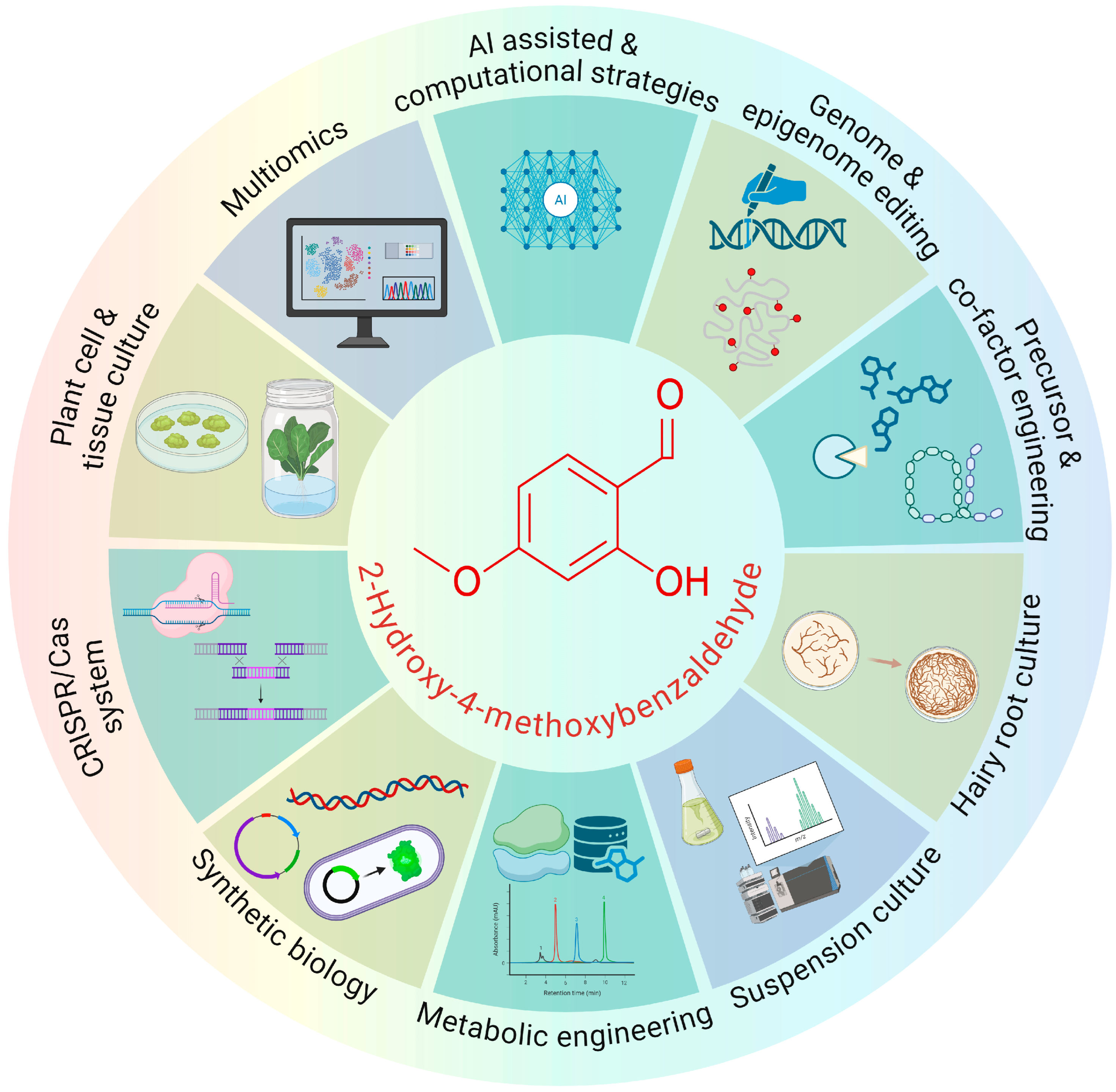
2-Hydroxy-4-Methoxybenzaldehyde (2H4MB): Integrating Cell Culture, Metabolic Engineering, and Intelligent Genome Editing
Abstract
1. Introduction
| Plant | Plant Parts | Biological Role | Extracting Solvent | Test Organism Models/Cell/Tissue | References |
|---|---|---|---|---|---|
| H. indicus | Root | Anti-biofilm | Methanol | Human cell lines | [13] |
| Mondia whytei | Root | Ovicidal | NS | Anopheles gambiae Eggs | [14] |
| H. indicus | Root | TLR2 inhibition | Methanol | Synovial fibroblast and endothelial | [15] |
| H. indicus | Root | Anti-virulence | Methanol | Human peripheral blood mononuclear cells (PBMCs) | [16] |
| H. indicus | Root | Anti-carcinogenic | Methanol | Breast cancer | [17] |
| D. arayalpathra | Root | Antioxidant and anticancer | Dichloromethane | Breast cancer cells | [18] |
| D. hamiltonii | Tuber | Antioxidant | Aqueous | C. elegans | [19] |
| Janakia arayalpatra | Root | Anti venom | Steam distilled | Swiss albino mice and Wistar rats | [20] |
| H. indicus | Root | Acetylcholinesterase Inhibitory Activities | Methanol | Neurotoxic | [21] |
| V. planifolia | Pod | Acetylcholinesterase Inhibitory Activities | Methanol | Neurotoxic | [21] |
| D. hamiltonii | Rhizome | Antimycotic, antiaflatoxigenic and antibiodetriorative activity | Petroleum ether | A. niger, A. columnaris and A. tamari, F. oxysporum, F. proliferatum, Drechslera tetramera and A. ochraceus | [22] |
| Periploca sepium | Root bark | Antimicrobial and antioxidant | Hexane, h n-hexane-ethyl acetate, acetone | B. subtilis, S. aureus, A. tumefaciens, E. coli, P. lachrymans, S. typhimurium, X. vesicatoria | [6] |
| Decalepis hamiltonii | Rhizomes | Antifungal | Petroleum ether | Alternaria alternata, Drechslera tetramera, Fusarium oxysporum, F. proliferatum, Pyricularia oryzae and Trichoconis padwickii | [23] |
2. Plant-Level Physiology and Development of 2H4MB
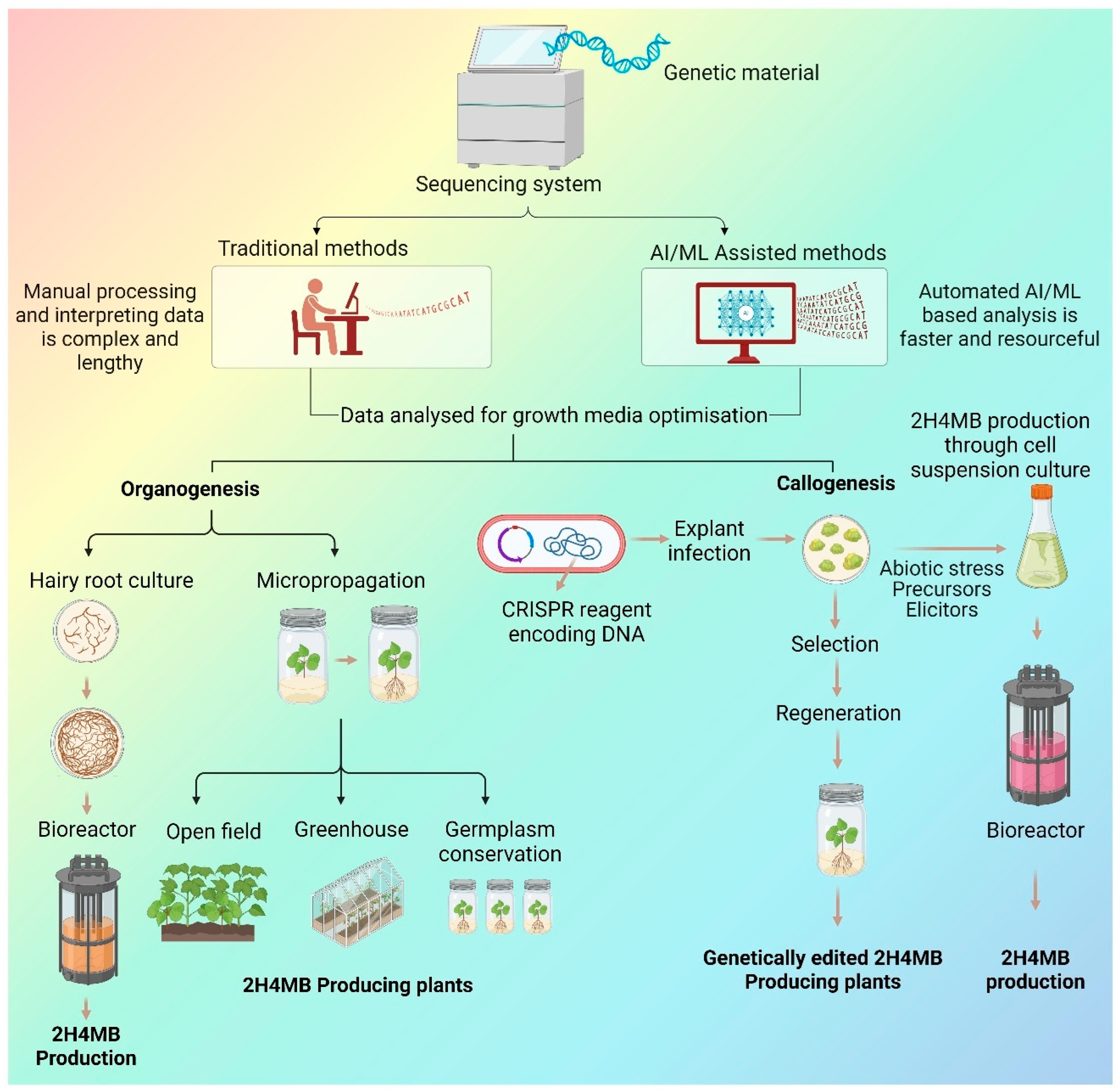
3. In Vitro Production of 2H4MB
3.1. 2H4MB Production Through Elicitation and Cell Suspension Culture
3.2. 2H4MB Production Through Root and Hairy Root Culture
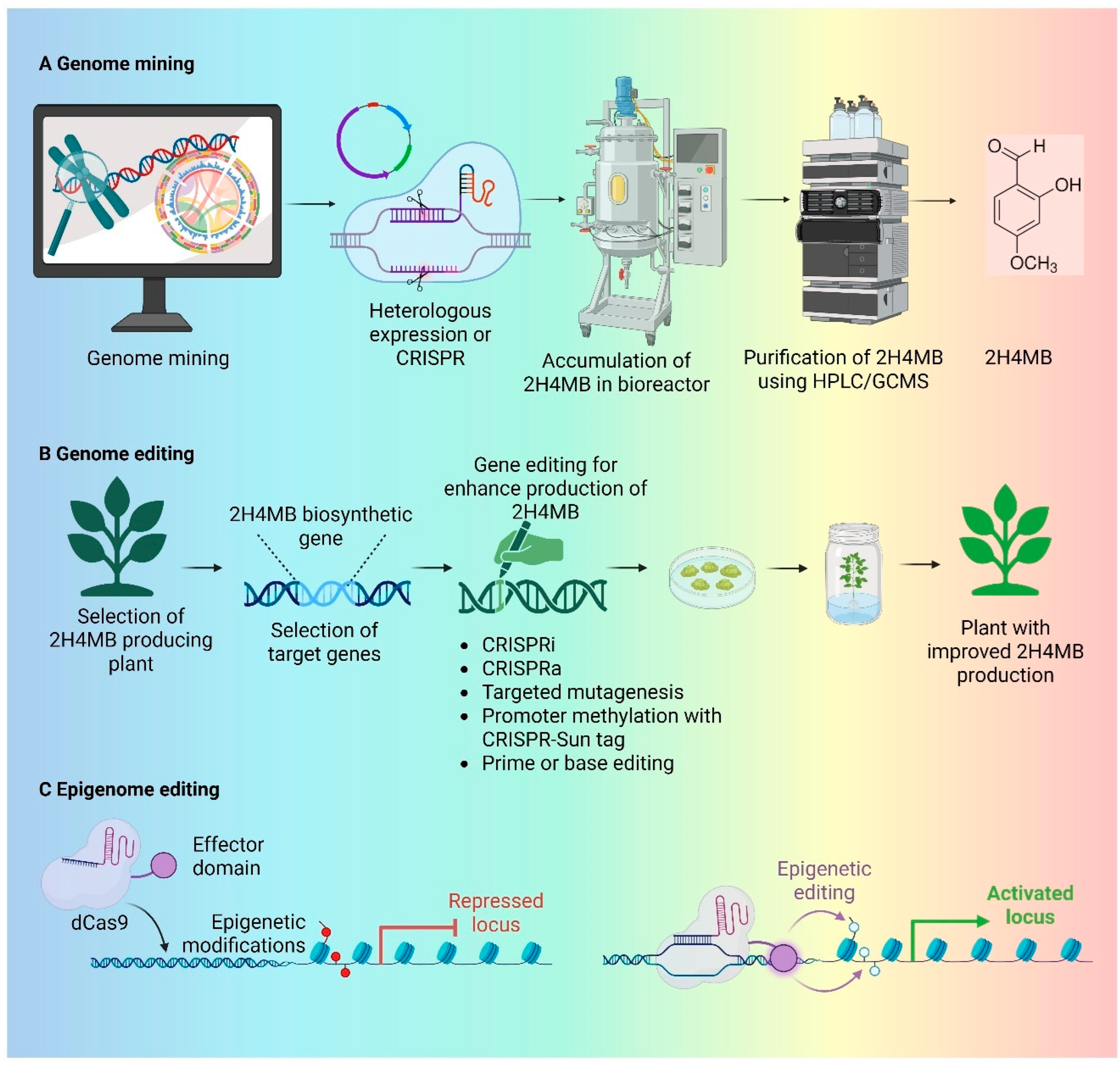
4. 2H4MB Production Through Metabolic Engineering and Genome Mining
4.1. 2H4MB Production Through Microbial System
4.2. Next-Generation Microbial Platforms for 2H4MB Biosynthesis
5. AI-Assisted Pathway Prediction and Genome Editing Strategies for 2H4MB Enhancement
5.1. Pathway Prediction and Optimisation of In Vitro Production System
5.2. Genome Editing Strategies for 2H4MB Enhancement
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vieira, S.F.; Reis, R.L.; Ferreira, H.; Neves, N.M. Plant-Derived Bioactive Compounds as Key Players in the Modulation of Immune-Related Conditions. Phytochem. Rev. 2025, 24, 343–460. [Google Scholar] [CrossRef]
- Luo, P.; Feng, X.; Liu, S.; Jiang, Y. Traditional Uses, Phytochemistry, Pharmacology and Toxicology of Ruta Graveolens L.: A Critical Review and Future Perspectives. DDDT 2024, 18, 6459–6485. [Google Scholar] [CrossRef] [PubMed]
- Parvin, S.; Reza, A.; Das, S.; Miah, M.M.U.; Karim, S. Potential Role and International Trade of Medicinal and Aromatic Plants in the World. Eur. J. Agric. Food Sci. 2023, 5, 89–99. [Google Scholar] [CrossRef]
- Wu, Z.; Shangguan, D.; Huang, Q.; Wang, Y.-K. Drug Metabolism and Transport Mediated the Hepatotoxicity of Pleuropterus Multiflorus Root: A Review. Drug Metab. Rev. 2024, 56, 349–358. [Google Scholar] [CrossRef]
- Gong, H.; Tan, X.; Hou, J.; Gong, Z.; Qin, X.; Nie, J.; Zhu, H.; Zhong, S. Separation, Purification, Structure Characterization, and Immune Activity of a Polysaccharide from Alocasia Cucullata Obtained by Freeze-Thaw Treatment. Int. J. Biol. Macromol. 2024, 282, 137232. [Google Scholar] [CrossRef]
- Wang, J.; Liu, H.; Zhao, J.; Gao, H.; Zhou, L.; Liu, Z.; Chen, Y.; Sui, P. Antimicrobial and Antioxidant Activities of the Root Bark Essential Oil of Periploca Sepium and Its Main Component 2-Hydroxy-4-Methoxybenzaldehyde. Molecules 2010, 15, 5807–5817. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, Z.; Babich, O.; Sukhikh, S.; Yadav, V.; Ramakrishnan, M.; Firdaus, F.; Shahzad, A. Unlocking the Biotechnological Potential of Decalepis Arayalpathra: Exploring Synthetic Seed Production, Metabolic Profiling, Genetic Stability, and the Impact of Photosynthetic Photon Flux Density on Acclimatization. BMC Plant Biol. 2025, 25, 189. [Google Scholar] [CrossRef]
- EFSA Panel on Food Additives and Flavourings (FAF); Younes, M.; Aquilina, G.; Castle, L.; Engel, K.; Fowler, P.; Frutos Fernandez, M.J.; Fürst, P.; Gürtler, R.; Gundert-Remy, U.; et al. Scientific Opinion on Flavouring Group Evaluation 414 (FGE.414): 2-Hydroxy-4-Methoxybenzaldehyde. EFSA J. 2021, 19, e06883. [Google Scholar] [CrossRef]
- Takma, D.K.; Korel, F. Microbial Production of Aromatic Benzaldehydes (Cherry and Fruit Flavors). In Microbial Production of Food Bioactive Compounds; Jafari, S.M., Harzevili, F.D., Karaca, A.C., Eds.; Springer: Cham, Switzerland, 2025; pp. 809–829. ISBN 978-3-031-90973-3. [Google Scholar]
- Ahmad, Z.; Shahzad, A.; Sharma, S. Chitosan versus Yeast Extract Driven Elicitation for Enhanced Production of Fragrant Compound 2-Hydroxy-4-Methoxybenzaldehyde (2H4MB) in Root Tuber Derived Callus of Decalepis Salicifolia (Bedd. Ex Hook.f.) Venter. Plant Cell Tiss. Organ. Cult. 2019, 136, 29–40. [Google Scholar] [CrossRef]
- Yadav, V.; Ahmad, Z.; Shahzad, A.; Upadhyay, A. Advancing Hemidesmus Indicus Propagation and Conservation: Somatic Embryogenesis, Histology, Metabolite Assessment and Genetic Stability. S. Afr. J. Bot. 2024, 168, 394–405. [Google Scholar] [CrossRef]
- Kamireddy, K.; Matam, P.; Parvatam, G. Biochemical Characterization of a Key Step Involved in 2H4MB Production in Decalepis Hamiltonii. J. Plant Physiol. 2017, 214, 74–80. [Google Scholar] [CrossRef]
- Kannappan, A.; Durgadevi, R.; Srinivasan, R.; Lagoa, R.J.L.; Packiavathy, I.A.S.V.; Pandian, S.K.; Veera Ravi, A. 2-Hydroxy-4-Methoxybenzaldehyde from Hemidesmus indicus Is Antagonistic to Staphylococcus epidermidis Biofilm Formation. Biofouling 2020, 36, 549–563. [Google Scholar] [CrossRef]
- Andati, R.E.; Omolo, M.O.; Ndiege, I.O. Ovicidal Activity of 2-Hydroxy-4-Methoxybenzaldehyde, Derivatives and Structural Analogues on Anopheles gambiae Eggs. J. Chem. 2025, 2025, 3387088. [Google Scholar] [CrossRef]
- Durgam, M.K.; Bodiga, V.L.; Vemuri, P.K.; Aenugu, V.R.; Bodiga, S. 2-Hydroxy-4-Methoxybenzaldehyde from Hemidesmus Indicus Root Extract Suppresses Toll-like Receptor2-Mediated Migration and Invasive Mechanisms in Rheumatoid Arthritis. J. Herb. Med. 2023, 42, 100820. [Google Scholar] [CrossRef]
- Kannappan, A.; Srinivasan, R.; Nivetha, A.; Annapoorani, A.; Pandian, S.K.; Ravi, A.V. Anti-Virulence Potential of 2-Hydroxy-4-Methoxybenzaldehyde against Methicillin-Resistant Staphylococcus Aureus and Its Clinical Isolates. Appl. Microbiol. Biotechnol. 2019, 103, 6747–6758. [Google Scholar] [CrossRef] [PubMed]
- Bansod, A.A.; Ramasamy, G.; Nathan, B.; Kandhasamy, R.; Palaniappan, M.; Vichangal Pridiuldi, S. Exploring the Endogenous Potential of Hemidesmus Indicus against Breast Cancer Using in Silico Studies and Quantification of 2-Hydroxy-4-Methoxybenzaldehyde through RP-HPLC. 3 Biotech 2021, 11, 235. [Google Scholar] [CrossRef] [PubMed]
- Thangam, R.; Gokul, S.; Sathuvan, M.; Suresh, V.; Sivasubramanian, S. A Novel Antioxidant Rich Compound 2-Hydoxy 4-Methylbenzaldehyde from Decalepis Arayalpathra Induces Apoptosis in Breast Cancer Cells. Biocatal. Agric. Biotechnol. 2019, 21, 101339. [Google Scholar] [CrossRef]
- Kamireddy, K.; Chinnu, S.; Priyanka, P.S.; Rajini, P.S.; Giridhar, P. Neuroprotective Effect of Decalepis Hamiltonii Aqueous Root Extract and Purified 2-Hydroxy-4-Methoxybenzaldehyde on 6-OHDA Induced Neurotoxicity in Caenorhabditis Elegans. Biomed. Pharmacother. 2018, 105, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.I.; Gomes, A. Viper Venom-Induced Inflammation and Inhibition of Free Radical Formation by Pure Compound (2-Hydroxy-4-Methoxy Benzoic Acid) Isolated and Purified from Anantamul (Hemidesmus Indicus R. BR) Root Extract. Toxicon 1998, 36, 207–215. [Google Scholar] [CrossRef]
- Kundu, A.; Mitra, A. Flavoring Extracts of Hemidesmus Indicus Roots and Vanilla Planifolia Pods Exhibit In Vitro Acetylcholinesterase Inhibitory Activities. Plant Foods Hum. Nutr. 2013, 68, 247–253. [Google Scholar] [CrossRef]
- Mohana, D.C.; Raveesha, K.A. Antimycotic, Antibiodeteriorative and Antiaflatoxigenic Potency of 2-Hydroxy-4-Methoxybenzaldehyde Isolated from Decalepis Hamiltonii on Fungi Causing Biodeterioration of Maize and Sorghum Grains. J. Mycol. Plant Pathol. 2010, 40, 197–206. [Google Scholar]
- Mohana, D.C.; Satish, S.; Raveesha, K.A.A. Antifungal Activity of 2-Hydroxy-4-Methoxybenzaldehyde Isolated from Decalepis hamiltonii (Wight & Arn.) on Seed-Borne Fungi Causing Biodeterioration of Paddy. J. Plant Prot. Res. 2009, 49. [Google Scholar]
- Ahmad, Z.; Shahzad, A.; Sharma, S.; Parveen, S. Ex Vitro Rescue, Physiochemical Evaluation, Secondary Metabolite Production and Assessment of Genetic Stability Using DNA Based Molecular Markers in Regenerated Plants of Decalepis Salicifolia (Bedd. Ex Hook.f.) Venter. Plant Cell Tiss. Organ. Cult. 2018, 132, 497–510. [Google Scholar] [CrossRef]
- Giridhar, P.; Rajasekaran, T.; Ravishankar, G.A. Improvement of Growth and Root Specific Flavour Compound 2-Hydroxy-4-Methoxybenzaldehyde of Micropropagated Plants of Decalepis Hamiltonii Wight & Arn., under Triacontanol Treatment. Sci. Hortic. 2005, 106, 228–236. [Google Scholar] [CrossRef]
- Guo, M.; Lv, H.; Chen, H.; Dong, S.; Zhang, J.; Liu, W.; He, L.; Ma, Y.; Yu, H.; Chen, S. Strategies on Biosynthesis and Production of Bioactive Compounds in Medicinal Plants. Chin. Herb. Med. 2024, 16, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Leal, C.A.; Puente-Garza, C.A.; García-Lara, S. In Vitro Plant Tissue Culture: Means for Production of Biological Active Compounds. Planta 2018, 248, 1–18. [Google Scholar] [CrossRef]
- Rodrigues, V.; Kumar, A.; Prabhu, K.N.; Pragadheesh, V.S.; Shukla, A.K.; Sundaresan, V. Adventitious Root Cultures of Decalepis Salicifolia for the Production of 2-Hydroxy-4-Methoxybenzaldehyde, a Vanillin Isomer Flavor Metabolite. Appl. Microbiol. Biotechnol. 2021, 105, 3087–3099. [Google Scholar] [CrossRef]
- Nandy, S.; Hazra, A.K.; Pandey, D.K.; Ray, P.; Dey, A. Elicitation of Industrially Promising Vanillin Type Aromatic Compound 2-Hydroxy 4-Methoxy Benzaldehyde (MBAlD) Yield in the in-Vitro Raised Medicinal Crop Hemidesmus Indicus (L) R. Br. by Methyl Jasmonate and Salicylic Acid. Ind. Crops Prod. 2021, 164, 113375. [Google Scholar] [CrossRef]
- Kiran, K.; Sonbarse, P.P.; Veeresh, L.; Shetty, N.P.; Parvatam, G. Expression Profile of Phenylpropanoid Pathway Genes in Decalepis Hamiltonii Tuberous Roots during Flavour Development. 3 Biotech 2018, 8, 365. [Google Scholar] [CrossRef]
- Ahmad, Z.; Shahzad, A.; Sharma, S. Enhanced Multiplication and Improved Ex Vitro Acclimatization of Decalepis Arayalpathra. Biol. Plant. 2018, 62, 1–10. [Google Scholar] [CrossRef]
- Sudha, C.G.; Sherina, T.V.; Anu Anand, V.P.; Reji, J.V.; Padmesh, P.; Soniya, E.V. Agrobacterium Rhizogenes Mediated Transformation of the Medicinal Plant Decalepis Arayalpathra and Production of 2-Hydroxy-4-Methoxybenzaldehyde. Plant Cell Tiss. Organ. Cult. 2013, 112, 217–226. [Google Scholar] [CrossRef]
- Kundu, A.; Jawali, N.; Mitra, A. Shikimate Pathway Modulates the Elicitor-Stimulated Accumulation of Fragrant 2-Hydroxy-4-Methoxybenzaldehyde in Hemidesmus Indicus Roots. Plant Physiol. Biochem. 2012, 56, 104–108. [Google Scholar] [CrossRef]
- Chakraborty, D.; Sircar, D.; Mitra, A. Phenylalanine Ammonia-Lyase-Mediated Biosynthesis of 2-Hydroxy-4-Methoxybenzaldehyde in Roots of Hemidesmus Indicus. J. Plant Physiol. 2008, 165, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Sreekumar, S.; Seeni, S.; Pushpangadan, P. Micropropagation of Hemidesmus Indicus for Cultivation and Production of 2-Hydroxy 4-Methoxy Benzaldehyde. Plant Cell Tissue Organ. Cult. 2000, 62, 211–218. [Google Scholar] [CrossRef]
- Selwal, N.; Supriadi, K.; Rahayu, F.; Sukmadjaja, D.; Khamidah, A.; Budiaarto, K.; Antarlina, S.S.; Tripatmasari, M.; Wani, A.K. Elicitation Strategies for Enhanced Secondary Metabolite Synthesis in Plant Cell Cultures and Its Role in Plant Defense Mechanism. Plant Gene 2025, 41, 100485. [Google Scholar] [CrossRef]
- Mishra, V.K.; Goswami, R.; Naidu, R.T. Establishment of in Vitro Cell Suspension Culture, Kinetics of Cell Growth, pH, Nutrient Uptake and Production of 2-Hydroxy-4-Methoxybenzaldehyde from the Germinated Root of Decalepis Hamiltonii Wight & Arn.-an Endangered Plant. Curr. Appl. Sci. Technol. 2022, 22. [Google Scholar] [CrossRef]
- Wu, T.; Kerbler, S.M.; Fernie, A.R.; Zhang, Y. Plant Cell Cultures as Heterologous Bio-Factories for Secondary Metabolite Production. Plant Commun. 2021, 2, 100235. [Google Scholar] [CrossRef] [PubMed]
- Matam, P.; Parvatam, G.; Shetty, N.P. Enhanced Production of Vanillin Flavour Metabolites by Precursor Feeding in Cell Suspension Cultures of Decalepis hamiltonii Wight & Arn., in Shake Flask Culture. 3 Biotech 2017, 7, 376. [Google Scholar] [CrossRef]
- Namdeo, A.G.; Meti, N.T. Effect of Ferulic Acid and Chitosan on Cell Suspension Cultures of Decalepis hamiltonii Wight & Arn. Ecol. Environ. Conserv. 2024, 30, S90–S96. [Google Scholar] [CrossRef]
- Mirmazloum, I.; Slavov, A.K.; Marchev, A.S. The Untapped Potential of Hairy Root Cultures and Their Multiple Applications. Int. J. Mol. Sci. 2024, 25, 12682. [Google Scholar] [CrossRef]
- Atabaki, N.; Shaharuddin, N.A.; Ahmad, S.A.; Nulit, R.; Malik, S.; Vahedi, M.; Kalhori, N.; Abiri, R. Hairy Root Culture: A Reliable Bioreactor from Transgenic Plants. Pept. Protein Drug Deliv. Using. Polysacch. 2024, 25–50. [Google Scholar]
- Srivastava, S.; Nalla, V.K.; Singh, V.P.; Prasad, R. Optimization of in Vitro Micropropagation and Root Establishment through Combinatorial Approaches for Enhanced Production of Secondary Metabolites in the Endangered Species Decalepis Arayalpathra KMA 05 Clones. Braz. J. Bot. 2022, 45, 869–881. [Google Scholar] [CrossRef]
- Xu, M.; Xu, D. Advanced Systems and Bioreactors for Large-Scale Secondary Metabolite Production in Medicinal Plants Using Suspension Cultured Cells. In Biosynthesis of Natural Products in Plants; Kumar, N., Ed.; Springer Nature Singapore: Singapore, 2024; pp. 293–313. ISBN 978-981-97-2165-8. [Google Scholar]
- Han, T.; Miao, G. Strategies, Achievements, and Potential Challenges of Plant and Microbial Chassis in the Biosynthesis of Plant Secondary Metabolites. Molecules 2024, 29, 2106. [Google Scholar] [CrossRef]
- Smit, S.J.; Lichman, B.R. Plant Biosynthetic Gene Clusters in the Context of Metabolic Evolution. Nat. Prod. Rep. 2022, 39, 1465–1482. [Google Scholar] [CrossRef]
- Wang, M.; Chen, L.; Zhang, Z.; Wang, Q. Recent Advances in Genome Mining and Synthetic Biology for Discovery and Biosynthesis of Natural Products. Crit. Rev. Biotechnol. 2025, 45, 236–256. [Google Scholar] [CrossRef] [PubMed]
- Scherlach, K.; Hertweck, C. Mining and Unearthing Hidden Biosynthetic Potential. Nat. Commun. 2021, 12, 3864. [Google Scholar] [CrossRef] [PubMed]
- Dinglasan, J.L.N.; Otani, H.; Doering, D.T.; Udwary, D.; Mouncey, N.J. Microbial Secondary Metabolites: Advancements to Accelerate Discovery towards Application. Nat. Rev. Microbiol. 2025, 23, 338–354. [Google Scholar] [CrossRef] [PubMed]
- Bauman, K.D.; Butler, K.S.; Moore, B.S.; Chekan, J.R. Genome Mining Methods to Discover Bioactive Natural Products. Nat. Prod. Rep. 2021, 38, 2100–2129. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X.; Dai, G.; Zhang, Y.; Bian, X. Microbial Chassis Engineering Drives Heterologous Production of Complex Secondary Metabolites. Biotechnol. Adv. 2022, 59, 107966. [Google Scholar] [CrossRef]
- Zhang, Z.; Lin, Q. Production of P-Anisaldehyde via Whole-Cell Transformation Using Recombinant E. Coli Expressing Trans-Anethole Oxygenase. J. Gen. Appl. Microbiol. 2024, 70. [Google Scholar] [CrossRef]
- Kunjapur, A.M.; Tarasova, Y.; Prather, K.L.J. Synthesis and Accumulation of Aromatic Aldehydes in an Engineered Strain of Escherichia coli. J. Am. Chem. Soc. 2014, 136, 11644–11654. [Google Scholar] [CrossRef]
- Kim, H.-S.; Choi, J.-A.; Kim, B.-Y.; Ferrer, L.; Choi, J.-M.; Wendisch, V.F.; Lee, J.-H. Engineered Corynebacterium Glutamicum as the Platform for the Production of Aromatic Aldehydes. Front. Bioeng. Biotechnol. 2022, 10, 880277. [Google Scholar] [CrossRef] [PubMed]
- Tazon, A.W.; Awwad, F.; Meddeb-Mouelhi, F.; Desgagné-Penix, I. Biotechnological Advances in Vanillin Production: From Natural Vanilla to Metabolic Engineering Platforms. BioChem 2024, 4, 323–349. [Google Scholar] [CrossRef]
- Santos, L.D.; Lautru, S.; Pernodet, J.-L. Genetic Engineering Approaches for the Microbial Production of Vanillin. Biomolecules 2024, 14, 1413. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, P.; Smolke, C.D. Biosynthesis of Medicinal Tropane Alkaloids in Yeast. Nature 2020, 585, 614–619. [Google Scholar] [CrossRef]
- Galanie, S.; Thodey, K.; Trenchard, I.J.; Filsinger Interrante, M.; Smolke, C.D. Complete Biosynthesis of Opioids in Yeast. Science 2015, 349, 1095–1100. [Google Scholar] [CrossRef]
- He, Y.; Guo, Y.; Liang, X.; Hu, H.; Xiong, X.; Zhou, X. Single-Cell Transcriptome and Microbiome Profiling Uncover Ileal ImmuneImpairment in Intrauterine Growth-Retarded Piglets. CPD 2025, 32. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Wang, M.; Gao, H.; Li, L.; Wu, M.; Li, Q.; Bu, F.; Dong, H.; Han, J.; Ying, J. The Interplay between Post-Transcriptional RNA Regulation and Wnt/β-Catenin Signaling in Cancer: A Review. Int. J. Biol. Macromol. 2025, 328, 147657. [Google Scholar] [CrossRef]
- Gu, S.; Shao, Y.; Rehm, K.; Bigler, L.; Zhang, D.; He, R.; Xu, R.; Shao, J.; Jousset, A.; Friman, V.-P. Feature Sequence-Based Genome Mining Uncovers the Hidden Diversity of Bacterial Siderophore Pathways. eLife 2024, 13, RP96719. [Google Scholar] [CrossRef]
- Rekadwad, B.N.; Sudhakar, K.P. antiSMASH: Genome Mining and Genome-Wide Analysis of Gene Clusters in Actinopolyspora Species. In Protocols of Actinomycetes; CRC Press: Boca Raton, FL, USA, 2024; pp. 303–307. [Google Scholar]
- Blin, K.; Shaw, S.; Vader, L.; Szenei, J.; Reitz, Z.L.; Augustijn, H.E.; Cediel-Becerra, J.D.; de Crécy-Lagard, V.; Koetsier, R.A.; Williams, S.E. antiSMASH 8.0: Extended Gene Cluster Detection Capabilities and Analyses of Chemistry, Enzymology, and Regulation. Nucleic Acids Res. 2025, 53, W32–W38. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, J.; Wang, C.; Hua, Y.; Wang, H.; Chen, J. The Deep Mining Era: Genomic, Metabolomic, and Integrative Approaches to Microbial Natural Products from 2018 to 2024. Mar. Drugs 2025, 23, 261. [Google Scholar] [CrossRef]
- Singh, S.; Praveen, A.; Dudha, N.; Sharma, V.K.; Bhadrecha, P. Single-Cell Transcriptomics: A New Frontier in Plant Biotechnology Research. Plant Cell Rep. 2024, 43, 294. [Google Scholar] [CrossRef]
- Wang, Y.-X.; Yu, T.-F.; Wang, C.-X.; Wei, J.-T.; Zhang, S.-X.; Liu, Y.-W.; Chen, J.; Zhou, Y.-B.; Chen, M.; Ma, Y.-Z. Heat Shock Protein TaHSP17. 4, a TaHOP Interactor in Wheat, Improves Plant Stress Tolerance. Int. J. Biol. Macromol. 2023, 246, 125694. [Google Scholar] [CrossRef]
- Anderton, C.R.; Uhrig, R.G. The Promising Role of Proteomes and Metabolomes in Defining the Single-cell Landscapes of Plants. New Phytol. 2025, 245, 945–948. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Veličković, M.; Burnum-Johnson, K.E. From Single Cell to Spatial Multi-Omics: Unveiling Molecular Mechanisms in Dynamic and Heterogeneous Systems. Curr. Opin. Biotechnol. 2024, 89, 103174. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Tian, J.; Zhang, L.; He, H.; Song, D. A Multi-Omics Approach to Reveal Critical Mechanisms of Activator Protein 1 (AP-1). Biomed. Pharmacother. 2024, 178, 117225. [Google Scholar] [CrossRef]
- Winzer, T.; Gazda, V.; He, Z.; Kaminski, F.; Kern, M.; Larson, T.R.; Li, Y.; Meade, F.; Teodor, R.; Vaistij, F.E.; et al. A Papaver Somniferum 10-Gene Cluster for Synthesis of the Anticancer Alkaloid Noscapine. Science 2012, 336, 1704–1708. [Google Scholar] [CrossRef]
- He, B.; Zhu, R.; Yang, H.; Lu, Q.; Wang, W.; Song, L.; Sun, X.; Zhang, G.; Li, S.; Yang, J. Assessing the Impact of Data Preprocessing on Analyzing next Generation Sequencing Data. Front. Bioeng. Biotechnol. 2020, 8, 817. [Google Scholar] [CrossRef]
- Dugé De Bernonville, T.; Amor Stander, E.; Dugé De Bernonville, G.; Besseau, S.; Courdavault, V. Predicting Monoterpene Indole Alkaloid-Related Genes from Expression Data with Artificial Neural Networks. In Catharanthus roseus; Methods in Molecular Biology; Courdavault, V., Besseau, S., Eds.; Springer US: New York, NY, 2022; Volume 2505, pp. 131–140. ISBN 978-1-0716-2348-0. [Google Scholar]
- Tan, W.H.; Tong, C.Y.; Chua, M.X.; Derek, C.J.C. Modelling and Optimizing Secondary Metabolites Production in Spirodela Polyrhiza Using Machine Learning. J. Clean. Prod. 2024, 478, 143986. [Google Scholar] [CrossRef]
- Kong, D.; Qian, J.; Gao, C.; Wang, Y.; Shi, T.; Ye, C. Machine Learning Empowering Microbial Cell Factory: A Comprehensive Review. Appl. Biochem. Biotechnol. 2025, 197, 4897–4913. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Eun, H.; Prabowo, C.P.S.; Cho, S.; Lee, S.Y. Metabolic and Cellular Engineering for the Production of Natural Products. Curr. Opin. Biotechnol. 2022, 77, 102760. [Google Scholar] [CrossRef]
- Lamichhane, S.; Thapa, S. Advances from Conventional to Modern Plant Breeding Methodologies. Plant Breed. Biotechnol. 2022, 10, 1–14. [Google Scholar] [CrossRef]
- Bhojiya, A.A.; Joshi, H. Crispr Gene Editing for Secondary Metabolite Production: A Review. In Gene Editing in Plants; Kumar, A., Arora, S., Ogita, S., Yau, Y.-Y., Mukherjee, K., Eds.; Springer: Singapore, 2024; pp. 437–475. ISBN 978-981-99-8528-9. [Google Scholar]
- Wu, S.; Kyaw, H.; Tong, Z.; Yang, Y.; Wang, Z.; Zhang, L.; Deng, L.; Zhang, Z.; Xiao, B.; Quick, W.P. A Simple and Efficient CRISPR/Cas9 System Permits Ultra-Multiplex Genome Editing in Plants. Crop J. 2024, 12, 569–582. [Google Scholar] [CrossRef]
- Li, X.; Xie, Y.; Zhu, Q.; Liu, Y.-G. Targeted Genome Editing in Genes and Cis-Regulatory Regions Improves Qualitative and Quantitative Traits in Crops. Mol. Plant 2017, 10, 1368–1370. [Google Scholar] [CrossRef] [PubMed]
- Saeed, F.; Manzoor, H.; Bukhat, S.; Azeem, F.; Rasul, S. Multiplex Genome Editing in Plants. In Genome Editing for Crop Improvement; Al-Khayri, J.M., Sattar, M.N., Sopory, S.K., Jain, S.M., Eds.; CABI: Wallingford, UK, 2025. [Google Scholar]
- Das, S.; Kwon, M.; Kim, J.-Y. Enhancement of Specialized Metabolites Using CRISPR/Cas Gene Editing Technology in Medicinal Plants. Front. Plant Sci. 2024, 15, 1279738. [Google Scholar] [CrossRef]
- Lai, G.; Fu, P.; He, L.; Che, J.; Wang, Q.; Lai, P.; Lu, J.; Lai, C. CRISPR/Cas9-Mediated CHS2 Mutation Provides a New Insight into Resveratrol Biosynthesis by Causing a Metabolic Pathway Shift from Flavonoids to Stilbenoids in Vitis Davidii Cells. Hortic. Res. 2025, 12, uhae268. [Google Scholar] [CrossRef]
- Hicks, C.; Witte, T.E.; Sproule, A.; Hermans, A.; Shields, S.W.; Colquhoun, R.; Blackman, C.; Boddy, C.N.; Subramaniam, R.; Overy, D.P. CRISPR-Cas9 Gene Editing and Secondary Metabolite Screening Confirm Fusarium Graminearum C16 Biosynthetic Gene Cluster Products as Decalin-Containing Diterpenoid Pyrones. J. Fungi 2023, 9, 695. [Google Scholar] [CrossRef]
- Shi, M.; Zhang, S.; Zheng, Z.; Maoz, I.; Zhang, L.; Kai, G. Molecular Regulation of the Key Specialized Metabolism Pathways in Medicinal Plants. JIPB 2024, 66, 510–531. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Shao, Y.; Pan, B.; Liu, C.; Tan, H. Regulation of Important Natural Products Biosynthesis by WRKY Transcription Factors in Plants. J. Adv. Res. 2025, 77, 119–135. [Google Scholar] [CrossRef]
- Pratyusha, D.S.; Sarada, D.V.L. MYB Transcription Factors—Master Regulators of Phenylpropanoid Biosynthesis and Diverse Developmental and Stress Responses. Plant Cell Rep. 2022, 41, 2245–2260. [Google Scholar] [CrossRef]
- Subramanian, A.T.; Roy, P.; Aravind, B.; Kumar, A.P.; Mohannath, G. Epigenome Editing Strategies for Plants: A Mini Review. Nucleus 2024, 67, 75–87. [Google Scholar] [CrossRef]
- Leech, D.; Previtera, D.A.; Zhang, Y.; Botella, J.R.; Crisp, P.A. Precision Plant Epigenome Editing: What, How, and Why. Trends Plant Sci. 2025, 6, 1360–1385. [Google Scholar] [CrossRef] [PubMed]
| Plant | Explant | In Vitro/Biological Approaches | Elicitors/Phytohormones/Treatments | 2H4MB Content [Reported Yield] | Quantification Method | Reference |
|---|---|---|---|---|---|---|
| Decalepis salicifolia | Leaf and Stem | Adventitious root culture | Woody plant medium, NAA (0.5 mg/L), Kn (1.0 mg/L), IBA (0.3 mg/L) and sucrose (2%) | Total production of 2H4MB increases to 4.9-fold (139.54 µg) | RP-HPLC | [28] |
| Hemidesmus indicus | Node, Internode, Leaf | Direct and Indirect Organogenesis | Methyl jasmonate (75µM) and Salicylic acid (2 mg/mL) with BAP (1.5 mg/L), TDZ (2.0 mg/L) and IBA (1.5 mg/L) | Root extracts produced 3.41 mg/g 2H4MB | HPTLC | [29] |
| D. salicifolia | Root tuber | Suspension culture (Callus) | Chitosan (CH, 200 µM) and Yeast extract (YE, 200 µM) | Maximum content of 2H4MB at 72 h increases to 1.4-fold (14.8 g/g) | HPLC | [10] |
| D. hamiltonii | Tuberous roots | Developmental (tuber maturation) accumulation | Expression profile of DhPAL, DhC4H, DhCOMT and DhVAN) | 2H4MB increase with maturation of tuber (10, 170, 500 µg/g in 3-month, 18 month and 60-month-old mature plant, respectively) | HPLC | [30] |
| D. arayalpathra | Nodal segment and Shoot tip | Direct organogenesis | Murashige and Skoog’s medium, BA (5.0 μM), IAA (0.5 μM), NAA (2.5 μM) and Adenine sulphate (20.0 μM) | Maximum 2H4MB content which is 9.22 μg/cm3 (root extract) recorded | HPLC | [31] |
| D. salicifolia | Nodal segment | Direct organogenesis | Murashige and Skoog’s medium, BA (5.0 μM), IBA (2.5 μM), NAA (0.5 μM) and Adenine sulphate (30.0 μM) | Maximum 2H4MB content which is 6.8 μg/mL (root extract) recorded | HPLC | [24] |
| D. arayalpathra | Cotyledons and Hypocotyls | Hairy root culture | Agrobacterium rhizogenes (different strains A4, MTCC 532, TR105 and LBA 5402) | Maximum accumulation of 2H4MB (0.22% dw) recorded at 6th week of growth | TLC | [32] |
| H. indicus | Roots | Roots culture with elicitation | Chitosan, Methyl jasmonate and Yeast extract | Yeast extract for 18 h showed maximum accumulation of 2H4MB (2.7 mg/g) | HPLC | [33] |
| H.indicus | Roots | Roots culture with inhibitor/elicitation treatment | Aqueous Chitosan solution (100 and 200 mg/L) and Aminooxyacetic acid (AOAA) solution (50 and 100 μg/L) | Maximum accumulation of 2H4MB (0.89 mg/g) was detected | TLC | [34] |
| H.indicus | Shoot tip and young roots | Micropropagation (Callogenesis) | Murashige and Skoogs Medium, BA (4.4 µM), IBA (9.8 µM) and NAA (2.69 µM) | Concentration of 2H4MB increases to 2.2-fold (0.12%/g dw) | HPLC | [35] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Firdaus, F.; Yadav, V.; Ramakrishnan, M.; Wasi, A.; Ganie, I.B.; Upadhyay, A.; Shahzad, A.; Ahmad, Z. 2-Hydroxy-4-Methoxybenzaldehyde (2H4MB): Integrating Cell Culture, Metabolic Engineering, and Intelligent Genome Editing. Int. J. Mol. Sci. 2026, 27, 503. https://doi.org/10.3390/ijms27010503
Firdaus F, Yadav V, Ramakrishnan M, Wasi A, Ganie IB, Upadhyay A, Shahzad A, Ahmad Z. 2-Hydroxy-4-Methoxybenzaldehyde (2H4MB): Integrating Cell Culture, Metabolic Engineering, and Intelligent Genome Editing. International Journal of Molecular Sciences. 2026; 27(1):503. https://doi.org/10.3390/ijms27010503
Chicago/Turabian StyleFirdaus, Fatima, Vikas Yadav, Muthusamy Ramakrishnan, Adla Wasi, Irfan Bashir Ganie, Anamica Upadhyay, Anwar Shahzad, and Zishan Ahmad. 2026. "2-Hydroxy-4-Methoxybenzaldehyde (2H4MB): Integrating Cell Culture, Metabolic Engineering, and Intelligent Genome Editing" International Journal of Molecular Sciences 27, no. 1: 503. https://doi.org/10.3390/ijms27010503
APA StyleFirdaus, F., Yadav, V., Ramakrishnan, M., Wasi, A., Ganie, I. B., Upadhyay, A., Shahzad, A., & Ahmad, Z. (2026). 2-Hydroxy-4-Methoxybenzaldehyde (2H4MB): Integrating Cell Culture, Metabolic Engineering, and Intelligent Genome Editing. International Journal of Molecular Sciences, 27(1), 503. https://doi.org/10.3390/ijms27010503







