Transcriptional Response of Durum Wheat During Interaction with Debaryomyces hansenii and Fusarium graminearum
Abstract
1. Introduction
2. Results
2.1. Gene Expression Analysis
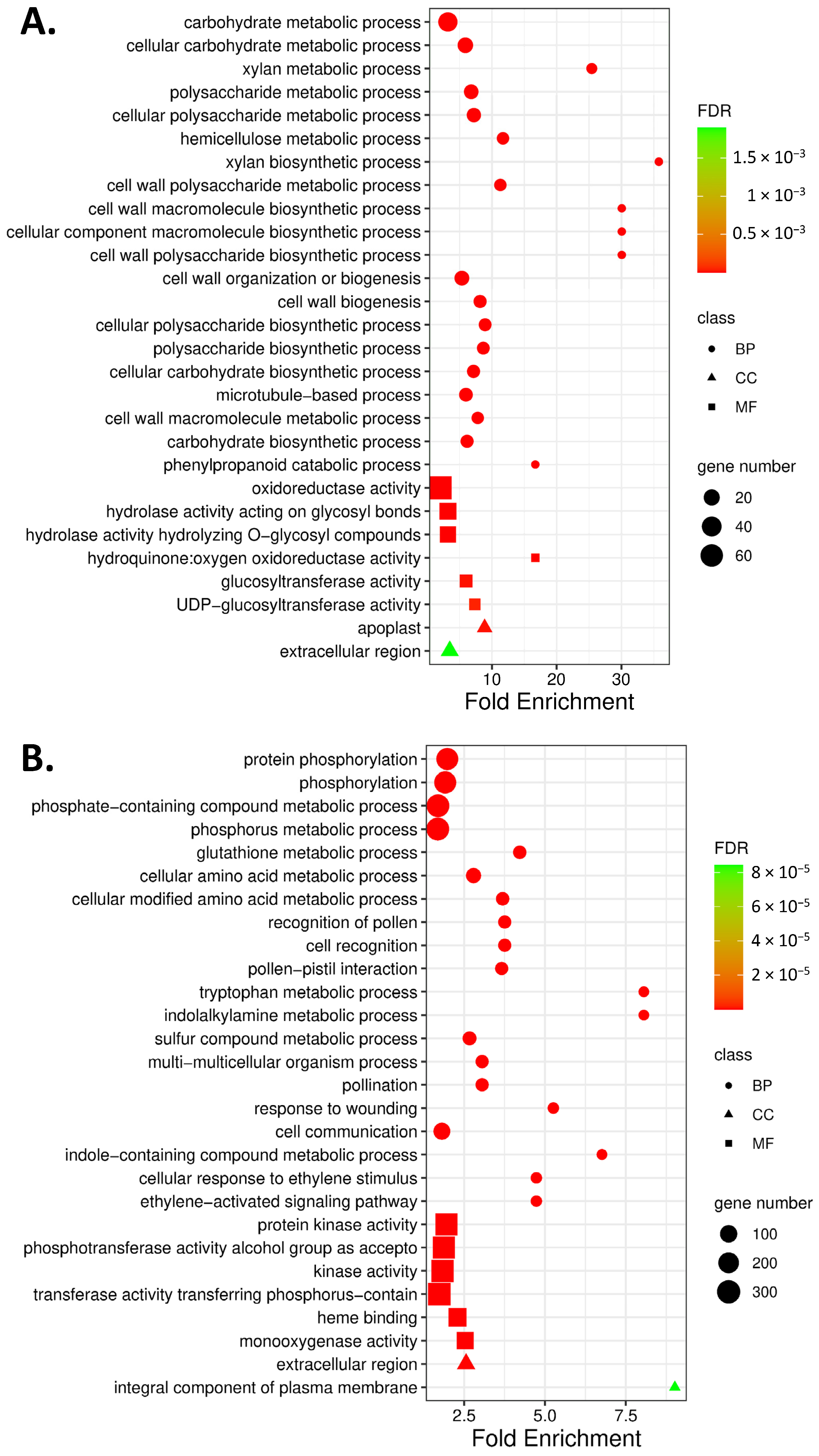
2.2. GO Enrichment and KEGG Pathway Analysis of DEG
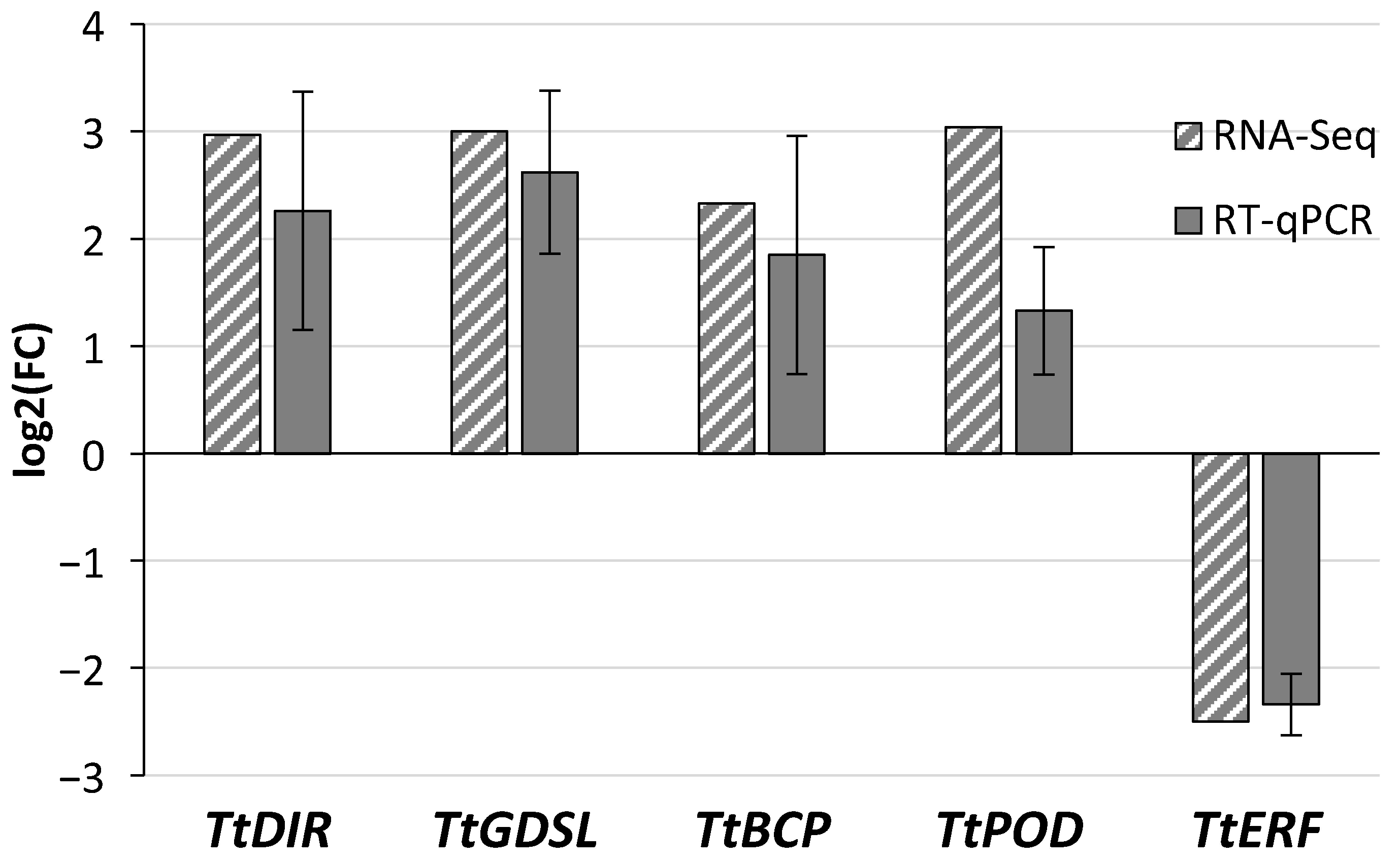
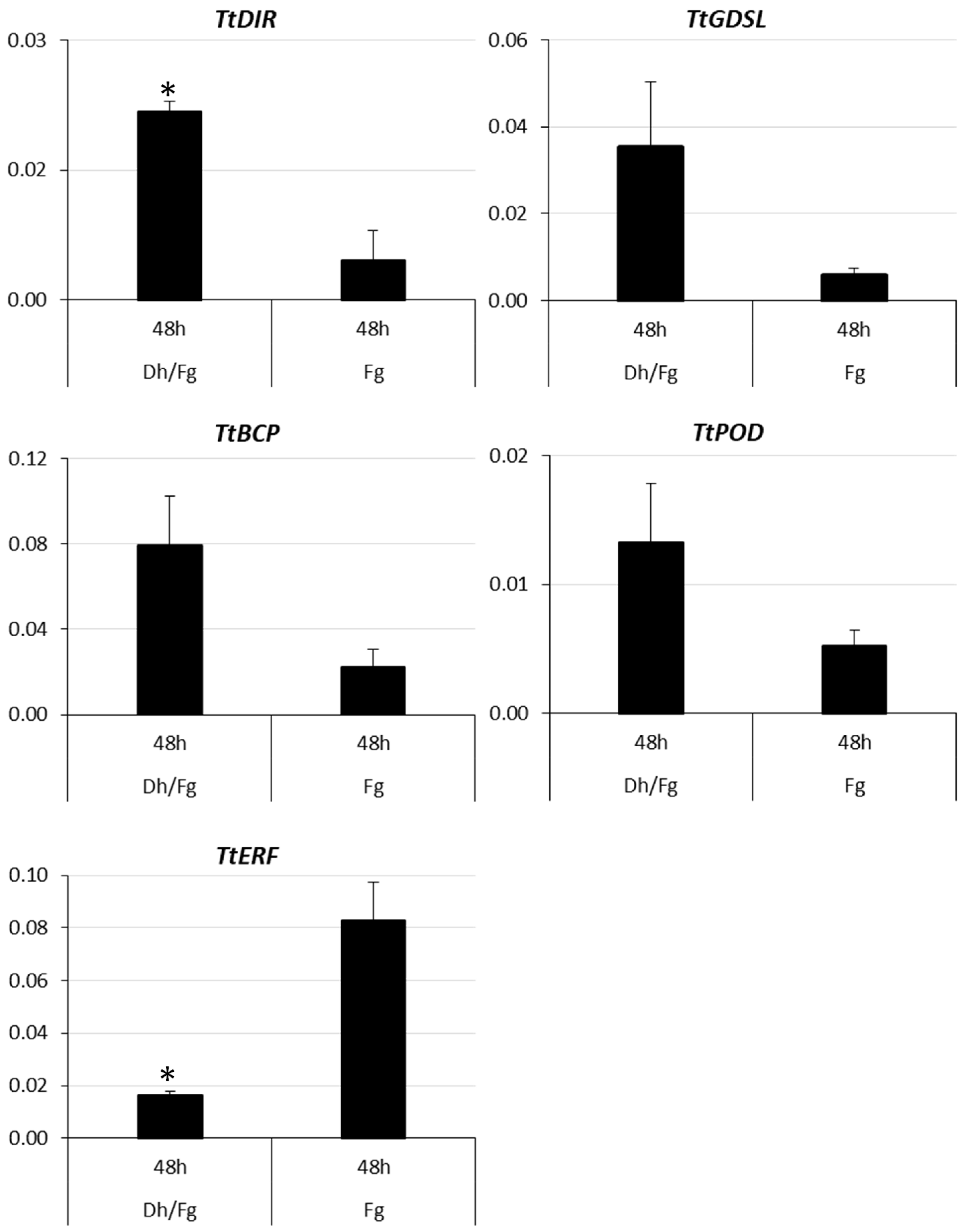
2.3. Quantitative Real-Time Expression Analysis
2.4. Expression of D. hansenii Gene
3. Discussion
3.1. Impact of Wheat Priming with D. hansenii on the Expression of Genes Involved in Signal Transduction
3.2. Impact of Wheat Priming with D. hansenii on the Expression of Genes Involved in Cell Wall Modification
3.3. Impact of Wheat Priming with D. hansenii on the Expression of Genes Involved in the Structure of the Cell Wall
3.4. Potential Direct Antagonism of D. hansenii Toward F. graminearum
4. Materials and Methods
4.1. Plant Material, Growth Conditions, and Biological Protection
4.2. Inoculation of Wheat Spike with Fusarium graminearum
4.3. RNA Extraction, Sequencing, and In Silico Analysis
4.4. Quantitative Real-Time PCR (RT-qPCR) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, S.; Sirikhachornkit, A.; Su, X.; Faris, J.; Gill, B.; Haselkorn, R.; Gornicki, P. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 2002, 99, 8133–8138. [Google Scholar] [CrossRef]
- FAO. FAOSTAT: Production: Crop and Livestock Products. 2023. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 4 June 2025).
- Blanco, A. Structure and Trends of Worldwide Research on Durum Wheat by Bibliographic Mapping. Int. J. Plant Biol. 2024, 15, 132–160. [Google Scholar] [CrossRef]
- Gai, X.T.; Xuan, Y.H.; Gao, Z.G. Diversity and pathogenicity of Fusarium graminearum species complex from maize stalk and ear rot strains in northeast China. Plant Pathol. 2017, 66, 1267–1275. [Google Scholar] [CrossRef]
- O’Donnell, K. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 2000, 92, 919. [Google Scholar] [CrossRef]
- O’Donnell, K.; Sutton, D.A.; Fothergill, A.; McCarthy, D.; Rinaldi, M.G.; Brandt, M.E.; Zhang, N.; Geiser, D.M. Molecular Phylogenetic Diversity, Multilocus Haplotype Nomenclature, and In Vitro Antifungal Resistance within the Fusarium solani Species Complex. J Clin. Microbiol. 2008, 46, 2477–2490. [Google Scholar] [CrossRef]
- Starkey, D.E.; Ward, T.J.; Aoki, T.; Gale, L.R.; Kistler, H.C.; Geiser, D.M.; Suga, H.; Tóth, B.; Varga, J.; O’Donnell, K. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol. 2007, 44, 1191–1204. [Google Scholar] [CrossRef]
- Yli-Mattila, T.; Gagkaeva, T.; Ward, T.J.; Aoki, T.; Kistler, H.C.; O’Donnell, K. A novel Asian clade within the Fusarium graminearum species complex includes a newly discovered cereal head blight pathogen from the Russian Far East. Mycologia 2009, 101, 841–852. [Google Scholar] [CrossRef]
- Sarver, B.A.; Ward, T.J.; Gale, L.R.; Broz, K.; Kistler, H.C.; Aoki, T.; Nicholson, P.; Carter, J.; O’Donnell, K. Novel Fusarium head blight pathogens from Nepal and Louisiana revealed by multilocus genealogical concordance. Fungal Genet. Biol. 2011, 48, 1096–1107. [Google Scholar] [CrossRef]
- Garofalo, M.; Payros, D.; Penary, M.; Oswald, E.; Nougayrède, J.-P.; Oswald, I.P. A novel toxic effect of foodborne trichothecenes: The exacerbation of genotoxicity. Environ. Pollut. 2023, 317, 120625. [Google Scholar] [CrossRef]
- Simsek, S.; Ovando-Martínez, M.; Ozsisli, B.; Whitney, K.; Ohm, J.-B. Occurrence of Deoxynivalenol and Deoxynivalenol-3-glucoside in Hard Red Spring Wheat Grown in the USA. Toxins 2013, 5, 2656–2670. [Google Scholar] [CrossRef]
- McMullen, M.; Bergstrom, G.; De Wolf, E.; Dill-Macky, R.; Hershman, D.; Shaner, G.; Van Sanford, D. A Unified Effort to Fight an Enemy of Wheat and Barley: Fusarium Head Blight. Plant Dis. 2012, 96, 1712–1728. [Google Scholar] [CrossRef]
- Franco, M.F.; Lori, G.A.; Cendoya, G.; Alonso, M.P.; Panelo, J.S.; Malbrán, I.; Mirabella, N.E.; Pontaroli, A.C. Spike architecture traits associated with type II resistance to fusarium head blight in bread wheat. Euphytica 2021, 217, 209. [Google Scholar] [CrossRef]
- Felici, L.; Francesconi, S.; Sestili, F.; Balestra, G.M. Physiological and morphological traits associated with Fusarium head blight response in a flavonoid-rich durum wheat genotype. J. Plant Pathol. 2024. [Google Scholar] [CrossRef]
- Lionetti, V.; Giancaspro, A.; Fabri, E.; Giove, S.L.; Reem, N.; A Zabotina, O.; Blanco, A.; Gadaleta, A.; Bellincampi, D. Cell wall traits as potential resources to improve resistance of durum wheat against Fusarium graminearum. BMC Plant Biol. 2015, 15, 6. [Google Scholar] [CrossRef]
- Giancaspro, A.; Lionetti, V.; Giove, S.L.; Zito, D.; Fabri, E.; Reem, N.; Zabotina, O.A.; De Angelis, E.; Monaci, L.; Bellincampi, D.; et al. Cell wall features transferred from common into durum wheat to improve Fusarium Head Blight resistance. Plant Sci. 2018, 274, 121–128. [Google Scholar] [CrossRef]
- Chen, C.; Guo, Q.; He, Q.; Tian, Z.; Hao, W.; Shan, X.; Lu, J.; Barkla, B.J.; Ma, C.; Si, H. Comparative transcriptomic analysis of wheat cultivars differing in their resistance to Fusarium head blight infection during grain-filling stages reveals unique defense mechanisms at play. BMC Plant Biol. 2023, 23, 433. [Google Scholar] [CrossRef]
- Ribichich, K.F.; Lopez, S.E.; Vegetti, A.C. Histopathological Spikelet Changes Produced by Fusarium graminearum in Susceptible and Resistant Wheat Cultivars. Plant Dis. 2000, 84, 794–802. [Google Scholar] [CrossRef]
- Prat, N.; Buerstmayr, M.; Steiner, B.; Robert, O.; Buerstmayr, H. Current knowledge on resistance to Fusarium head blight in tetraploid wheat. Mol. Breed. 2014, 34, 1689–1699. [Google Scholar] [CrossRef]
- Haile, J.K.; Sertse, D.; N’dIaye, A.; Klymiuk, V.; Wiebe, K.; Ruan, Y.; Chawla, H.S.; Henriquez, M.-A.; Wang, L.; Kutcher, H.R.; et al. Multi-locus genome-wide association studies reveal the genetic architecture of Fusarium head blight resistance in durum wheat. Front. Plant Sci. 2023, 14, 1182548. [Google Scholar] [CrossRef]
- Miedaner, T.; Longin, C.F.H. Genetic variation for resistance to Fusarium head blight in winter durum material. Crop. Pasture Sci. 2014, 65, 46–51. [Google Scholar] [CrossRef]
- Miedaner, T.; Sieber, A.; Desaint, H.; Buerstmayr, H.; Longin, C.F.H.; Würschum, T. The potential of genomic-assisted breeding to improve Fusarium head blight resistance in winter durum wheat. Plant Breed. 2017, 136, 610–619. [Google Scholar] [CrossRef]
- Buerstmayr, M.; Steiner, B.; Buerstmayr, H. Breeding for Fusarium head blight resistance in wheat—Progress and challenges. Plant Breed. 2020, 139, 429–454. [Google Scholar] [CrossRef]
- Fabre, F.; Rocher, F.; Alouane, T.; Langin, T.; Bonhomme, L. Searching for FHB Resistances in Bread Wheat: Susceptibility at the Crossroad. Front. Plant Sci. 2020, 11, 731. [Google Scholar] [CrossRef]
- Buerstmayr, M.; Huber, K.; Heckmann, J.; Steiner, B.; Nelson, J.C.; Buerstmayr, H. Mapping of QTL for Fusarium head blight resistance and morphological and developmental traits in three backcross populations derived from Triticum dicoccum × Triticum durum. Theor. Appl. Genet. 2012, 125, 1751–1765. [Google Scholar] [CrossRef]
- Zhang, Z.; Belcram, H.; Gornicki, P.; Charles, M.; Just, J.; Huneau, C.; Magdelenat, G.; Couloux, A.; Samain, S.; Gill, B.S.; et al. Duplication and partitioning in evolution and function of homoeologous Q loci governing domestication characters in polyploid wheat. Proc. Natl. Acad. Sci. USA 2011, 108, 18737–18742, Correction in Proc. Natl. Acad. Sci. USA 2012, 109, 1353. https://doi.org/10.1073/pnas.1121066109. [Google Scholar] [CrossRef]
- Kirana, R.P.; Gaurav, K.; Arora, S.; Wiesenberger, G.; Doppler, M.; Michel, S.; Zimmerl, S.; Matic, M.; Eze, C.E.; Kumar, M.; et al. Identification of a UDP-glucosyltransferase conferring deoxynivalenol resistance in Aegilops tauschii and wheat. Plant Biotechnol. J. 2023, 21, 109–121. [Google Scholar] [CrossRef]
- Kirana, R.P.; Michel, S.; Moreno-Amores, J.; Prat, N.; Lemmens, M.; Buerstmayr, M.; Buerstmayr, H.; Steiner, B. Pyramiding Fusarium head blight resistance QTL from T. aestivum, T. dicoccum and T. dicoccoides in durum wheat. Theor. Appl. Genet. 2023, 136, 201. [Google Scholar] [CrossRef]
- Ding, L.; Xu, H.; Yi, H.; Yang, L.; Kong, Z.; Zhang, L.; Xue, S.; Jia, H.; Ma, Z. Resistance to Hemi-Biotrophic F. graminearum Infection Is Associated with Coordinated and Ordered Expression of Diverse Defense Signaling Pathways. PLoS ONE 2011, 6, e19008. [Google Scholar] [CrossRef]
- Qi, P.-F.; Balcerzak, M.; Rocheleau, H.; Leung, W.; Wei, Y.-M.; Zheng, Y.-L.; Ouellet, T. Jasmonic acid and abscisic acid play important roles in host–pathogen interaction between Fusarium graminearum and wheat during the early stages of fusarium head blight. Physiol. Mol. Plant Pathol. 2016, 93, 39–48. [Google Scholar] [CrossRef]
- Brauer, E.K.; Rocheleau, H.; Balcerzak, M.; Pan, Y.; Fauteux, F.; Liu, Z.; Wang, L.; Zheng, W.; Ouellet, T. Transcriptional and hormonal profiling of Fusarium graminearum-infected wheat reveals an association between auxin and susceptibility. Physiol. Mol. Plant Pathol. 2019, 107, 33–39. [Google Scholar] [CrossRef]
- Klocke, B.; Sommerfeldt, N.; Wagner, C.; Schwarz, J.; Baumecker, M.; Ellmer, F.; Jacobi, A.; Matschiner, K.; Petersen, J.; Wehling, P.; et al. Disease threshold-based fungicide applications: Potential of multi-disease resistance in winter wheat cultivars in Germany. Eur. J. Plant Pathol. 2023, 165, 363–383. [Google Scholar] [CrossRef]
- Talas, F.; McDonald, B.A. Genome-wide analysis of Fusarium graminearum field populations reveals hotspots of recombination. BMC Genom. 2015, 16, 996. [Google Scholar] [CrossRef]
- Matelionienė, N.; Žvirdauskienė, R.; Kadžienė, G.; Zavtrikovienė, E.; Supronienė, S. In Vitro Sensitivity Test of Fusarium Species from Weeds and Non-Gramineous Plants to Triazole Fungicides. Pathogens 2024, 13, 160. [Google Scholar] [CrossRef]
- Ivic, D.; Sever, Z.; Kuzmanovska, B. In vitro sensitivity of Fusarium graminearum, F. avenaceum and F. verticillioides to carbendazim, tebuconazole, flutriafol, metconazole and prochloraz. Pestic. Fitomedicina 2011, 26, 35–42. [Google Scholar] [CrossRef]
- Anonymous. Directive 2009/128/EC of the European Parliament and of the Council of 21 October 2009 establishing a framework for Community action to achieve the sustainable use of pesticides. Off. J. Eur. Union 2009, 309, 71–86. [Google Scholar]
- Fröhlich-Wyder, M.; Arias-Roth, E.; Jakob, E. Cheese yeasts. Yeast 2019, 36, 129–141. [Google Scholar] [CrossRef]
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Hilbert, F.; Lindqvist, R.; et al. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 12: Suitability of taxonomic units notified to EFSA until March 2020. EFSA J. 2020, 18, e06174. [Google Scholar]
- Grzegorczyk, M.; Żarowska, B.; Restuccia, C.; Cirvilleri, G. Postharvest biocontrol ability of killer yeasts against Monilinia fructigena and Monilinia fructicola on stone fruit. Food Microbiol. 2017, 61, 93–101. [Google Scholar] [CrossRef]
- Wachowska, U.; Sulyok, M.; Wiwart, M.; Suchowilska, E.; Kandler, W.; Krska, R. The application of antagonistic yeasts and bacteria: An assessment of in vivo and under field conditions pattern of Fusarium mycotoxins in winter wheat grain. Food Control 2022, 138, 109039. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef]
- Medina-Córdova, N.; López-Aguilar, R.; Ascencio, F.; Castellanos, T.; Campa-Córdova, A.I.; Angulo, C. Biocontrol activity of the marine yeast Debaryomyces hansenii against phytopathogenic fungi and its ability to inhibit mycotoxins production in maize grain (Zea mays L.). Biol. Control 2016, 97, 70–79. [Google Scholar] [CrossRef]
- Rivas-Garcia, T.; Murillo-Amador, B.; Reyes-Pérez, J.J.; Chiquito-Contreras, R.G.; Preciado-Rangel, P.; Ávila-Quezada, G.D.; Lara-Capistran, L.; Hernandez-Montiel, L.G. Debaryomyces hansenii, Stenotrophomonas rhizophila, and Ulvan as Biocontrol Agents of Fruit Rot Disease in Muskmelon (Cucumis melo L.). Plants 2022, 11, 184. [Google Scholar] [CrossRef]
- Payne, C.; Bruce, A. The Yeast Debaryomyces hansenii as a Short-Term Biological Control Agent against Fungal Spoilage of Sawn Pinus sylvestris Timber. Biol. Control 2001, 22, 22–28. [Google Scholar] [CrossRef]
- Wachowska, U.; Pluskota, W.; Jastrzębski, J.P.; Głowacka, K.; Szablewska-Stuper, K.; Balcerzak, M. A method for reducing the concentrations of Fusarium graminearum trichothecenes in durum wheat grain with the use of Debaryomyces hansenii. Int. J. Food Microbiol. 2023, 397, 110211. [Google Scholar] [CrossRef]
- Giedrojć, W.; Wachowska, U. Mycobiome and Pathogenic Fusarium Fungi in the Rhizosphere of Durum Wheat After Seed Dressing with Debaryomyces hansenii. Agriculture 2025, 15, 639. [Google Scholar] [CrossRef]
- Sevillano-Caño, J.; García, M.J.; Córdoba-Galván, C.; Luque-Cruz, C.; Agustí-Brisach, C.; Lucena, C.; Ramos, J.; Pérez-Vicente, R.; Romera, F.J. Exploring the Role of Debaryomyces hansenii as Biofertilizer in Iron-Deficient Environments to Enhance Plant Nutrition and Crop Production Sustainability. Int. J. Mol. Sci. 2024, 25, 5729. [Google Scholar] [CrossRef]
- Núñez-Cano, J.; Ruiz-Castilla, F.J.; Ramos, J.; Romera, F.J.; Lucena, C. Debaryomyces hansenii Enhances Growth, Nutrient Uptake, and Yield in Rice Plants (Oryza sativa L.) Cultivated in Calcareous Soil. Agronomy 2025, 15, 1696. [Google Scholar] [CrossRef]
- Narusaka, M.; Minami, T.; Iwabuchi, C.; Hamasaki, T.; Takasaki, S.; Kawamura, K.; Narusaka, Y. Yeast Cell Wall Extract Induces Disease Resistance against Bacterial and Fungal Pathogens in Arabidopsis thaliana and Brassica Crop. PLoS ONE 2015, 10, e0115864. [Google Scholar] [CrossRef]
- Yaguchi, T.; Kinami, T.; Ishida, T.; Yasuhara, T.; Takahashi, K.; Matsuura, H. Induction of plant disease resistance upon treatment with yeast cell wall extract. Biosci. Biotechnol. Biochem. 2017, 81, 2071–2078. [Google Scholar] [CrossRef]
- Gao, X.; Li, F.; Sun, Y.; Jiang, J.; Tian, X.; Li, Q.; Duan, K.; Lin, J.; Liu, H.; Wang, Q. Basal defense is enhanced in a wheat cultivar resistant to Fusarium head blight. J. Integr. Agric. 2024, 23, 1238–1258. [Google Scholar] [CrossRef]
- del Carmen Orozco-Mosqueda, M.; Glick, B.R.; Santoyo, G. ACC deaminase in plant growth-promoting bacteria (PGPB): An efficient mechanism to counter salt stress in crops. Microbiol. Res. 2020, 235, 126439. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.-J.; Zhang, J.-Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef]
- Chen, X.; Steed, A.; Travella, S.; Keller, B.; Nicholson, P. Fusarium graminearum exploits ethylene signalling to colonize dicotyledonous and monocotyledonous plants. New Phytol. 2009, 182, 975–983. [Google Scholar] [CrossRef]
- Su, P.; Zhao, L.; Li, W.; Zhao, J.; Yan, J.; Ma, X.; Li, A.; Wang, H.; Kong, L. Integrated metabolo-transcriptomics and functional characterization reveals that the wheat auxin receptor TIR1 negatively regulates defense against Fusarium graminearum. J. Integr. Plant Biol. 2021, 63, 340–352, Correction in J. Integr. Plant Biol. 2025, 67, 1685–1686. https://doi.org/10.1111/jipb.13870. [Google Scholar] [CrossRef]
- Pan, Y.; Liu, Z.; Rocheleau, H.; Fauteux, F.; Wang, Y.; McCartney, C.; Ouellet, T. Transcriptome dynamics associated with resistance and susceptibility against fusarium head blight in four wheat genotypes. BMC Genom. 2018, 19, 642. [Google Scholar] [CrossRef]
- Rocher, F.; Dou, S.; Philippe, G.; Martin, M.-L.; Label, P.; Langin, T.; Bonhomme, L. Integrative systems biology of wheat susceptibility to Fusarium graminearum uncovers a conserved gene regulatory network and identifies master regulators targeted by fungal core effectors. BMC Biol. 2024, 22, 53. [Google Scholar] [CrossRef]
- Thapa, G.; Gunupuru, L.R.; Hehir, J.G.; Kahla, A.; Mullins, E.; Doohan, F.M. A Pathogen-Responsive Leucine Rich Receptor Like Kinase Contributes to Fusarium Resistance in Cereals. Front. Plant Sci. 2018, 9, 867. [Google Scholar] [CrossRef]
- Yan, J.; Su, P.; Meng, X.; Liu, P. Phylogeny of the plant receptor-like kinase (RLK) gene family and expression analysis of wheat RLK genes in response to biotic and abiotic stresses. BMC Genom. 2023, 24, 224. [Google Scholar] [CrossRef]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef]
- Paniagua, C.; Bilkova, A.; Jackson, P.; Dabravolski, S.; Riber, W.; Didi, V.; Houser, J.; Gigli-Bisceglia, N.; Wimmerova, M.; Budínská, E.; et al. Dirigent proteins in plants: Modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 2017, 68, 3287–3301. [Google Scholar] [CrossRef]
- Sun, Z.; Zhou, Y.; Hu, Y.; Jiang, N.; Hu, S.; Li, L.; Li, T. Identification of Wheat LACCASEs in Response to Fusarium graminearum as Potential Deoxynivalenol Trappers. Front. Plant Sci. 2022, 13, 832800. [Google Scholar] [CrossRef]
- Ursache, R.; Teixeira, C.D.J.V.; Tendon, V.D.; Gully, K.; De Bellis, D.; Schmid-Siegert, E.; Andersen, T.G.; Shekhar, V.; Calderon, S.; Pradervand, S.; et al. GDSL-domain proteins have key roles in suberin polymerization and degradation. Nat. Plants 2021, 7, 353–364. [Google Scholar] [CrossRef]
- Basheer, J.; Vadovič, P.; Šamajová, O.; Melicher, P.; Komis, G.; Křenek, P.; Králová, M.; Pechan, T.; Ovečka, M.; Takáč, T.; et al. Knockout of MITOGEN-ACTIVATED PROTEIN KINASE 3 causes barley root resistance against Fusarium graminearum. Plant Physiol. 2022, 190, 2847–2867. [Google Scholar] [CrossRef]
- Eldakak, M.; Das, A.; Zhuang, Y.; Rohila, J.S.; Glover, K.; Yen, Y. A Quantitative Proteomics View on the Function of Qfhb1, a Major QTL for Fusarium Head Blight Resistance in Wheat. Pathogens 2018, 7, 58. [Google Scholar] [CrossRef]
- Schweiger, W.; Steiner, B.; Vautrin, S.; Nussbaumer, T.; Siegwart, G.; Zamini, M.; Jungreithmeier, F.; Gratl, V.; Lemmens, M.; Mayer, K.F.X.; et al. Suppressed recombination and unique candidate genes in the divergent haplotype encoding Fhb1, a major Fusarium head blight resistance locus in wheat. Theor. Appl. Genet. 2016, 129, 1607–1623. [Google Scholar] [CrossRef]
- Bischoff, V.; Nita, S.; Neumetzler, L.; Schindelasch, D.; Urbain, A.; Eshed, R.; Persson, S.; Delmer, D.; Scheible, W.-R. TRICHOME BIREFRINGENCE and Its Homolog AT5G01360 Encode Plant-Specific DUF231 Proteins Required for Cellulose Biosynthesis in Arabidopsis. Plant Physiol. 2010, 153, 590–602. [Google Scholar] [CrossRef]
- Huang, Y.; Li, L.; Smith, K.P.; Muehlbauer, G.J. Differential transcriptomic responses to Fusarium graminearum infection in two barley quantitative trait loci associated with Fusarium head blight resistance. BMC Genom. 2016, 17, 387. [Google Scholar] [CrossRef]
- Gille, S.; Pauly, M. O-Acetylation of Plant Cell Wall Polysaccharides. Front. Plant Sci. 2012, 3, 12. [Google Scholar] [CrossRef]
- Lin, S.; Miao, Y.; Huang, H.; Zhang, Y.; Huang, L.; Cao, J. Arabinogalactan Proteins: Focus on the Role in Cellulose Synthesis and Deposition during Plant Cell Wall Biogenesis. Int. J. Mol. Sci. 2022, 23, 6578. [Google Scholar] [CrossRef]
- Medina-Córdova, N.; Rosales-Mendoza, S.; Hernández-Montiel, L.G.; Angulo, C. The potential use of Debaryomyces hansenii for the biological control of pathogenic fungi in food. Biol. Control 2018, 121, 216–222. [Google Scholar] [CrossRef]
- Hernandez-Montiel, L.G.; Gutierrez-Perez, E.D.; Murillo-Amador, B.; Vero, S.; Chiquito-Contreras, R.G.; Rincon-Enriquez, G. Mechanisms employed by Debaryomyces hansenii in biological control of anthracnose disease on papaya fruit. Postharvest Biol. Technol. 2018, 139, 31–37. [Google Scholar] [CrossRef]
- Li, Z.; Chen, Y.; Liu, D.; Zhao, N.; Cheng, H.; Ren, H.; Guo, T.; Niu, H.; Zhuang, W.; Wu, J.; et al. Involvement of glycolysis/gluconeogenesis and signaling regulatory pathways in Saccharomyces cerevisiae biofilms during fermentation. Front. Microbiol. 2015, 6, 139. [Google Scholar] [CrossRef]
- Czarnecka, M.; Żarowska, B.; Połomska, X.; Restuccia, C.; Cirvilleri, G. Role of biocontrol yeasts Debaryomyces hansenii and Wickerhamomyces anomalus in plants’ defence mechanisms against Monilinia fructicola in apple fruits. Food Microbiol. 2019, 83, 1–8. [Google Scholar] [CrossRef]
- Lukša, J.; Podoliankaitė, M.; Vepštaitė, I.; Strazdaitė-Žielienė, Ž.; Urbonavičius, J.; Servienė, E. Yeast β-1,6-Glucan Is a Primary Target for the Saccharomyces cerevisiae K2 Toxin. Eukaryot. Cell 2015, 14, 406–414. [Google Scholar] [CrossRef]
- Billerbeck, S.; Walker, R.S.; Pretorius, I.S. Killer yeasts: Expanding frontiers in the age of synthetic biology. Trends Biotechnol. 2024, 42, 1081–1096. [Google Scholar] [CrossRef]
- Liu, G.-L.; Chi, Z.; Wang, G.-Y.; Wang, Z.-P.; Li, Y.; Chi, Z.-M. Yeast killer toxins, molecular mechanisms of their action and their applications. Crit. Rev. Biotechnol. 2015, 35, 222–234. [Google Scholar] [CrossRef]
- Meier, U. Phenological Growth Stages. In Phenology: An Integrative Environmental Science; Tasks for Vegetation Science; Schwartz, M.D., Ed.; Springer: Dordrecht, The Netherlands, 2003; pp. 269–283. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 October 2019).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A Program for Genome-wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Pluskota, W.E.; Jastrzębski, J.P.; Paukszto, Ł.; Wachowska, U.B. Transcriptional Response of Durum Wheat During Interaction with Debaryomyces hansenii and Fusarium graminearum. Int. J. Mol. Sci. 2026, 27, 457. https://doi.org/10.3390/ijms27010457
Pluskota WE, Jastrzębski JP, Paukszto Ł, Wachowska UB. Transcriptional Response of Durum Wheat During Interaction with Debaryomyces hansenii and Fusarium graminearum. International Journal of Molecular Sciences. 2026; 27(1):457. https://doi.org/10.3390/ijms27010457
Chicago/Turabian StylePluskota, Wioletta E., Jan P. Jastrzębski, Łukasz Paukszto, and Urszula B. Wachowska. 2026. "Transcriptional Response of Durum Wheat During Interaction with Debaryomyces hansenii and Fusarium graminearum" International Journal of Molecular Sciences 27, no. 1: 457. https://doi.org/10.3390/ijms27010457
APA StylePluskota, W. E., Jastrzębski, J. P., Paukszto, Ł., & Wachowska, U. B. (2026). Transcriptional Response of Durum Wheat During Interaction with Debaryomyces hansenii and Fusarium graminearum. International Journal of Molecular Sciences, 27(1), 457. https://doi.org/10.3390/ijms27010457








