A Novel CFEM Effector in Fusarium verticillioides Required for Virulence Involved in Plant Immunity Suppression and Fungal Cell Wall Integrity
Abstract
1. Introduction
2. Results
2.1. Bioinformatic Identification of CFEM Proteins in F. verticillioides
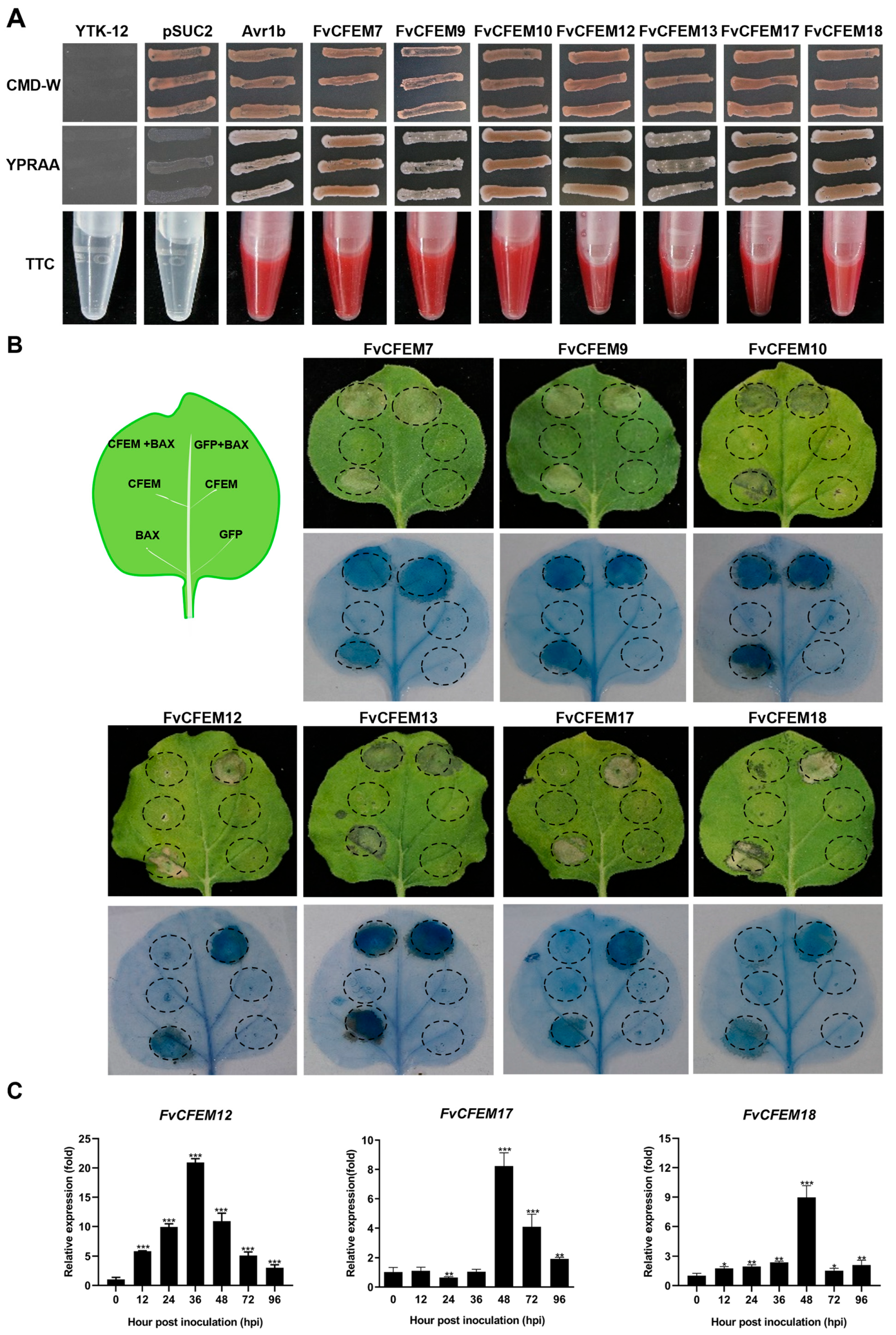
2.2. FvCFEM12 Is an Important Candidate Effector in F. verticillioides CFEM Family
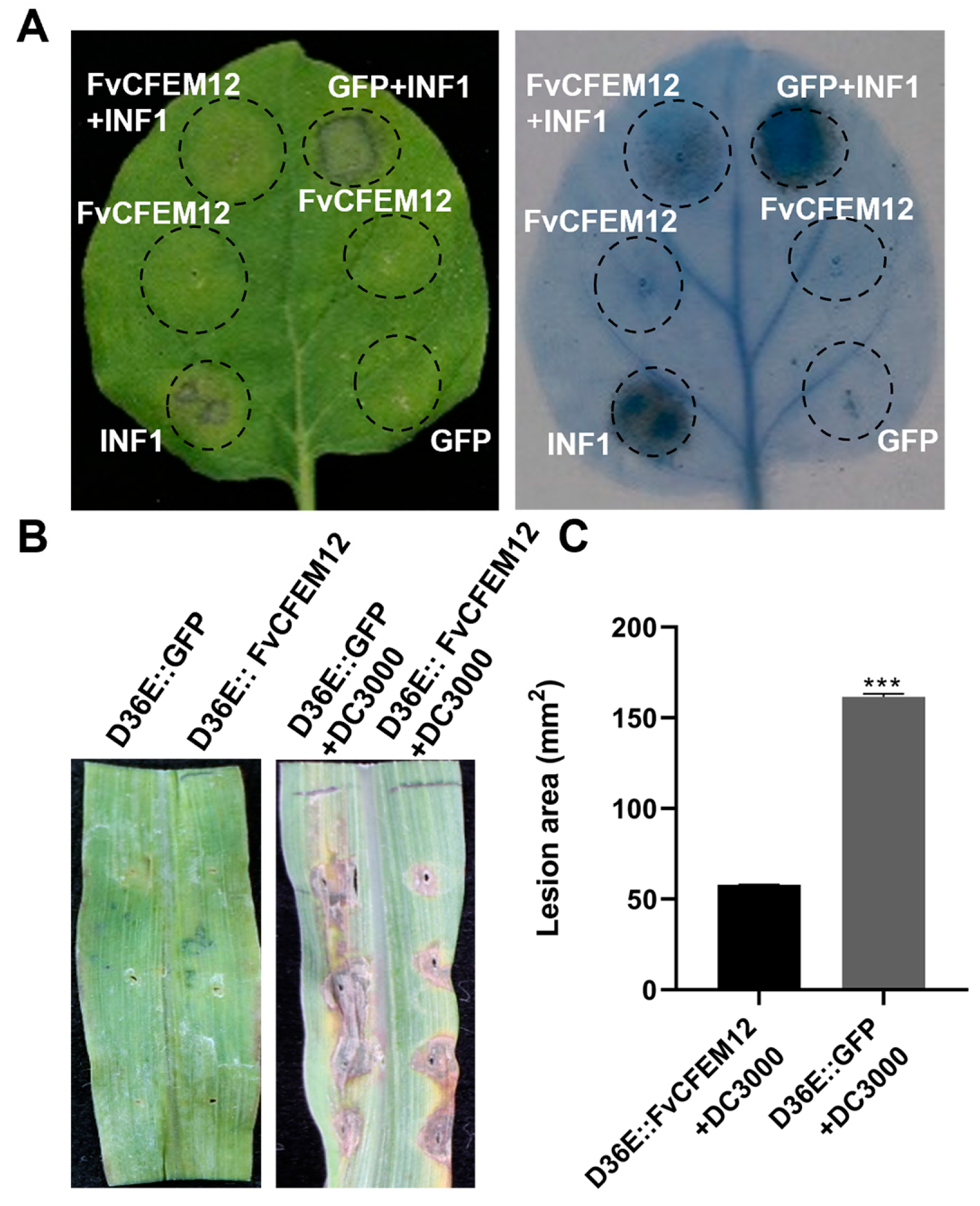
2.3. FvCFEM12 Suppresses Cell Death Induced by INF1 and P. syringae DC3000
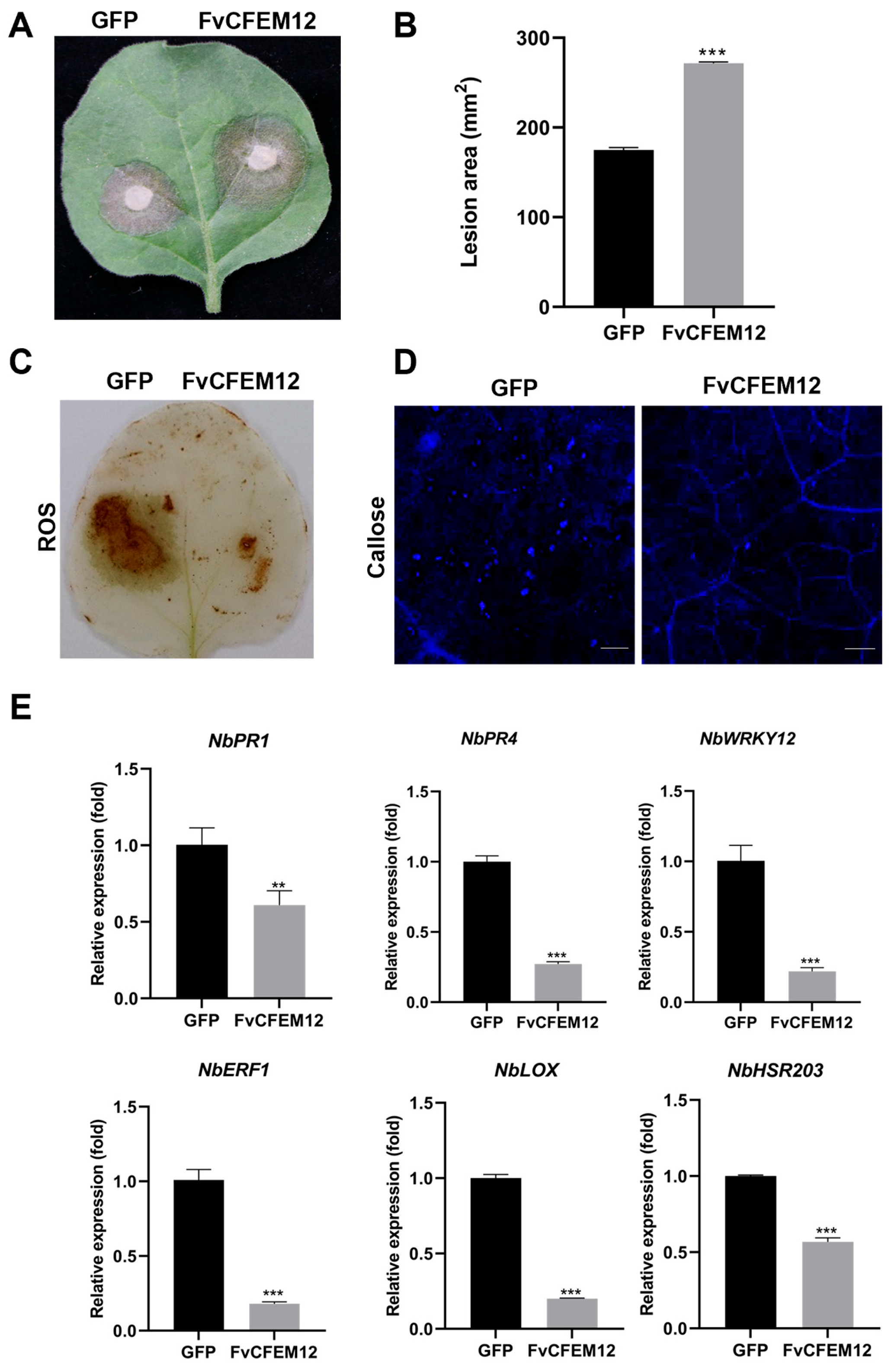
2.4. Transient Expression of FvCFEM12 Enhances Susceptibility to B.cinerea and Suppresses Plant Immunity
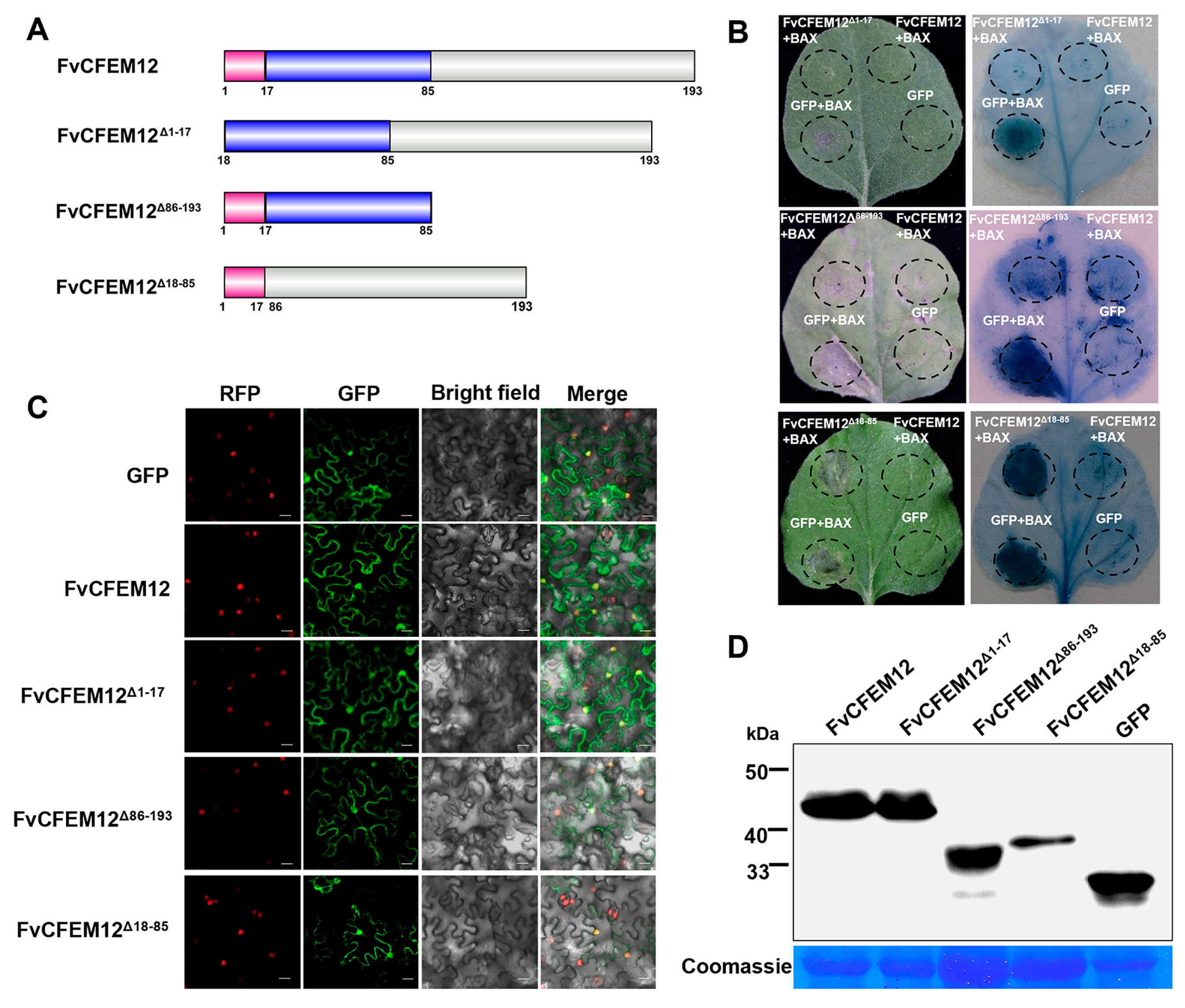
2.5. CFEM Domain Required for FvCFEM12 Cell Death Suppression, Independent of Subcellular Localization
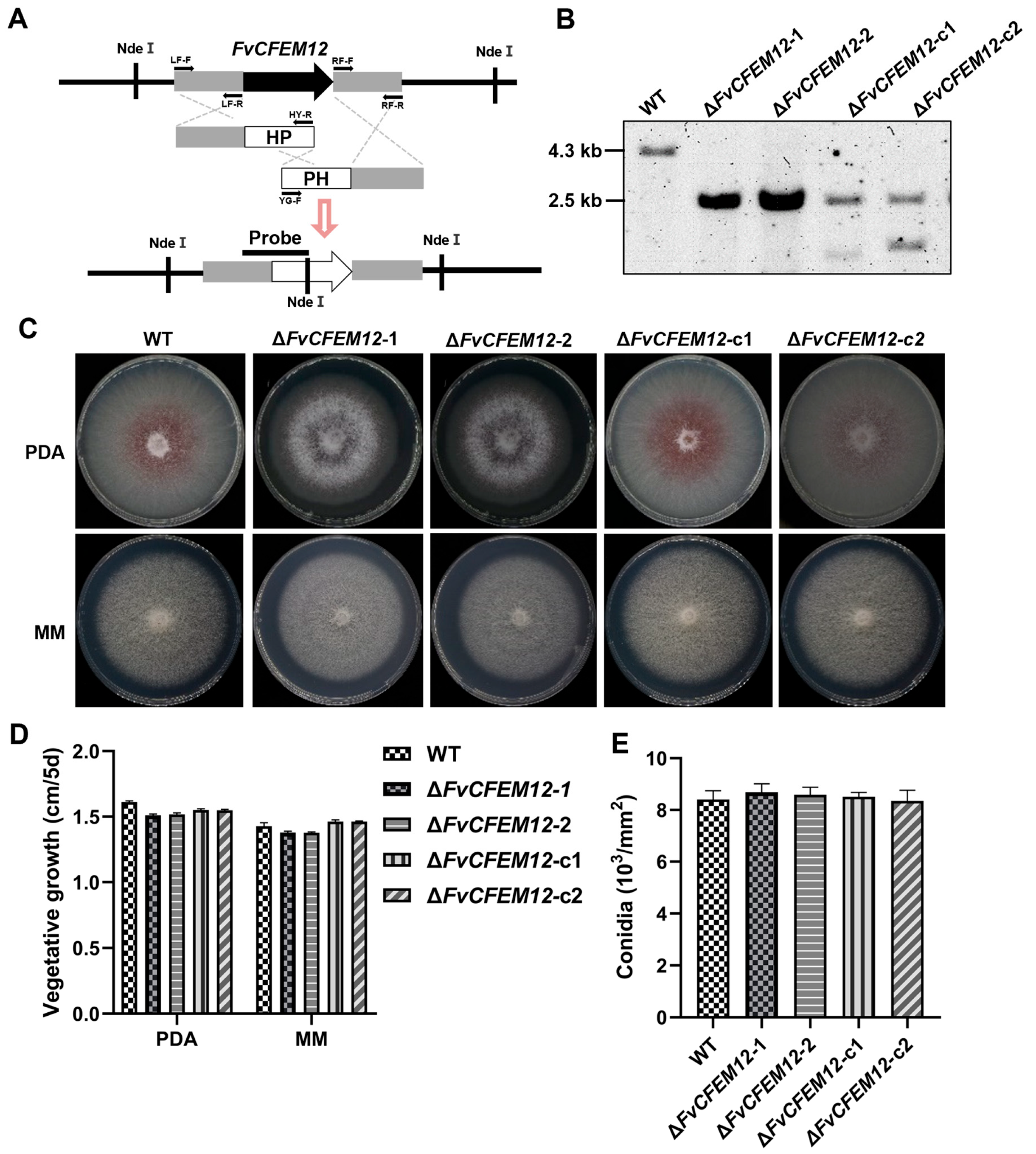
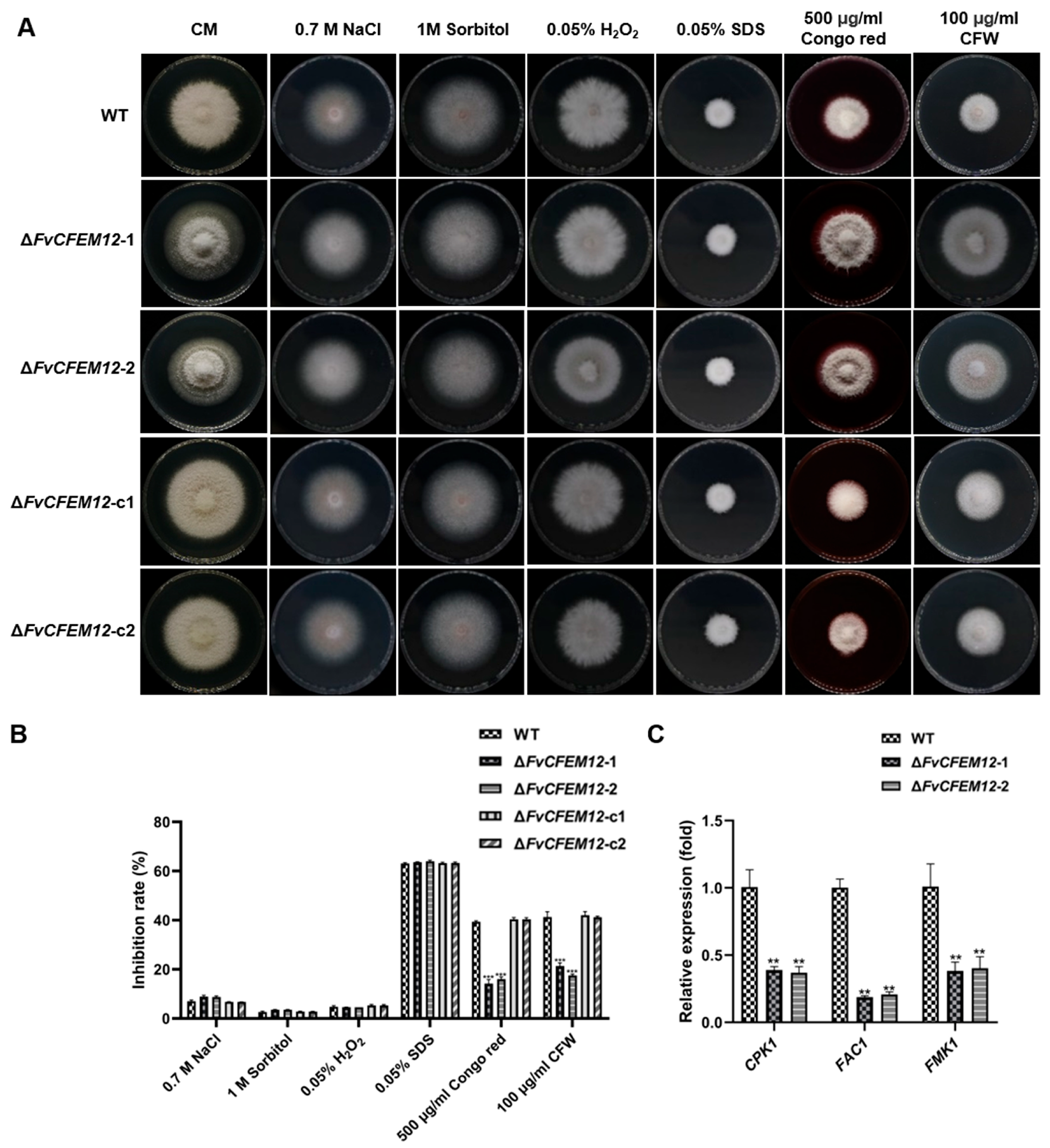
2.6. FvCFEM12 Is Involved in Phenotype Development and Cell Wall Stress Responses
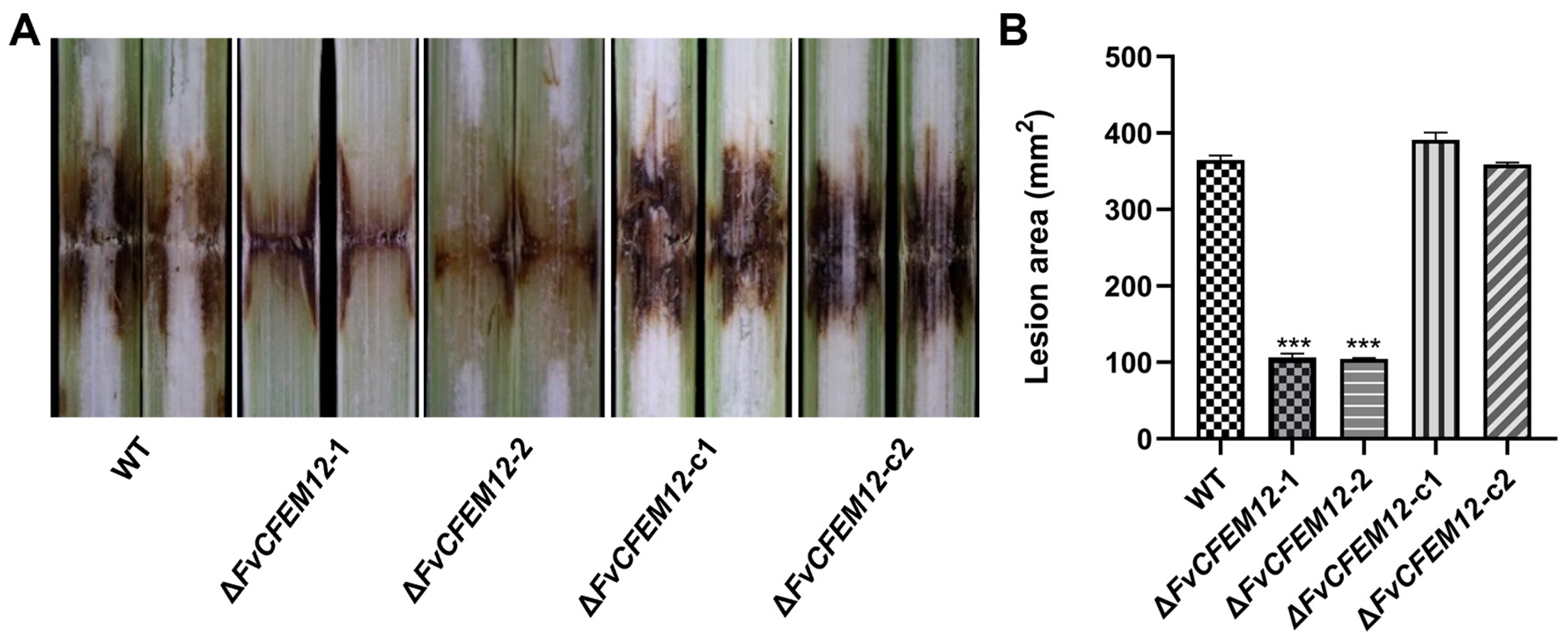
2.7. FvCFEM12 Is Required for the Full Virulence of F. verticillioides
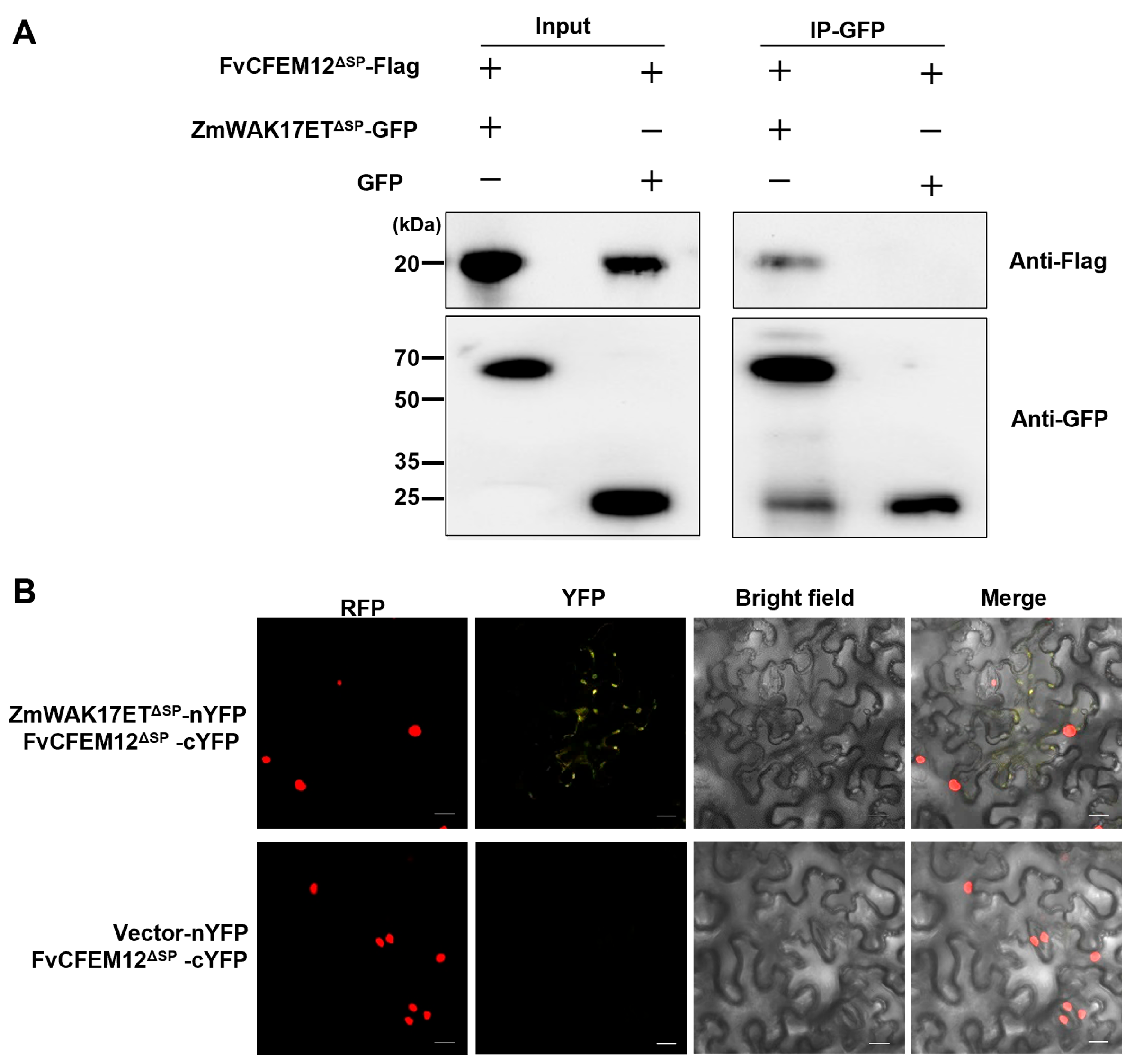
2.8. FvCFEM12 Interacts with Wall-Associated Receptor Kinase ZmWAK17
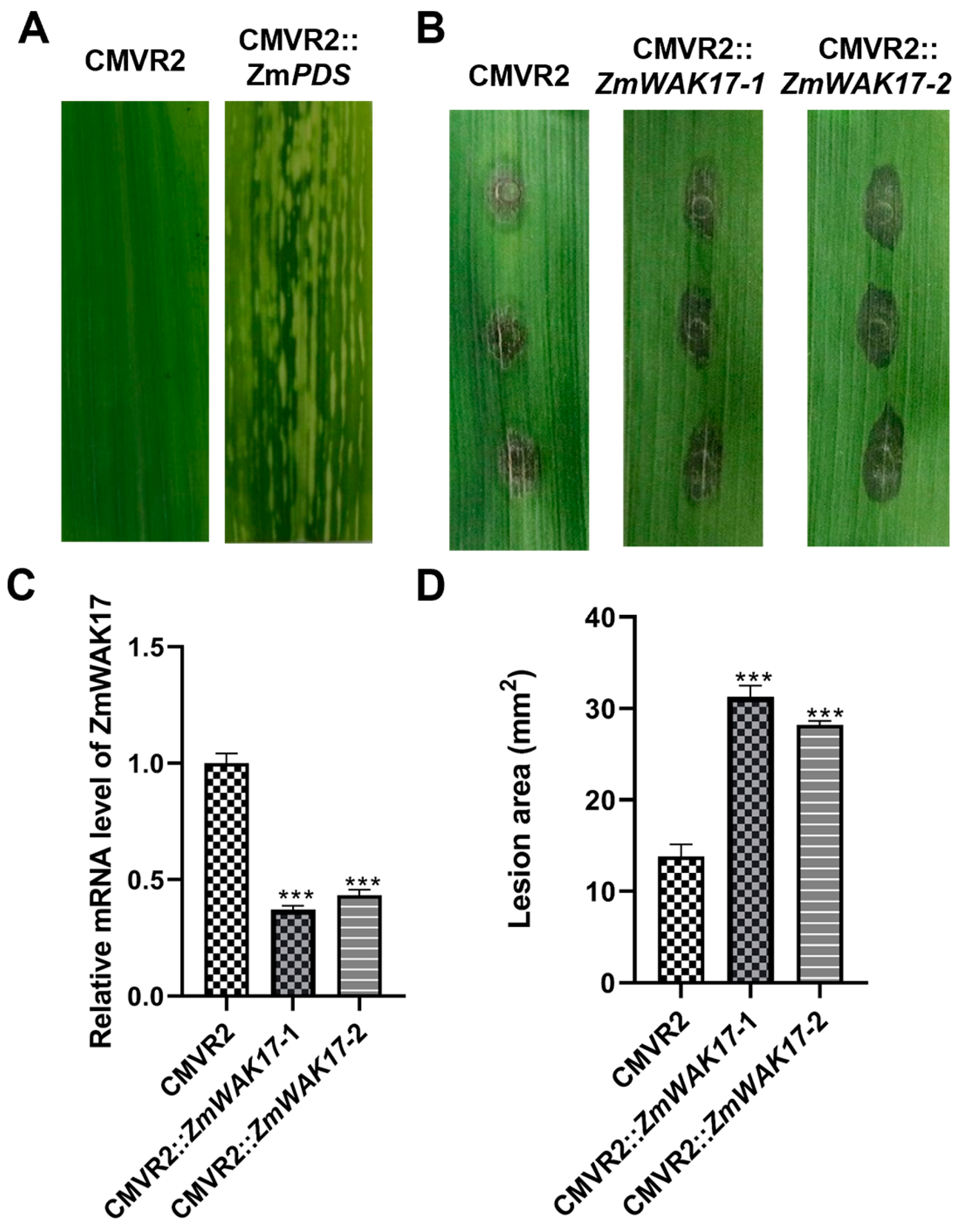
2.9. Silencing ZmWAK17 in Maize Reduce Resistance to F. verticillioides
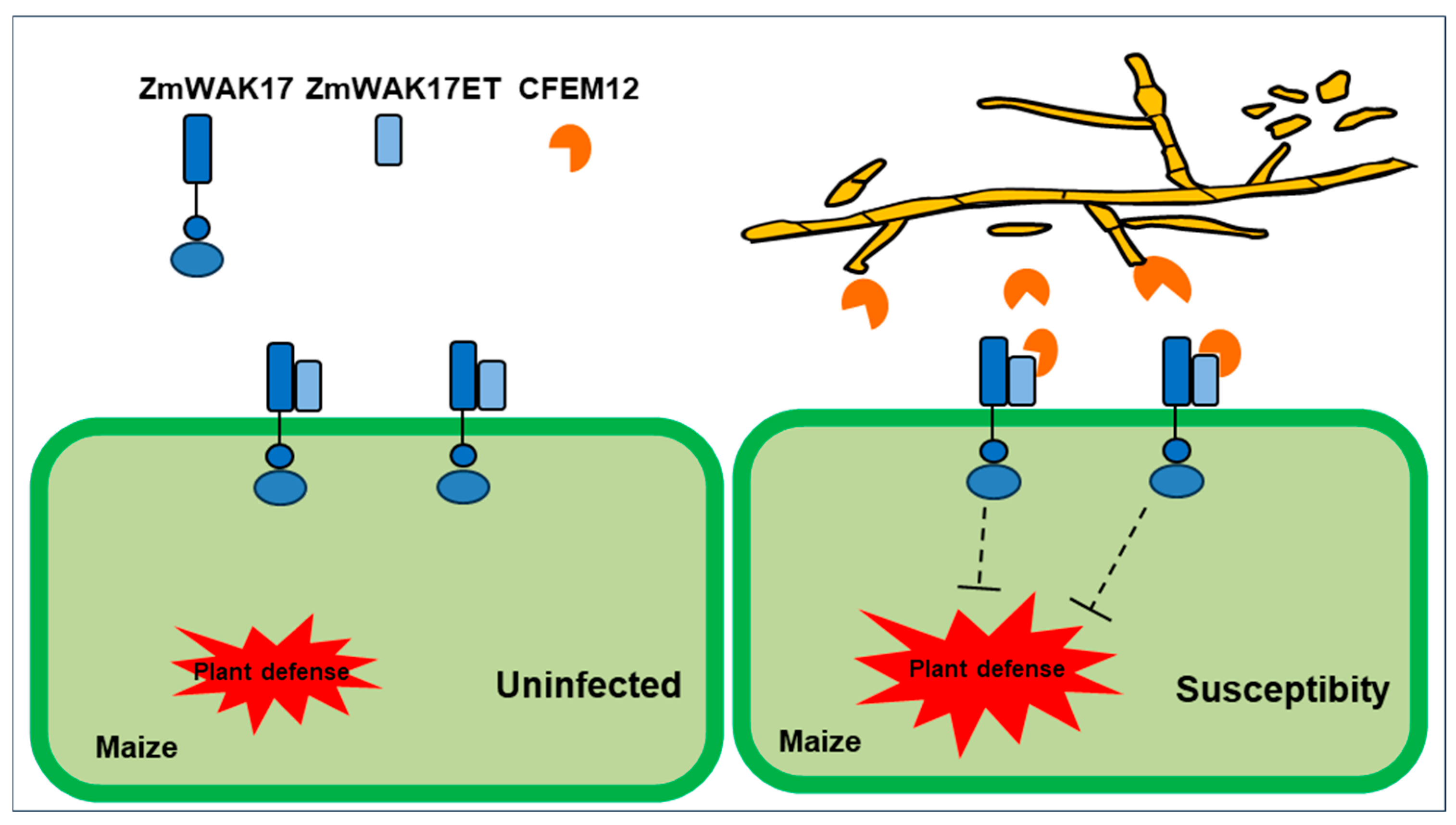
3. Discussion
4. Materials and Methods
4.1. Fungal Strains and Plant Materials
4.2. Bioinformatics Identification of CFEM Proteins in F. verticillioides
4.3. Phylogenetic Analysis and Multiple Sequence Alignment
4.4. Analysis of Chromosomal Distribution of CFEM Genes
4.5. Yeast Signal Sequence Trap Assays
4.6. Transient Expression of CFEM Proteins in N. benthamiana and Z. mays
4.7. RT-qPCR Analysis
4.8. Trypan Blue Staining
4.9. Detection of Oxidative Burst and Callose Deposition
4.10. F. verticillioides Gene Knockout and Complementation
4.11. Pathogenicity Assay
4.12. Co-IP Assay
4.13. BiFC Assay
4.14. Virus-Induced Gene Silencing (VIGS) Assay in Maize
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Katagiri, F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 2010, 13, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Feng, B.; He, P.; Shan, L. From chaos to harmony: Responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 2017, 55, 109–137. [Google Scholar] [CrossRef] [PubMed]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef]
- Dou, D.; Zhou, J.-M. Phytopathogen effectors subverting host immunity: Different foes, similar battleground. Cell Host Microbe 2012, 12, 484–495. [Google Scholar] [CrossRef]
- Munkvold, G.P.; Desjardins, A.E. Fumonisins in maize: Can we reduce their occurrence? Plant Dis. 1997, 81, 556–565. [Google Scholar] [CrossRef]
- Cotten, T.K.; Munkvold, G.P. Survival of Fusarium moniliforme, F. proliferatum, and F. subglutinans in maize stalk residue. Phytopathology 1998, 88, 550–555. [Google Scholar] [CrossRef]
- Blacutt, A.A.; Gold, S.E.; Voss, K.A.; Gao, M.; Glenn, A.E. Fusarium verticillioides: Advancements in understanding the toxicity, virulence, and niche adaptations of a model mycotoxigenic pathogen of maize. Phytopathology 2018, 108, 312–326. [Google Scholar] [CrossRef]
- Ferrigo, D.; Raiola, A.; Causin, R. Fusarium toxins in cereals: Occurrence, legislation, factors promoting the appearance and their management. Molecules 2016, 21, 627. [Google Scholar] [CrossRef]
- Munkvold, G.P. Fusarium species and their associated mycotoxins. In Mycotoxigenic Fungi: Methods and Protocols; Moretti, A., Susca, A., Eds.; Springer: New York, NY, USA, 2017; pp. 51–106. [Google Scholar]
- Schulz, B.; RÖMmert, A.-K.; Dammann, U.; Aust, H.-J.; Strack, D. The endophyte-host interaction: A balanced antagonism? Mycol. Res. 1999, 103, 1275–1283. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rafiqi, M.; Gan, P.H.P.; Hardham, A.R.; Jones, D.A.; Ellis, J.G. Effectors of biotrophic fungi and oomycetes: Pathogenicity factors and triggers of host resistance. New Phytol. 2009, 183, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Koeck, M.; Hardham, A.R.; Dodds, P.N. The role of effectors of biotrophic and hemibiotrophic fungi in infection. Cell. Microbiol. 2011, 13, 1849–1857. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-J.; van der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.-J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef]
- Zhang, H.; Kim, M.S.; Huang, J.; Yan, H.; Yang, T.; Song, L.; Yu, W.; Shim, W.B. Transcriptome analysis of maize pathogen Fusarium verticillioides revealed FvLcp1, a secreted protein with type-D fungal LysM and chitin-binding domains, that plays important roles in pathogenesis and mycotoxin production. Microbiol. Res. 2022, 265, 127195. [Google Scholar] [CrossRef]
- Naumann, T.A.; Wicklow, D.T.; Price, N.P.J. Identification of a chitinase-modifying protein from Fusarium verticillioides: Truncation of a host resistance protein by a fungalysin metalloprotease. J. Biol. Chem. 2011, 286, 35358–35366. [Google Scholar] [CrossRef]
- Kulkarni, R.D.; Kelkar, H.S.; Dean, R.A. An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 2003, 28, 118–121. [Google Scholar] [CrossRef]
- Zhang, Z.-N.; Wu, Q.-Y.; Zhang, G.-Z.; Zhu, Y.-Y.; Murphy, R.W.; Liu, Z.; Zou, C.-G. Systematic analyses reveal uniqueness and origin of the CFEM domain in fungi. Sci. Rep. 2015, 5, 13032. [Google Scholar] [CrossRef]
- Arya, G.C.; Srivastava, D.A.; Pandaranayaka, E.P.J.; Manasherova, E.; Prusky, D.B.; Elad, Y.; Frenkel, O.; Dvir, H.; Harel, A. Characterization of the role of a non-gpcr membrane-bound cfem protein in the pathogenicity and germination of Botrytis cinerea. Microorganisms 2020, 8, 1043. [Google Scholar] [CrossRef]
- Moukadiri, I.; Armero, J.; Abad, A.; Sentandreu, R.; Zueco, J. Identification of a mannoprotein present in the inner layer of the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 1997, 179, 2154–2162. [Google Scholar] [CrossRef]
- Nasser, L.; Weissman, Z.; Pinsky, M.; Amartely, H.; Dvir, H.; Kornitzer, D. Structural basis of haem-iron acquisition by fungal pathogens. Nat. Microbiol. 2016, 1, 16156. [Google Scholar] [CrossRef]
- Kou, Y.; Tan, Y.H.; Ramanujam, R.; Naqvi, N.I. Structure–function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytol. 2017, 214, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Sabnam, N.; Roy Barman, S. WISH, a novel CFEM GPCR is indispensable for surface sensing, asexual and pathogenic differentiation in rice blast fungus. Fungal Genet. Biol. 2017, 105, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Gong, A.-d.; Jing, Z.-y.; Zhang, K.; Tan, Q.-q.; Wang, G.-l.; Liu, W.-d. Bioinformatic analysis and functional characterization of the CFEM proteins in maize anthracnose fungus Colletotrichum graminicola. J. Integr. Agric. 2020, 19, 541–550. [Google Scholar] [CrossRef]
- Wang, J.-x.; Long, F.; Zhu, H.; Zhang, Y.; Wu, J.-y.; Shen, S.; Dong, J.-g.; Hao, Z.-m. Bioinformatic analysis and functional characterization of CFEM proteins in Setosphaeria turcica. J. Integr. Agric. 2021, 20, 2438–2449. [Google Scholar] [CrossRef]
- Qian, Y.; Zheng, X.; Wang, X.; Yang, J.; Zheng, X.; Zeng, Q.; Li, J.; Zhuge, Q.; Xiong, Q. Systematic identification and functional characterization of the CFEM proteins in poplar fungus Marssonina brunnea. Front. Cell. Infect. Microbiol. 2022, 12, 1045615. [Google Scholar] [CrossRef]
- Zhu, W.; Wei, W.; Wu, Y.; Zhou, Y.; Peng, F.; Zhang, S.; Chen, P.; Xu, X. BcCFEM1, a CFEM domain-containing protein with putative gpi-anchored site, is involved in pathogenicity, conidial production, and stress tolerance in Botrytis cinerea. Front. Microbiol. 2017, 8, 1807. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, D.-D.; Song, J.; Li, J.-J.; Wang, J.; Li, R.; Klosterman, S.J.; Kong, Z.-Q.; Lin, F.-Z.; Dai, X.-F.; et al. Verticillium dahliae CFEM proteins manipulate host immunity and differentially contribute to virulence. BMC Biol. 2022, 20, 55. [Google Scholar] [CrossRef]
- Bai, X.; Peng, H.; Goher, F.; Islam, M.A.; Xu, S.; Guo, J.; Kang, Z.; Guo, J. A candidate effector protein PstCFEM1 contributes to virulence of stripe rust fungus and impairs wheat immunity. Stress Biol. 2022, 2, 21. [Google Scholar] [CrossRef]
- Shang, S.; Liu, G.; Zhang, S.; Liang, X.; Zhang, R.; Sun, G. A fungal CFEM-containing effector targets NPR1 regulator NIMIN2 to suppress plant immunity. Plant Biotechnol. J. 2024, 22, 82–97. [Google Scholar] [CrossRef]
- Kulkarni, R.D.; Dean, R.A. Identification of proteins that interact with two regulators of appressorium development, adenylate cyclase and cAMP-dependent protein kinase A, in the rice blast fungus Magnaporthe grisea. Mol. Genet. Genom. 2004, 270, 497–508. [Google Scholar] [CrossRef]
- Giraldo, M.C.; Valent, B. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 2013, 11, 800–814. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, K.A.; Collins-Racie, L.A.; Colbert, M.; Duckett, M.; Golden-Fleet, M.; Kelleher, K.; Kriz, R.; LaVallie, E.R.; Merberg, D.; Spaulding, V.; et al. A genetic selection for isolating cDNAs encoding secreted proteins. Gene 1997, 198, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Abramovitch, R.B.; Martin, G.B. Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 2004, 7, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, M.S.; McCormack, M.E.; Argueso, C.T.; Pajerowska-Mukhtar, K.M. Pathogen tactics to manipulate plant cell death. Curr. Biol. 2016, 26, R608–R619. [Google Scholar] [CrossRef]
- Upadhyaya, N.M.; Mago, R.; Staskawicz, B.J.; Ayliffe, M.A.; Ellis, J.G.; Dodds, P.N. A bacterial type iii secretion assay for delivery of fungal effector proteins into wheat. Mol. Plant-Microbe Interact. 2014, 27, 255–264. [Google Scholar] [CrossRef]
- Liang, X.; Bao, Y.; Zhang, M.; Du, D.; Rao, S.; Li, Y.; Wang, X.; Xu, G.; Zhou, Z.; Shen, D.; et al. A Phytophthora capsici RXLR effector targets and inhibits the central immune kinases to suppress plant immunity. New Phytol. 2021, 232, 264–278. [Google Scholar] [CrossRef]
- Wu, S.; Shan, L.; He, P. Microbial signature-triggered plant defense responses and early signaling mechanisms. Plant Sci. 2014, 228, 118–126. [Google Scholar] [CrossRef]
- Rajput, N.A.; Zhang, M.; Ru, Y.; Liu, T.; Xu, J.; Liu, L.; Mafurah, J.J.; Dou, D. Phytophthora sojae Effector PsCRN70 Suppresses Plant Defenses in Nicotiana benthamiana. PLoS ONE 2014, 9, e98114. [Google Scholar] [CrossRef]
- Klinkert, B.; Narberhaus, F. Microbial thermosensors. Cell. Mol. Life Sci. 2009, 66, 2661–2676. [Google Scholar] [CrossRef]
- Zuo, N.; Bai, W.-Z.; Wei, W.-Q.; Yuan, T.-L.; Zhang, D.; Wang, Y.-Z.; Tang, W.-H. Fungal CFEM effectors negatively regulate a maize wall-associated kinase by interacting with its alternatively spliced variant to dampen resistance. Cell Rep. 2022, 41, 111877. [Google Scholar] [CrossRef]
- Cai, N.; Liu, R.; Yan, D.; Zhang, N.; Zhu, K.; Zhang, D.; Nong, X.; Tu, X.; Zhang, Z.; Wang, G. bioinformatics analysis and functional characterization of the CFEM Proteins of Metarhizium anisopliae. J. Fungi 2022, 8, 661. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Kinoshita, T. Structural remodeling, trafficking and functions of glycosylphosphatidylinositol-anchored proteins. Prog. Lipid Res. 2011, 50, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Plaine, A.; Walker, L.; Da Costa, G.; Mora-Montes, H.M.; McKinnon, A.; Gow, N.A.R.; Gaillardin, C.; Munro, C.A.; Richard, M.L. Functional analysis of Candida albicans GPI-anchored proteins: Roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 2008, 45, 1404–1414. [Google Scholar] [CrossRef]
- Pittet, M.; Conzelmann, A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2007, 1771, 405–420. [Google Scholar] [CrossRef]
- Muszkieta, L.; Fontaine, T.; Beau, R.; Mouyna, I.; Vogt, M.S.; Trow, J.; Cormack, B.P.; Essen, L.-O.; Jouvion, G.; Latgé, J.-P. The Glycosylphosphatidylinositol-anchored dfg family is essential for the insertion of galactomannan into the β-(1,3)-glucan–chitin core of the cell wall of Aspergillus fumigatus. mSphere 2019, 4, e00397-19. [Google Scholar] [CrossRef]
- Vaknin, Y.; Shadkchan, Y.; Levdansky, E.; Morozov, M.; Romano, J.; Osherov, N. The three Aspergillus fumigatus CFEM-domain GPI-anchored proteins (CfmA-C) affect cell-wall stability but do not play a role in fungal virulence. Fungal Genet. Biol. 2014, 63, 55–64. [Google Scholar] [CrossRef]
- Lamkanfi, M.; Dixit, V.M. Manipulation of host cell death pathways during microbial infections. Cell Host Microbe 2010, 8, 44–54. [Google Scholar] [CrossRef]
- Wagner, T.A.; Kohorn, B.D. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 2001, 13, 303–318. [Google Scholar] [CrossRef]
- Zhang, B.; Li, P.; Su, T.; Li, P.; Xin, X.; Wang, W.; Zhao, X.; Yu, Y.; Zhang, D.; Yu, S.; et al. Comprehensive analysis of wall-associated kinase genes and their expression under abiotic and biotic stress in chinese cabbage (Brassica rapa ssp. pekinensis). J. Plant Growth Regul. 2020, 39, 72–86. [Google Scholar] [CrossRef]
- Abdul Haseeb, H.; Zhang, J.; Guo, Y.-s.; Gao, M.-x.; Guo, W. Proteomic analysis of pathogen-responsive proteins from maize stem apoplast triggered by Fusarium verticillioides. J. Integr. Agric. 2022, 21, 446–459. [Google Scholar] [CrossRef]
- Han, Y.; Li, X.; Cheng, L.; Liu, Y.; Wang, H.; Ke, D.; Yuan, H.; Zhang, L.; Wang, L. Genome-wide analysis of soybean jmjc domain-containing proteins suggests evolutionary conservation following whole-genome duplication. Front. Plant Sci. 2016, 7, 1800. [Google Scholar] [CrossRef] [PubMed]
- Fang, A.; Han, Y.; Zhang, N.; Zhang, M.; Liu, L.; Li, S.; Lu, F.; Sun, W. Identification and characterization of plant cell death–inducing secreted proteins from Ustilaginoidea virens. Mol. Plant-Microbe Interact. 2016, 29, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Ren, Y.; Wang, D.; Farooq, T.; He, Z.; Zhang, C.; Li, S.; Yang, X.; Zhou, X. A group I WRKY transcription factor regulates mulberry mosaic dwarf-associated virus-triggered cell death in Nicotiana benthamiana. Mol. Plant Pathol. 2022, 23, 237–253. [Google Scholar] [CrossRef]
- Tang, C.; Xu, Q.; Zhao, J.; Yue, M.; Wang, J.; Wang, X.; Kang, Z.; Wang, X. A rust fungus effector directly binds plant pre-mRNA 769 splice site to reprogram alternative splicing and suppress host immunity. Plant Biotechnol. J. 2022, 20, 1167–1181. [Google Scholar] [CrossRef]
- Schenk, S.T.; Hernández-Reyes, C.; Samans, B.; Stein, E.; Neumann, C.; Schikora, M.; Reichelt, M.; Mithöfer, A.; Becker, A.; Kogel, K.-H.; et al. N-acyl-homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell 2014, 26, 2708–2723. [Google Scholar] [CrossRef]
- Choi, Y.-E.; Xu, J.-R. The cAMP Signaling Pathway in Fusarium verticillioides is important for conidiation, plant infection, and stress responses but not fumonisin production. Mol. Plant-Microbe Interact. 2010, 23, 522–533. [Google Scholar] [CrossRef]
- Ridenour, J.B.; Hirsch, R.L.; Bluhm, B.H. Identifying genes in Fusarium verticillioides through forward and reverse genetics. In Plant Fungal Pathogens: Methods and Protocols; Bolton, M.D., Thomma, B.P.H.J., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 457–479. [Google Scholar]
- Jiang, C.; Hei, R.; Yang, Y.; Zhang, S.; Wang, Q.; Wang, W.; Zhang, Q.; Yan, M.; Zhu, G.; Huang, P.; et al. An orphan protein of Fusarium graminearum modulates host immunity by mediating proteasomal degradation of TaSnRK1α. Nat. Commun. 2020, 11, 4382. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, X.; Xi, K.; Liu, Y.; Jijakli, M.H.; Guo, W. Development of an RPA-based CRISPR/Cas12a assay in combination with a lateral flow strip for rapid detection of toxigenic Fusarium verticillioides in maize. Food Control 2024, 157, 110172. [Google Scholar] [CrossRef]
- Li, H.; Zhang, D.; Xie, K.; Wang, Y.; Liao, Q.; Hong, Y.; Liu, Y. Efficient and high-throughput pseudorecombinant-chimeric Cucumber mosaic virus-based VIGS in maize. Plant Physiol. 2021, 187, 2865–2876. [Google Scholar] [CrossRef]
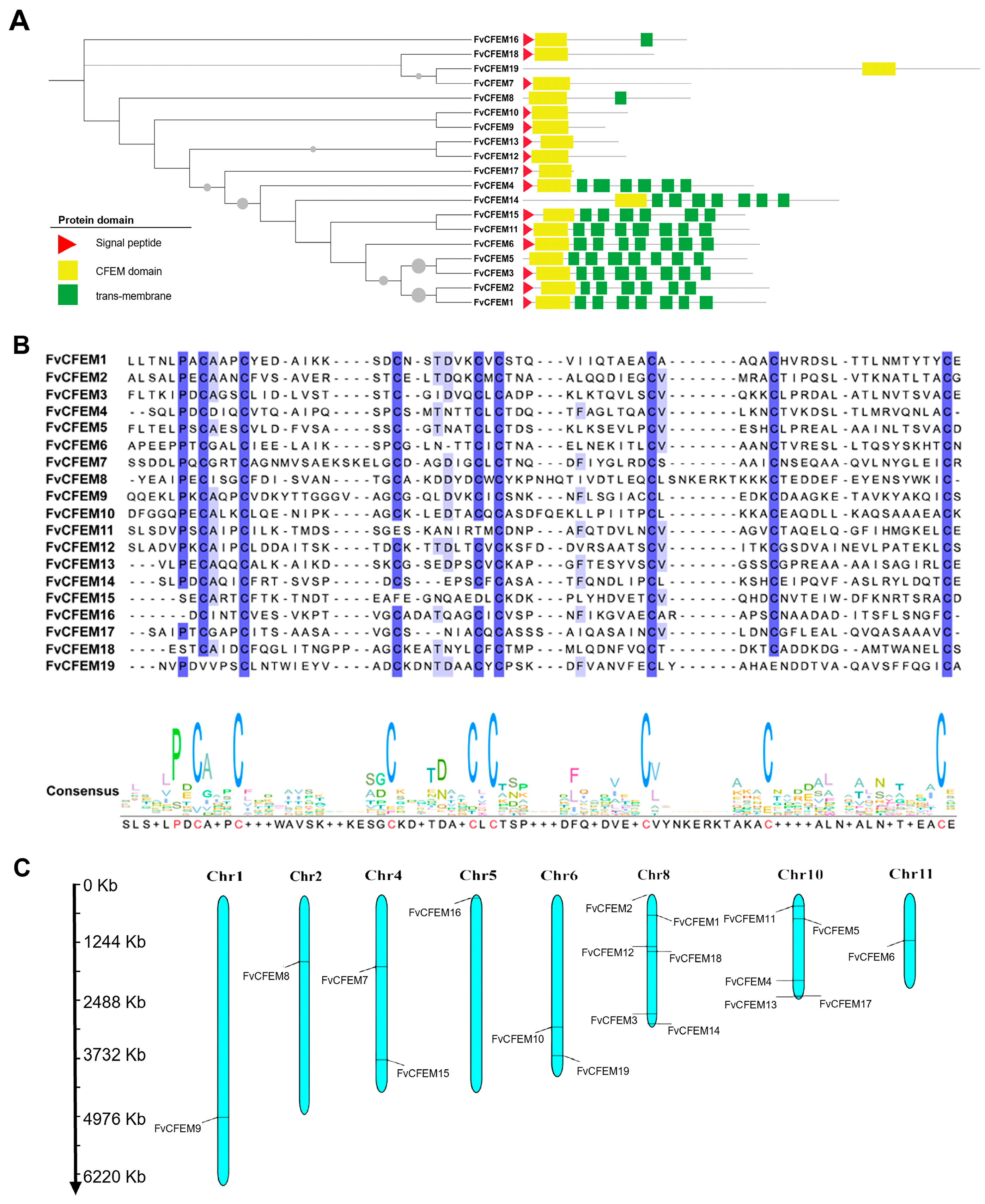
| Name | ID | SP a | TM b | Amino Acid | Position of CFEM Domain (aa) | GPI-Anchored |
|---|---|---|---|---|---|---|
| FvCFEM1 | FVEG_16152 | 1–17 | 7 | 454 | 24–88 | |
| FvCFEM2 | FVEG_07166 | 1–20 | 6 | 460 | 34–98 | |
| FvCFEM3 | FVEG_13472 | 1–19 | 7 | 429 | 25–88 | |
| FvCFEM4 | FVEG_08756 | 1–19 | 6 | 431 | 27–88 | |
| FvCFEM5 | FVEG_17608 | 7 | 419 | 12–75 | ||
| FvCFEM6 | FVEG_10583 | 1–22 | 7 | 442 | 23–86 | |
| FvCFEM7 | FVEG_04843 | 1–17 | 0 | 314 | 19–88 | A291/A314 |
| FvCFEM8 | FVEG_06284 | 1 | 313 | 10–82 | ||
| FvCFEM9 | FVEG_01052 | 1–17 | 0 | 154 | 18–85 | N133/A134 |
| FvCFEM10 | FVEG_02239 | 1–15 | 0 | 196 | 17–84 | S166/S167 |
| FvCFEM11 | FVEG_13696 | 1–18 | 7 | 423 | 20–84 | |
| FvCFEM12 | FVEG_07535 | 1–16 | 0 | 193 | 18–85 | G168/S173 |
| FvCFEM13 | FVEG_08884 | 1–18 | 0 | 179 | 33–94 | |
| FvCFEM14 | FVEG_13561 | 7 | 590 | 173–231 | ||
| FvCFEM15 | FVEG_17202 | 1–21 | 6 | 415 | 38–97 | |
| FvCFEM16 | FVEG_02591 | 1–21 | 1 | 306 | 23–82 | |
| FvCFEM17 | FVEG_08871 | 1–18 | 0 | 95 | 31–91 | |
| FvCFEM18 | FVEG_07573 | 1–19 | 0 | 245 | 21–84 | D219 |
| FvCFEM19 | FVEG_02431 | 0 | 853 | 633–695 | A773/G774 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Ishfaq, S.; Liang, X.; Wang, R.; Wei, H.; Guo, W. A Novel CFEM Effector in Fusarium verticillioides Required for Virulence Involved in Plant Immunity Suppression and Fungal Cell Wall Integrity. Int. J. Mol. Sci. 2025, 26, 4369. https://doi.org/10.3390/ijms26094369
Li H, Ishfaq S, Liang X, Wang R, Wei H, Guo W. A Novel CFEM Effector in Fusarium verticillioides Required for Virulence Involved in Plant Immunity Suppression and Fungal Cell Wall Integrity. International Journal of Molecular Sciences. 2025; 26(9):4369. https://doi.org/10.3390/ijms26094369
Chicago/Turabian StyleLi, Huan, Shumila Ishfaq, Xiaoyan Liang, Rui Wang, Hailei Wei, and Wei Guo. 2025. "A Novel CFEM Effector in Fusarium verticillioides Required for Virulence Involved in Plant Immunity Suppression and Fungal Cell Wall Integrity" International Journal of Molecular Sciences 26, no. 9: 4369. https://doi.org/10.3390/ijms26094369
APA StyleLi, H., Ishfaq, S., Liang, X., Wang, R., Wei, H., & Guo, W. (2025). A Novel CFEM Effector in Fusarium verticillioides Required for Virulence Involved in Plant Immunity Suppression and Fungal Cell Wall Integrity. International Journal of Molecular Sciences, 26(9), 4369. https://doi.org/10.3390/ijms26094369





