Optimal Reference Genes for Gene Expression Analysis of Overmating Stress-Induced Aging and Natural Aging in Male Macrobrachium rosenbergii
Abstract
1. Introduction
2. Results
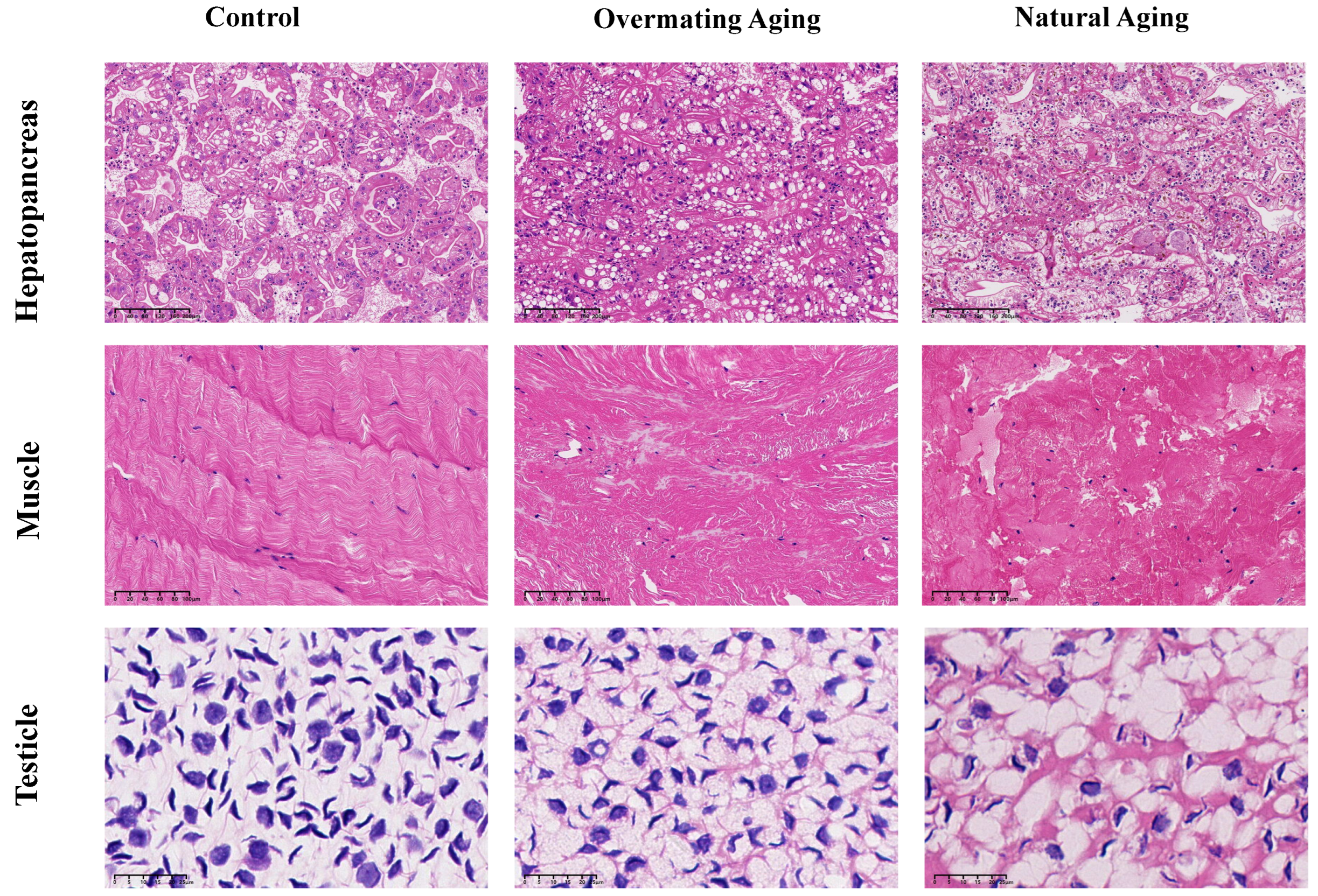
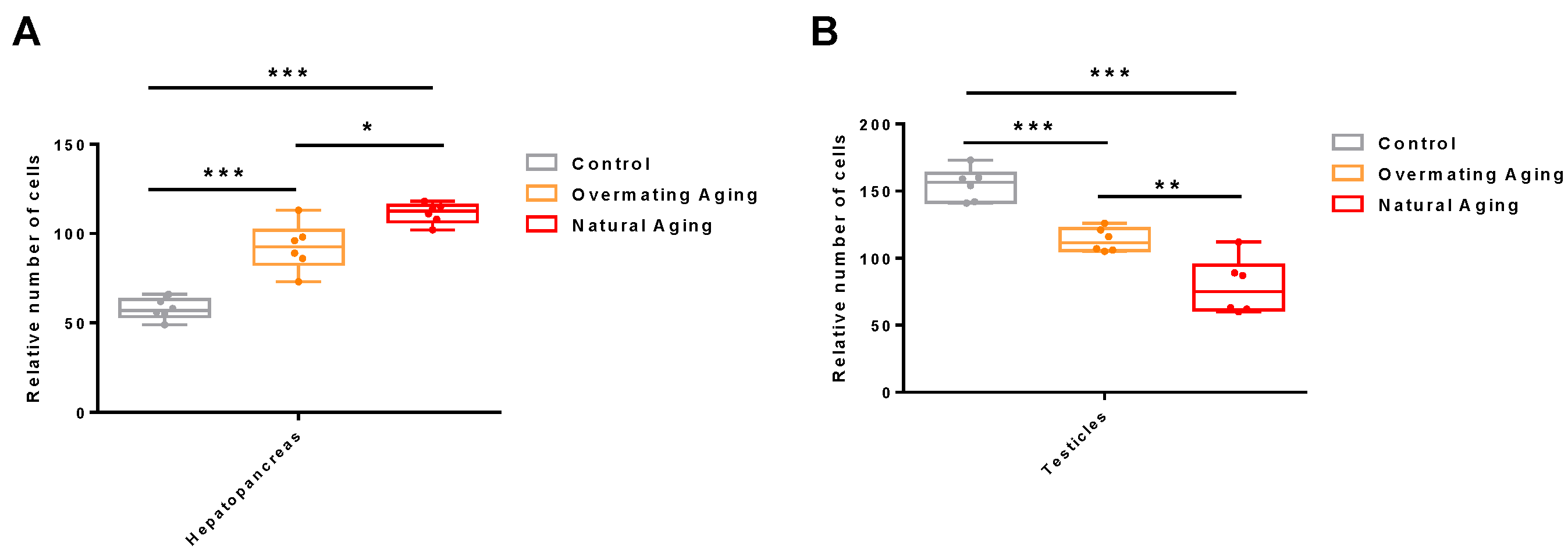
2.1. Analysis of Male M. rosenbergii in Three Different Physiological States
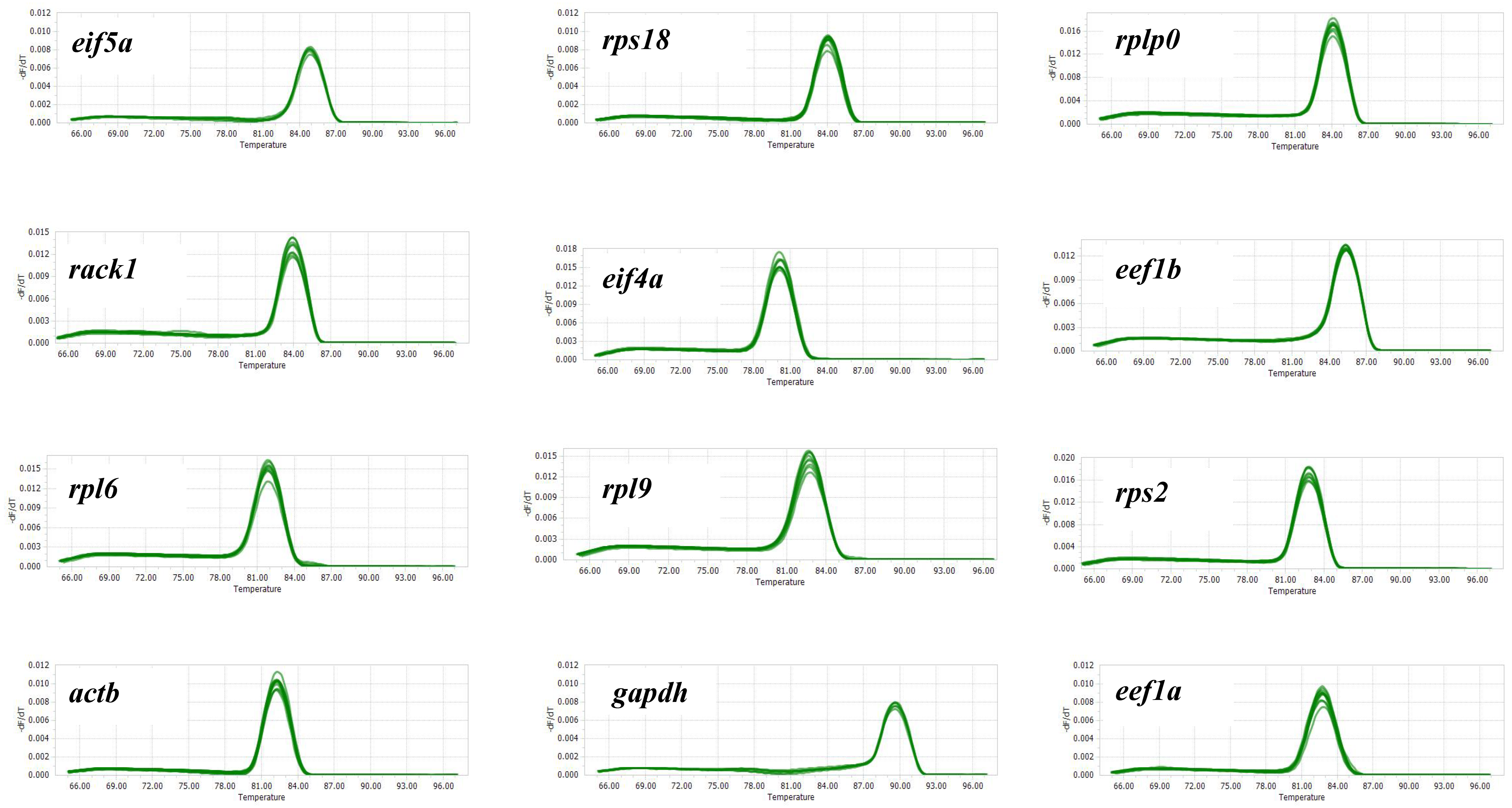
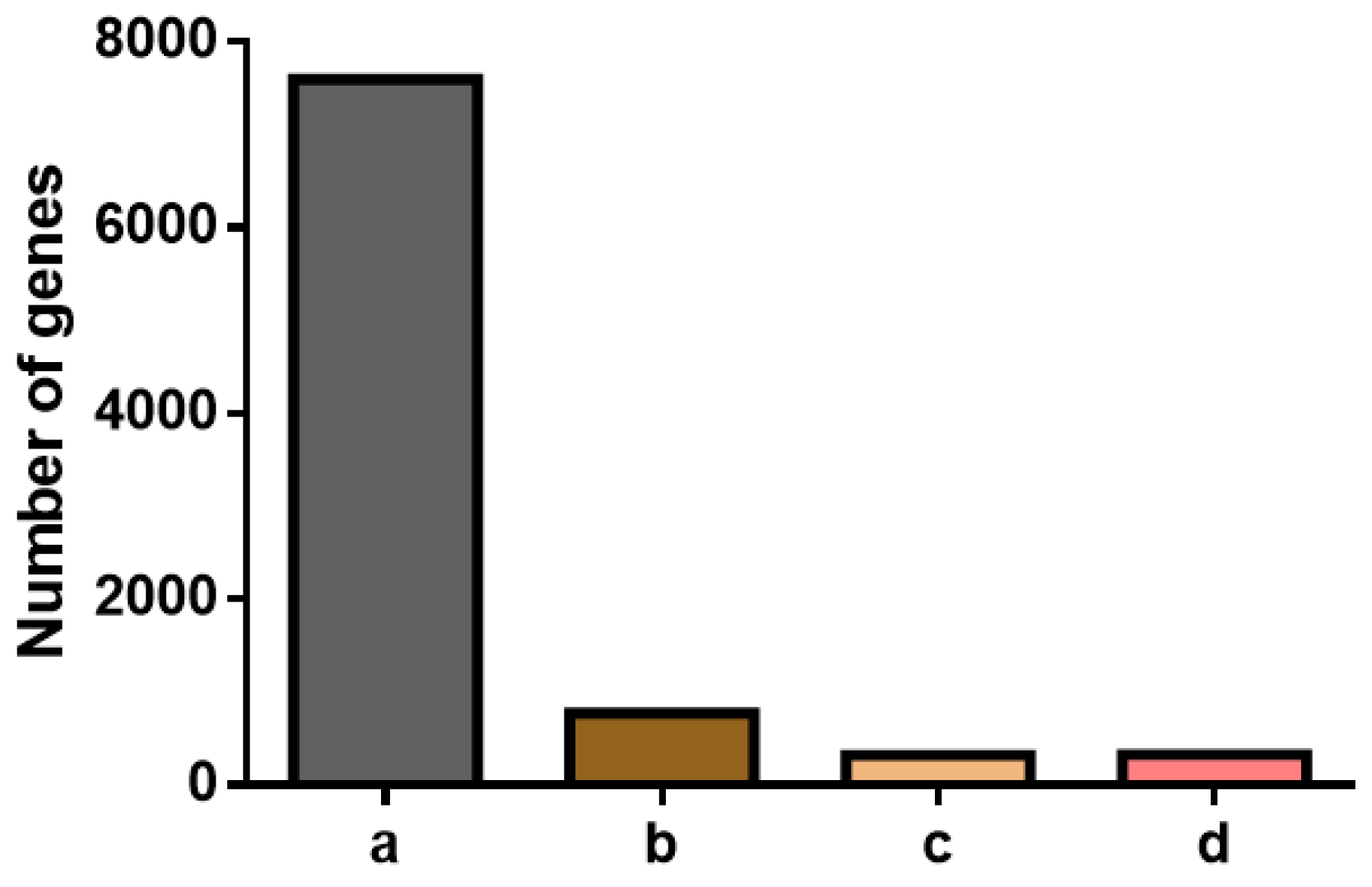
2.2. Selection of Candidate Reference Genes
2.3. Distribution of Cycle Threshold (Quantitative Cycle) Values
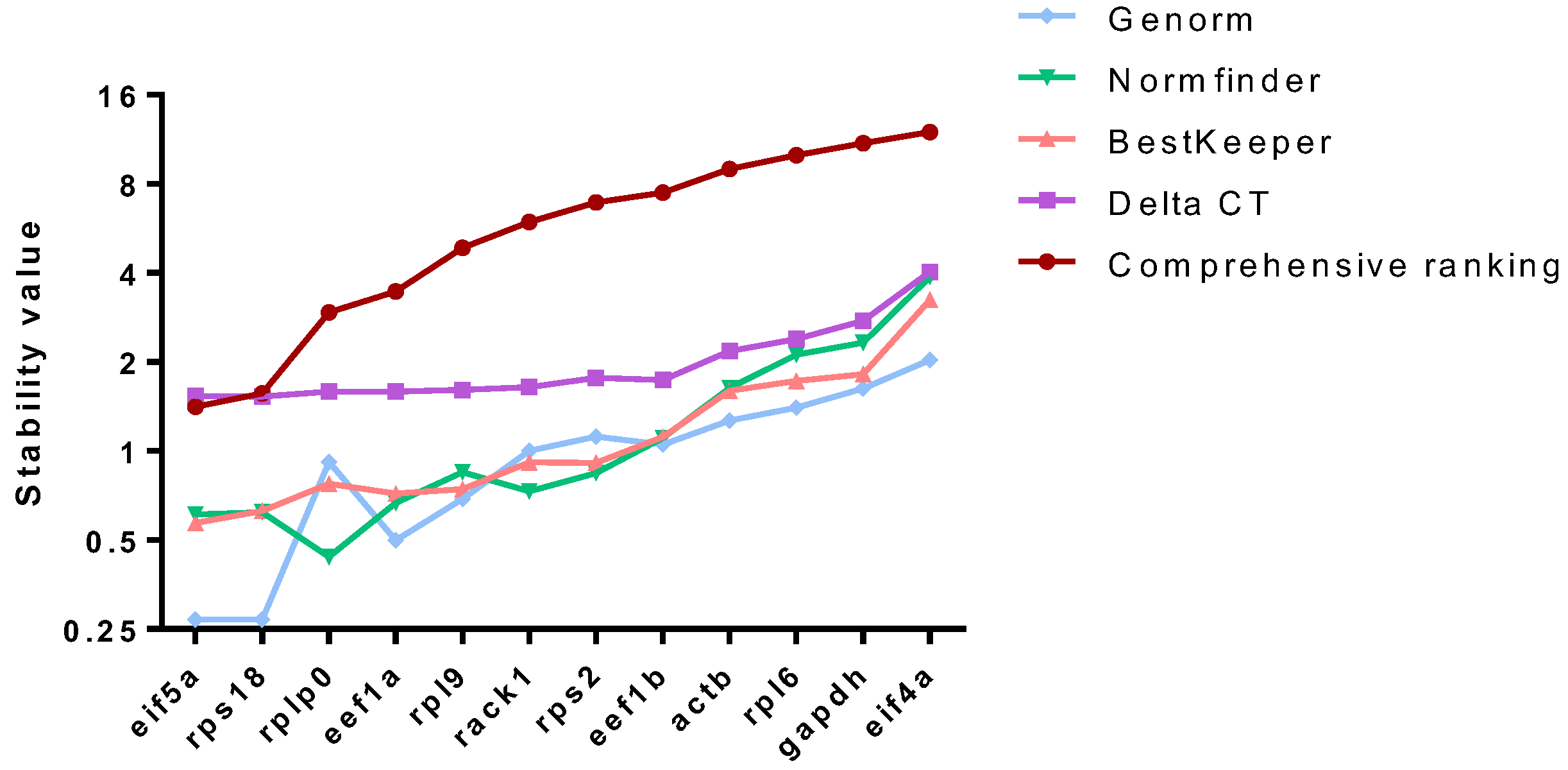
2.4. The Stability of eif5a and rps18 as Internal Controls Under Different Conditions Was Further Evaluated
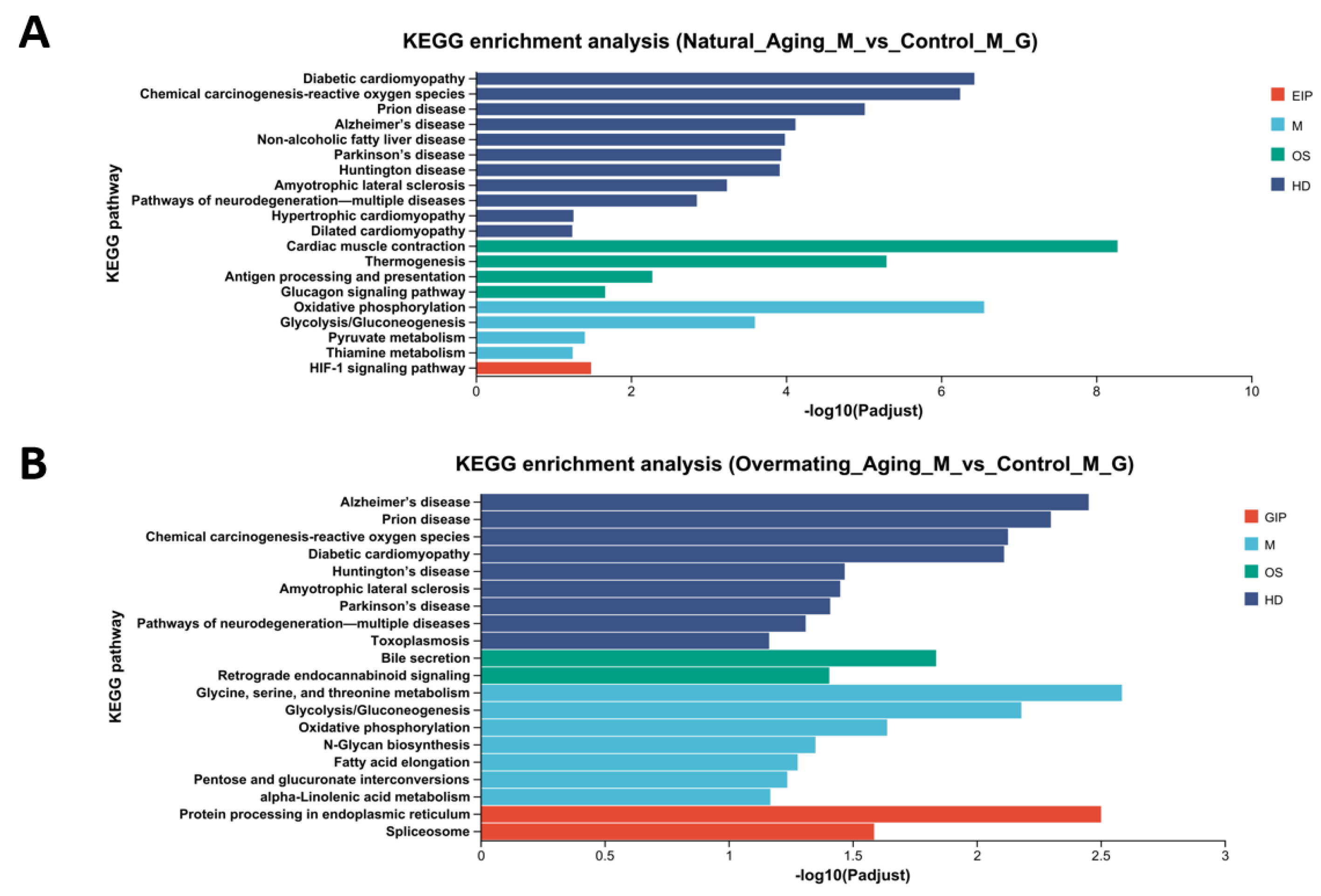
3. Discussion
4. Materials and Methods
4.1. Animals and Experiment Design
4.2. Histological Observation
4.3. RNA Extraction, Sequencing, and Synthesizing cDNA
4.4. Selection of Candidate Reference Genes from Transcriptome Data and Primer Design
4.5. Real-Time Quantitative PCR (qPCR) Analysis of Gene Expression
4.6. Stability Assessment of Reference Genes
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| M. rosenbergii | Macrobrachium rosenbergii |
| RT-qPCR | Real-Time Quantitative Reverse Transcription PCR |
| Ct | Quantitative Cycle/cycle threshold |
| rps2 | Ribosomal protein S2 |
| rack1 | Receptor for activated protein kinase c1 |
| rpl6 | Ribosomal protein L6 |
| eef1b | Eukaryotic translation elongation factor 1β |
| eif4a | Eukaryotic translation initiation factor 4a |
| rpl9 | Ribosomal protein L9 |
| rps18 | Ribosomal protein S18 |
| rplp0 | Ribosomal protein lateral stalk subunit P0 |
| eif5a | Eukaryotic translation initiation factor 5a |
| eef1a | Eukaryotic elongation factor 1α |
| actb | β-actin |
| gapdh | Glyceraldehyde-3-phosphate dehydrogenase |
| TPM | Transcripts per million |
| MIQE | Minimum Information for Publication of Quantitative Real-Time PCR Experiments |
Appendix A

References
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef]
- Legnini, I.; Emmenegger, L.; Zappulo, A.; Rybak-Wolf, A.; Wurmus, R.; Martinez, A.O.; Jara, C.C.; Boltengagen, A.; Hessler, T.; Mastrobuoni, G.; et al. Spatiotemporal, optogenetic control of gene expression in organoids. Nat. Methods 2023, 20, 1544–1552. [Google Scholar]
- Fan, Y.; Zhang, G.; Zhao, K.; Fu, W.; Chen, S.; Liu, J.; Liu, W.; Peng, L.; Ren, L.; Liu, S. Characteristics of SP600125 Induced Tetraploid Cells in Comparison With Diploid and Tetraploid Cells of Fish. Front. Genet. 2021, 12, 781007. [Google Scholar]
- Chen, X.; Sun, Y.; Zhang, P.; Li, J.; Li, H.; Wei, C.; Cao, Z.; Zhou, Y. Screening of stable internal reference genes by quantitative real-time PCR in humpback grouper Cromileptes altivelis. J. Oceanol. Limnol. 2021, 39, 1985–1999. [Google Scholar]
- Kumar, D.; Bansal, G.; Narang, A.; Basak, T.; Abbas, T.; Dash, D. Integrating transcriptome and proteome profiling: Strategies and applications. Proteomics 2016, 16, 2533–2544. [Google Scholar]
- Liu, W.; Yuan, X.; Yuan, S.; Dai, L.; Dong, S.; Liu, J.; Peng, L.; Wang, M.; Tang, Y.; Xiao, Y. Optimal reference genes for gene expression analysis in polyploid of Cyprinus carpio and Carassius auratus. BMC Genet. 2020, 21, 107. [Google Scholar] [CrossRef]
- Hu, Y.; Fu, H.; Qiao, H.; Sun, S.; Zhang, W.; Jin, S.; Jiang, S.; Gong, Y.; Xiong, Y.; Wu, Y. Validation and evaluation of reference genes for quantitative real-time PCR in Macrobrachium Nipponense. Int. J. Mol. Sci. 2018, 19, 2258. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef]
- Dhar, A.K.; Bowers, R.M.; Licon, K.S.; Veazey, G.; Read, B. Validation of reference genes for quantitative measurement of immune gene expression in shrimp. Mol. Immunol. 2009, 46, 1688–1695. [Google Scholar] [CrossRef]
- Mo, Y.; Fan, Y.; Fu, W.; Xu, W.; Chen, S.; Wen, Y.; Liu, S.; Peng, L.; Xiao, Y. Acute immune stress improves cell resistance to chemical poison damage in SP600125-induced polyploidy of fish cells in vitro. Fish Shellfish Immunol. 2019, 84, 656–663. [Google Scholar] [CrossRef]
- Krishna, G.; Marappan, M.; Chaudhari, A. Characterization and Evaluation of Selected Housekeeping Genes for Quantitative RT-PCR in Macrobrachium rosenbergii Morphotypes. Fish. Technol. 2015, 52, 177–183. [Google Scholar]
- Sang, J.; Wang, Z.; Li, M.; Cao, J.; Niu, G.; Xia, L.; Zou, D.; Wang, F.; Xu, X.; Han, X.; et al. ICG: A wiki-driven knowledgebase of internal control genes for RT-qPCR normalization. Nucleic Acids Res. 2017, 46, D121–D126. [Google Scholar] [CrossRef]
- Mao, H.; Chen, X.; Zhu, X.; Luo, Q.; Zhao, J.; Li, X.; Wu, H.; Xu, H. Identification of suitable reference genes for quantitative real-time PCR normalization in blotched snakehead Channa maculata. J. Fish Biol. 2017, 90, 2312–2322. [Google Scholar] [CrossRef]
- Zhang, G.; Tang, L.; Huang, J.; Wang, Y.; Wang, H.; Fan, Y.; Yuan, X.; Liu, W.; Peng, L.; Liu, J. Formation of asymmetric body color in the caudal fin of common carp (Cyprinus carpio). Aquaculture 2023, 577, 739970. [Google Scholar] [CrossRef]
- Hawkins, L.J.; Luu, B.E.; Storey, K.B. Selection of reference genes for accurate RT-qPCR analysis of dehydration tolerance in Xenopus laevis. Gene Rep. 2018, 13, 192–198. [Google Scholar] [CrossRef]
- Valenzuela-Castillo, A.; Mendoza-Cano, F.; Enríquez-Espinosa, T.; Grijalva-Chon, J.M.; Sánchez-Paz, A. Selection and validation of candidate reference genes for quantitative real-time PCR studies in the shrimp Penaeus vannamei under viral infection. Mol. Cell. Probes 2017, 33, 42–50. [Google Scholar] [CrossRef]
- Pillai, B.R.; Ponzoni, R.W.; Das Mahapatra, K.; Panda, D. Genetic improvement of giant freshwater prawn Macrobrachium rosenbergii: A review of global status. Rev. Aquac. 2022, 14, 1285–1299. [Google Scholar] [CrossRef]
- Li, X.; Xu, B.; Shen, P.; Cheng, H.; Fan, Y.; Gao, Q. Regulation and Response Mechanism of Acute Low-Salinity Stress during Larval Stages in Macrobrachium rosenbergii Based on Multi-Omics Analysis. Int. J. Mol. Sci. 2024, 25, 6809. [Google Scholar] [CrossRef]
- Hooper, C.; Debnath, P.P.; Stentiford, G.D.; Bateman, K.S.; Salin, K.R.; Bass, D. Diseases of the giant river prawn Macrobrachium rosenbergii: A review for a growing industry. Rev. Aquac. 2023, 15, 738–758. [Google Scholar] [CrossRef]
- Zhan, F.; Li, Y.; Shi, F.; Lu, Z.; Yang, M.; Li, Q.; Lin, L.; Qin, Z. Transcriptome analysis of Macrobrachium rosenbergii hemocytes reveals in-depth insights into the immune response to Vibrio parahaemolyticus infection. Fish Shellfish Immunol. 2023, 133, 108533. [Google Scholar] [CrossRef]
- Wang, J.; Yang, G.; Kong, J.; Xia, Z.; Sui, J.; Tang, Q.; Luo, K.; Dai, P.; Meng, X.; Chen, L. An analysis on genetic variation of threshold traits of female and male morphotypes in Macrobrachium rosenbergii. Oceanol. Limnol. Sin. 2022, 53, 710–717. [Google Scholar]
- Sui, J.; Luan, S.; Yang, G.; Xia, Z.; Tang, Q.; Luo, K.; Meng, X.; Kong, J. Effects of the individual rearing stage on the growth traits of candidate giant freshwater prawns (Macrobrachium rosenbergii). Aquac. Int. 2022, 30, 1659–1673. [Google Scholar] [CrossRef]
- Phuc, T.H.; Vu, N.T.; Nga, N.T.K.; Ky, N.T.; Nguyen, N.H. Assessment of a long-term selective breeding program for giant freshwater prawn Macrobrachium rosenbergii since 2007. Aquaculture 2021, 541, 736745. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Li, R.; Zhang, M.; Li, Y.; Wang, H.; Wang, S.; Bao, Z. Systematic identification and validation of the reference genes from 60 RNA-Seq libraries in the scallop Mizuhopecten yessoensis. BMC Genom. 2019, 20, 288. [Google Scholar]
- Han, P.; Chen, J.; Sun, Z.; Ren, S.; Wang, X. Evaluation of reference genes for gene expression analysis in Japanese flounder (Paralichthys olivaceus) under temperature stress. BMC Genom. 2025, 26, 117. [Google Scholar] [CrossRef]
- Jia, L.; Hui, D.; Tao, L.; Cheng, C.; Ying, Y.; Tianqiang, L.; Lei, L.; Erlong, W. Accurate reflection of hepatopancreas antioxidation and detoxification in Procambarus clarkii during virus infection and drug treatment: Reference gene selection, evaluation and expression analysis. Aquaculture 2022, 556, 738283. [Google Scholar]
- Mi, L.Y.; Hayoung, C.; Ok, K.R.; Soyeon, I.; Joo, K.S.; Ji, W.E. Validation of reference genes for quantitative real-time PCR in chemical exposed and at different age’s brackish water flea Diaphanosoma celebensis. Sci. Rep. 2021, 11, 23691. [Google Scholar]
- Sonia, T.; Carmen, L.; Lalia, M.; Emmanouil, F.; Carlos, I. Selection and validation of reference genes for quantitative real-time PCR in the green microalgae Tetraselmis chui. PLoS ONE 2021, 16, e0245495. [Google Scholar]
- Li, G.; Xu, G.; Lin, Z.; Li, H.; Liu, Z.; Xu, Y.; Zhang, H.; Ji, R.; Luo, W.; Qiu, Y.; et al. Selection of suitable reference genes for RT-qPCR normalisation in sweet potato (Ipomoea batatas L.) under different stresses. J. Hortic. Sci. Biotechnol. 2021, 96, 209–219. [Google Scholar]
- Tauc, M.; Cougnon, M.; Carcy, R.; Melis, N.; Hauet, T.; Pellerin, L.; Blondeau, N.; Pisani, D. The eukaryotic initiation factor 5A (eIF5A1), the molecule, mechanisms and recent insights into the pathophysiological roles. Cell Biosci. 2021, 11, 219. [Google Scholar] [CrossRef]
- Wolff, E.C.; Kang, K.R.; Kim, Y.S.; Park, M.H. Posttranslational synthesis of hypusine: Evolutionary progression and specificity of the hypusine modification. Amino Acids 2007, 33, 341–350. [Google Scholar] [CrossRef]
- Hukelmann, J.L.; Anderson, K.E.; Sinclair, L.V.; Grzes, K.M.; Cantrell, D.A. The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat. Immunol. 2015, 17, 104. [Google Scholar] [CrossRef]
- Moreira, V.S.; Soares, V.L.F.; Silva, R.J.S.; Sousa, A.O.; Otoni, W.C.; Costa, M.G.C. Selection and validation of reference genes for quantitative gene expression analyses in various tissues and seeds at different developmental stages in Bixa orellana L. Physiol. Mol. Biol. Plants 2018, 24, 369–378. [Google Scholar] [CrossRef]
- Xu, J.; Welker, D.L.; James, R.R. Variation in Expression of Reference Genes across Life Stages of a Bee, Megachile rotundata. Insects 2021, 12, 36. [Google Scholar] [CrossRef]
- Tan, K.; Wang, W. The early life culture and gonadal development of giant freshwater prawn, Macrobrachium rosenbergii: A review. Aquaculture 2022, 559, 738357. [Google Scholar] [CrossRef]
- Song, J.; Jian, Y.; Xie, Y.; Liang, J.; Shao, C.; Pan, X.; Chen, Z.; Gao, Q.; Kong, Y.; Xu, Q. The Dietary Lipid Requirement for Ovarian Maturation and Health in Female Giant River Prawn, Macrobrachium rosenbergii Broodstock. Aquac. Nutr. 2024, 2024, 7462841. [Google Scholar] [CrossRef]
- Jian, Y.; Song, J.; Chen, Z.; Kong, Y.; Qi, C.; Ding, Z.; Xu, Q. Positive effects of a mixture of fish oil and soybean oil as a dietary lipid source on the ovarian development and health of female giant river prawn, Macrobrachium rosenbergii broodstock. Aquac. Rep. 2024, 39, 102407. [Google Scholar] [CrossRef]
- Tinikul, Y.; Tinikul, R.; Engsusophon, A.; Sobhon, P. The effects of short neuropeptide F on ovarian maturation and spawning in female giant freshwater prawn, Macrobrachium rosenbergii, and associated regulatory mechanisms. Aquaculture 2023, 569, 739361. [Google Scholar] [CrossRef]
- Pillai, B.R.; Panda, D. Global Status of Giant Prawn, Macrobrachium rosenbergii Farming with Special Reference to India and Measures for Enhancing Production. J. Aquac. 2024, 33, 1–14. [Google Scholar] [CrossRef]
- Tu, H.; Peng, X.; Yao, X.; Tang, Q.; Xia, Z.; Li, J.; Yang, G.; Yi, S. Integrated transcriptomic and metabolomic analyses reveal low-temperature tolerance mechanism in giant freshwater prawn Macrobrachium rosenbergii. Animals 2023, 13, 1605. [Google Scholar] [CrossRef]
- Xing, Q.; Tu, H.; Yang, M.; Chen, G.; Tang, Q.; Yi, S.; Gao, Q.; Ibrahim, S.; Liu, Y.; Xia, Z. Evaluation of cold tolerance and gene expression patterns associated with low-temperature stress in giant freshwater prawn Macrobrachium rosenbergii. Aquac. Rep. 2022, 24, 101172. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
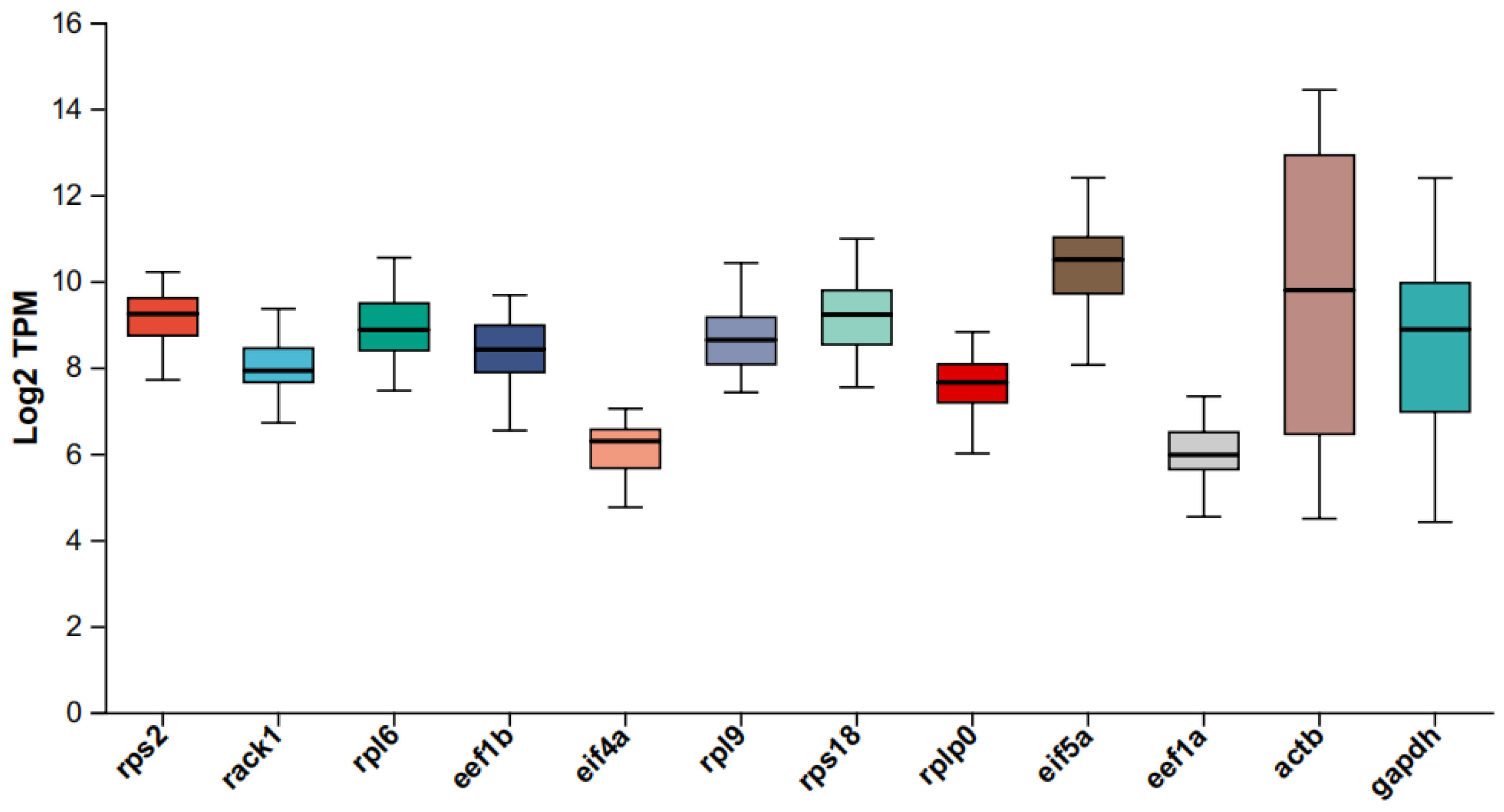
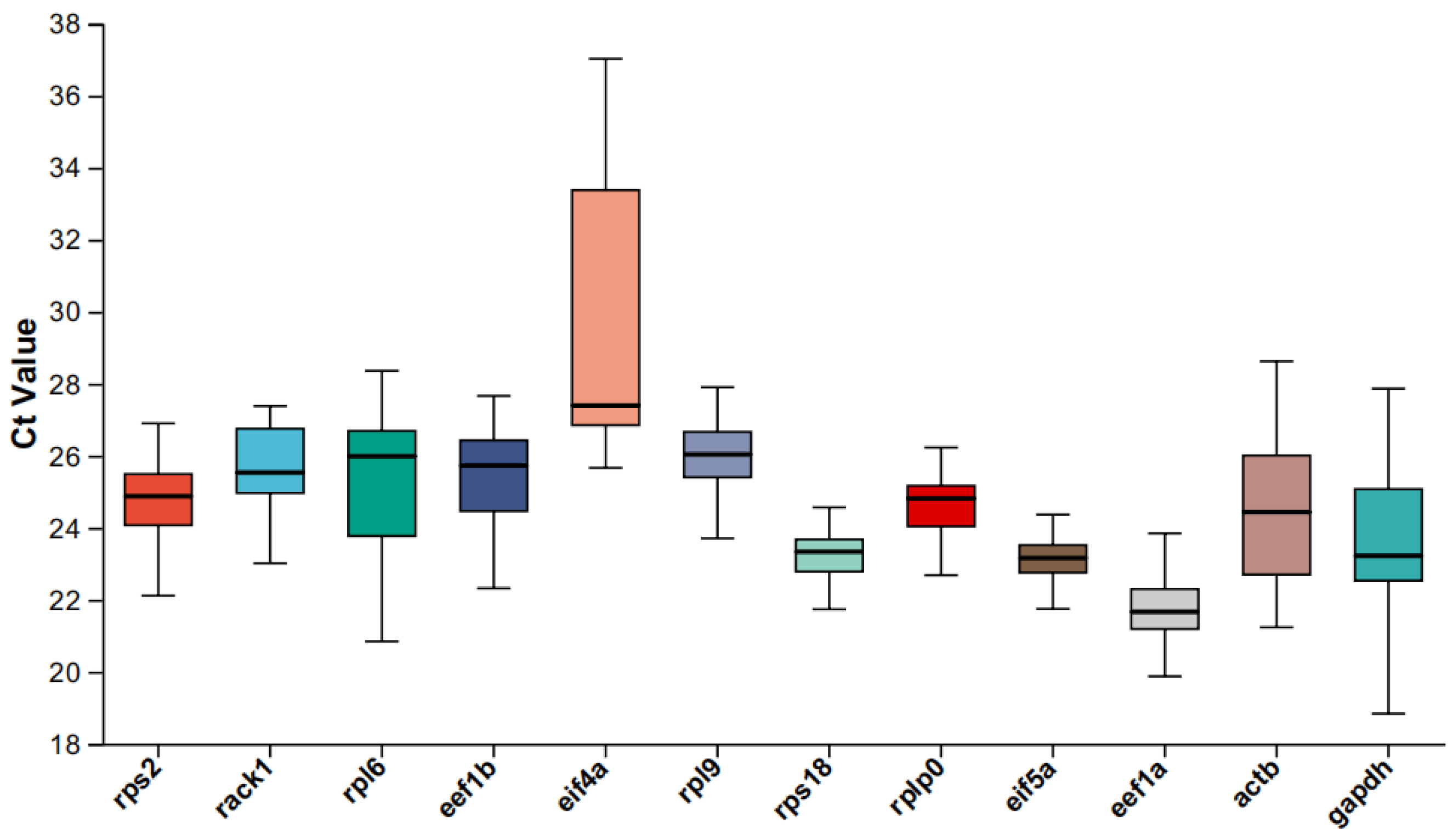

| Gene Name | Description | Mean | Stdev | CV Value |
|---|---|---|---|---|
| rps2 | ribosomal protein S2 | 9.20 | 0.57 | 0.062 |
| rack1 | receptor for activated protein kinase c1 | 7.99 | 0.65 | 0.081 |
| rpl6 | ribosomal protein L6 | 8.94 | 0.73 | 0.082 |
| eef1b | eukaryotic translation elongation factor 1β | 8.41 | 0.71 | 0.084 |
| eif4a | eukaryotic translation initiation factor 4a | 6.11 | 0.53 | 0.087 |
| rpl9 | ribosomal protein L9 | 8.62 | 0.75 | 0.087 |
| rps18 | ribosomal protein S18 | 9.17 | 0.87 | 0.095 |
| rplp0 | ribosomal protein lateral stalk subunit P0 | 7.57 | 0.72 | 0.095 |
| eif5a | eukaryotic translation initiation factor 5a | 6.02 | 0.60 | 0.099 |
| eef1a | eukaryotic elongation factor 1α | 10.32 | 1.06 | 0.103 |
| actb | β-actin | 9.77 | 3.42 | 0.350 |
| gapdh | glyceraldehyde-3-phosphate dehydrogenase | 8.55 | 2.10 | 0.246 |
| Gene | Primer Sequence (5′–3′) | Length (bp) | PCR Efficiency (%) | Correlation Coefficient |
|---|---|---|---|---|
| Forward/Reverse | ||||
| rps18 | TACCTACGACCCACACCCTT | 137 | 98.0 | 0.99 |
| TATCAACGCACCGCCAAGAT | ||||
| rack1 | TGTTTGGCCATCTGCAGACC | 177 | 97.5 | 1.00 |
| CTTTGCTTCAGTCCCAACCG | ||||
| rps2 | GCACGTGTCTGTTTCTGGAC | 176 | 97.5 | 1.00 |
| TGGGTGCCAATCACCAAACT | ||||
| rplp0 | ACATGTTGAGAAGCGTGGCT | 163 | 96.0 | 1.00 |
| CCTGCCCAGAACACTGGATT | ||||
| eif4a | TCACCACAGACTTGCTTGCT | 118 | 95.0 | 1.00 |
| AAACGTCCACCACGTCCAAT | ||||
| eef1b | GGCACTTTTCTTTGCAGCGT | 178 | 94.0 | 1.00 |
| AGCCGAGAAGTCCAAGTTCC | ||||
| rpl6 | CAAGTACGCACGAAGAAGCG | 107 | 96.5 | 1.00 |
| AAAGACTTGCGGAGAGGAGC | ||||
| rpl9 | AAACCGCAAGGAGGTAGCTG | 106 | 98.0 | 1.00 |
| AGCGTATACAGCACGCATCT | ||||
| eif5a | CATGGATGTACCTGTAGTGAAAC | 179 | 104.0 | 0.98 |
| CTGTCAGCAGAAGGTCCTCATTA | ||||
| actb | ACCACCATGTACCCAGGAATCGCTG | 133 | 101.5 | 0.98 |
| CCAAGATTGAACCGCCGATCCAG | ||||
| eef1a | ACTGCGCTGTGTTGATTGTAGCTG | 90 | 101.5 | 1.00 |
| ACAACAGTACGTGTTCACGGGTCT | ||||
| gapdh | TGAAGCCCGAGAACATTCCATG | 170 | 104.0 | 1.00 |
| GTTCACGCCGCAGACGAACATG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Y.; Gao, Q.; Cheng, H.; Li, X.; Xu, Y.; Yuan, H.; Yuan, X.; Bao, S.; Kuan, C.; Zhang, H. Optimal Reference Genes for Gene Expression Analysis of Overmating Stress-Induced Aging and Natural Aging in Male Macrobrachium rosenbergii. Int. J. Mol. Sci. 2025, 26, 3465. https://doi.org/10.3390/ijms26083465
Fan Y, Gao Q, Cheng H, Li X, Xu Y, Yuan H, Yuan X, Bao S, Kuan C, Zhang H. Optimal Reference Genes for Gene Expression Analysis of Overmating Stress-Induced Aging and Natural Aging in Male Macrobrachium rosenbergii. International Journal of Molecular Sciences. 2025; 26(8):3465. https://doi.org/10.3390/ijms26083465
Chicago/Turabian StyleFan, Yunpeng, Qiang Gao, Haihua Cheng, Xilian Li, Yang Xu, Huwei Yuan, Xiudan Yuan, Songsong Bao, Chu Kuan, and Haiqi Zhang. 2025. "Optimal Reference Genes for Gene Expression Analysis of Overmating Stress-Induced Aging and Natural Aging in Male Macrobrachium rosenbergii" International Journal of Molecular Sciences 26, no. 8: 3465. https://doi.org/10.3390/ijms26083465
APA StyleFan, Y., Gao, Q., Cheng, H., Li, X., Xu, Y., Yuan, H., Yuan, X., Bao, S., Kuan, C., & Zhang, H. (2025). Optimal Reference Genes for Gene Expression Analysis of Overmating Stress-Induced Aging and Natural Aging in Male Macrobrachium rosenbergii. International Journal of Molecular Sciences, 26(8), 3465. https://doi.org/10.3390/ijms26083465






