Overexpression of the Transcription Factor GmbZIP60 Increases Salt and Drought Tolerance in Soybean (Glycine max)
Abstract
1. Introduction
2. Results
2.1. Sequence and Domain Analyses of GmbZIP60
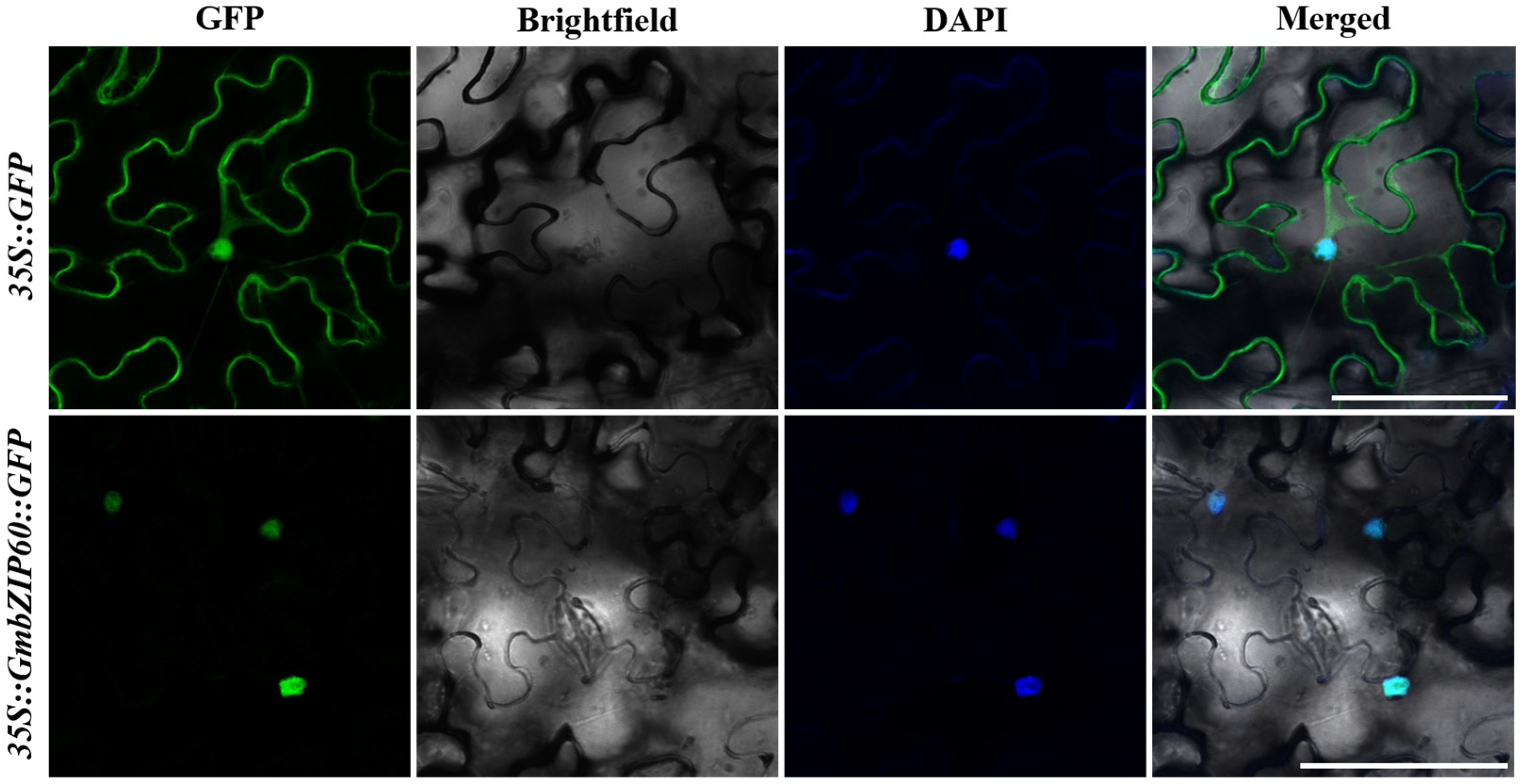
2.2. Subcellular Localization of GmbZIP60
2.3. Expression Patterns of GmbZIP60 in Response to Various Treatments
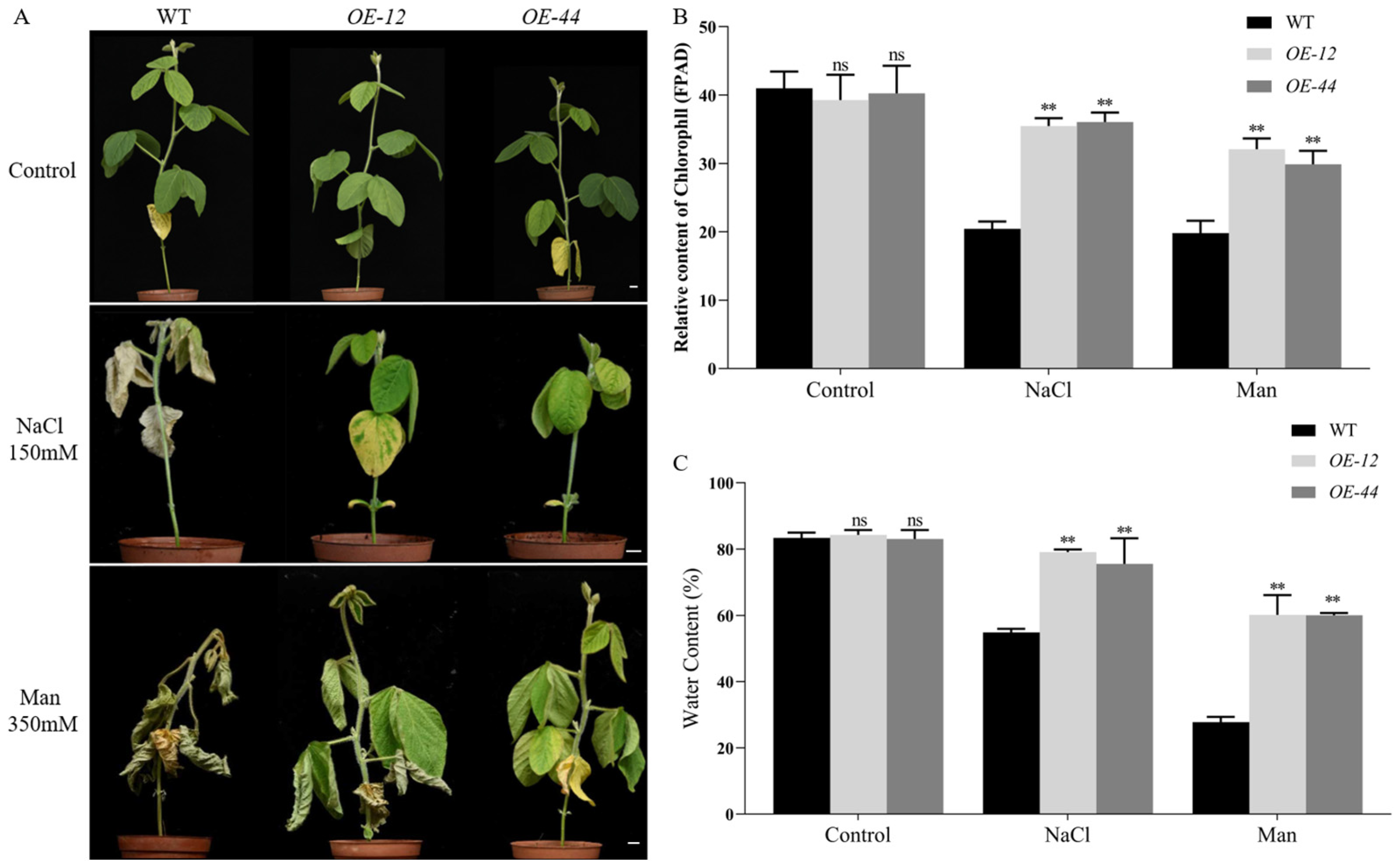
2.4. Overexpression of GmbZIP60 Enhances the Resistance of Transgenic Soybean Plants to Salt and Drought Stresses
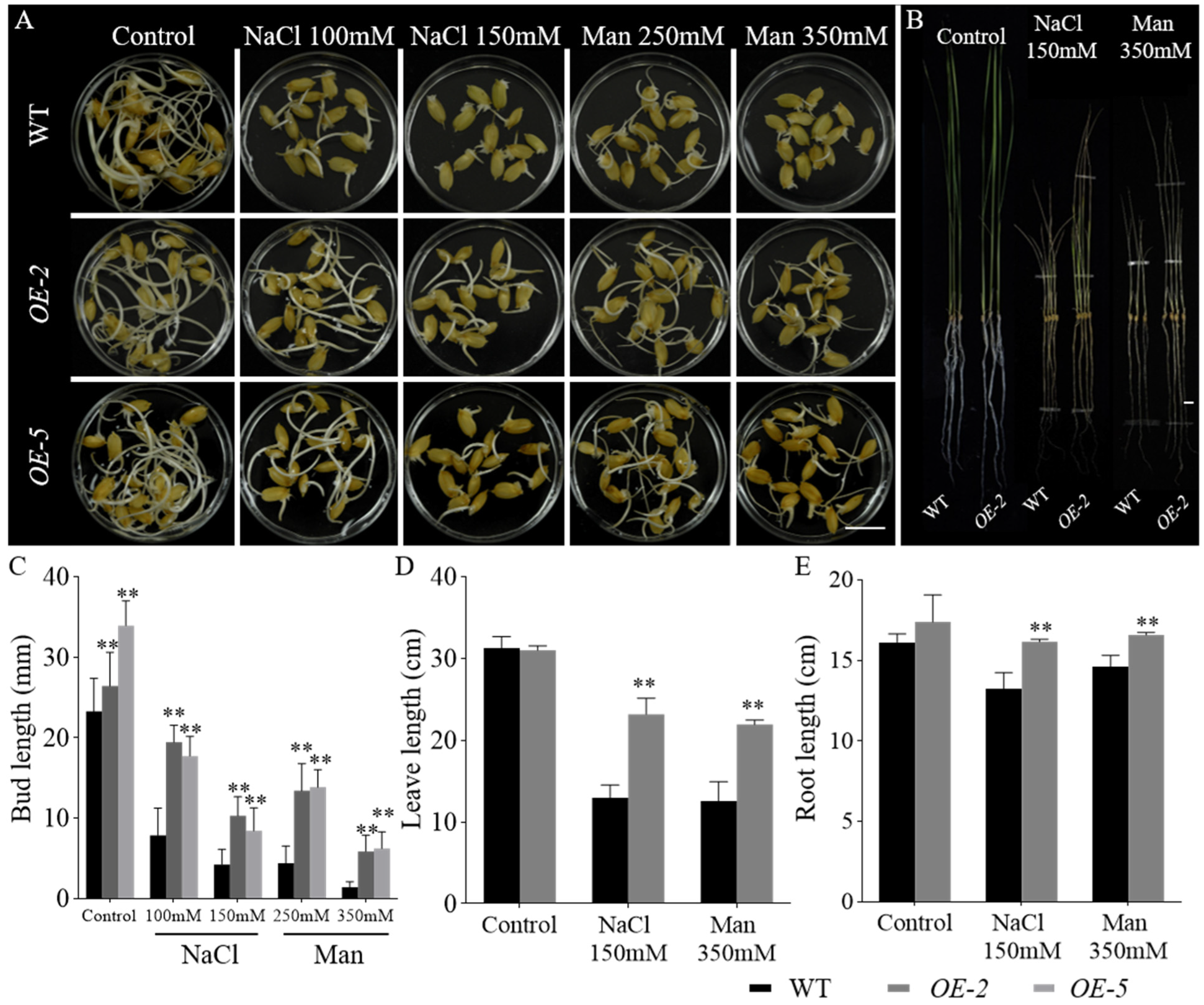
2.5. Overexpression of GmbZIP60 Enhances the Resistance of Transgenic Rice Plants to Salt and Drought Stresses
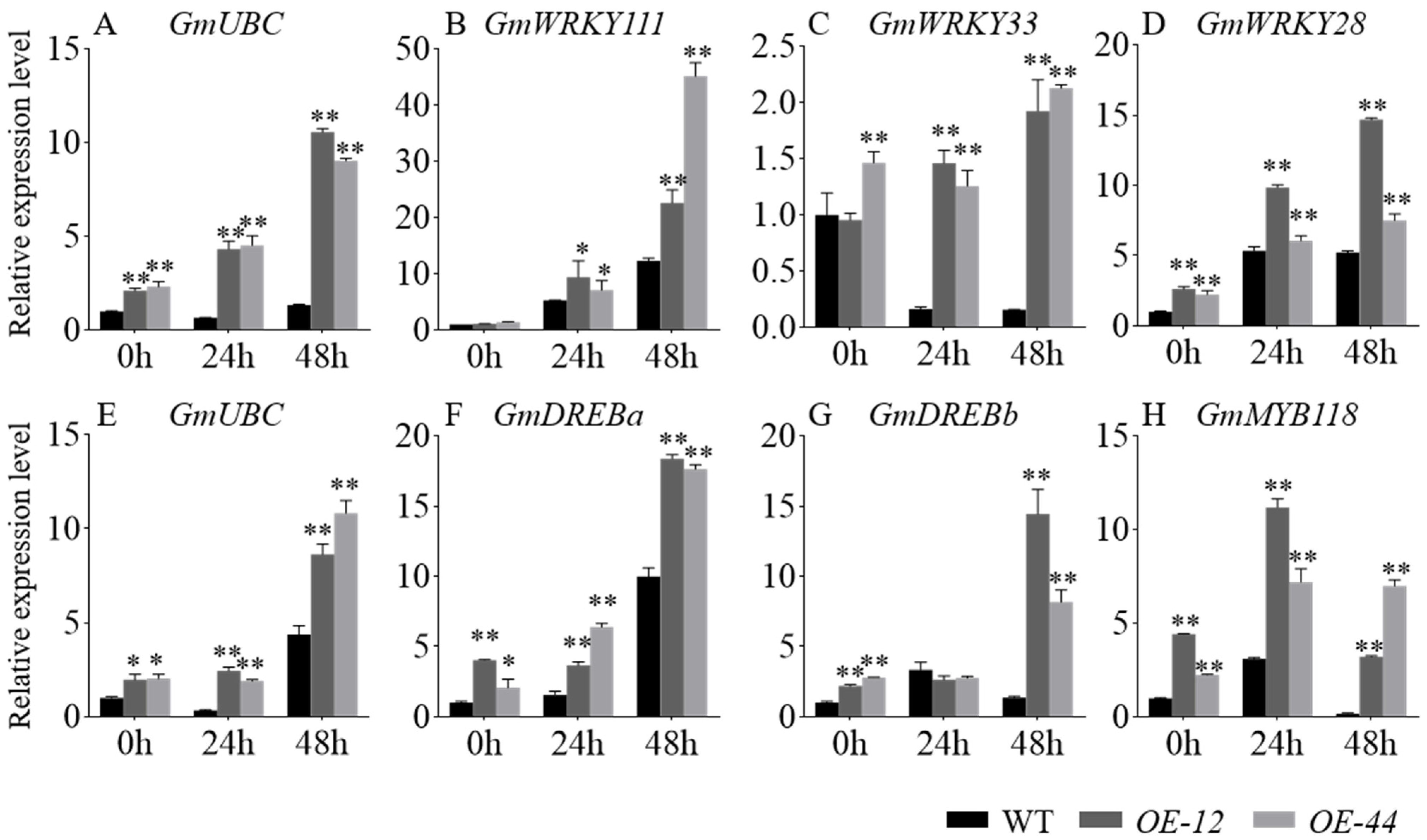
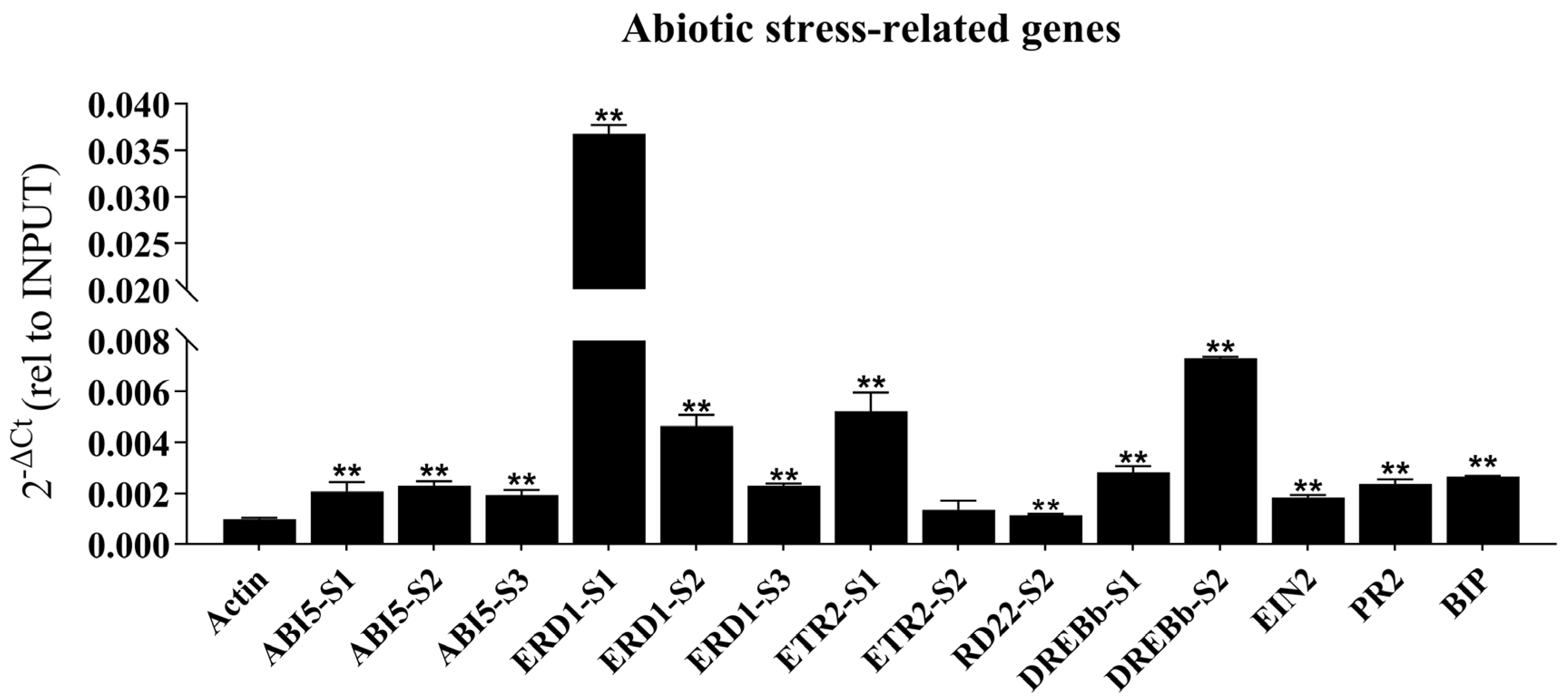
2.6. Expression Analysis of Stress-Related Genes in OE-GmbZIP60 Transgenic Rice and Soybean Under Salt and Drought Stress
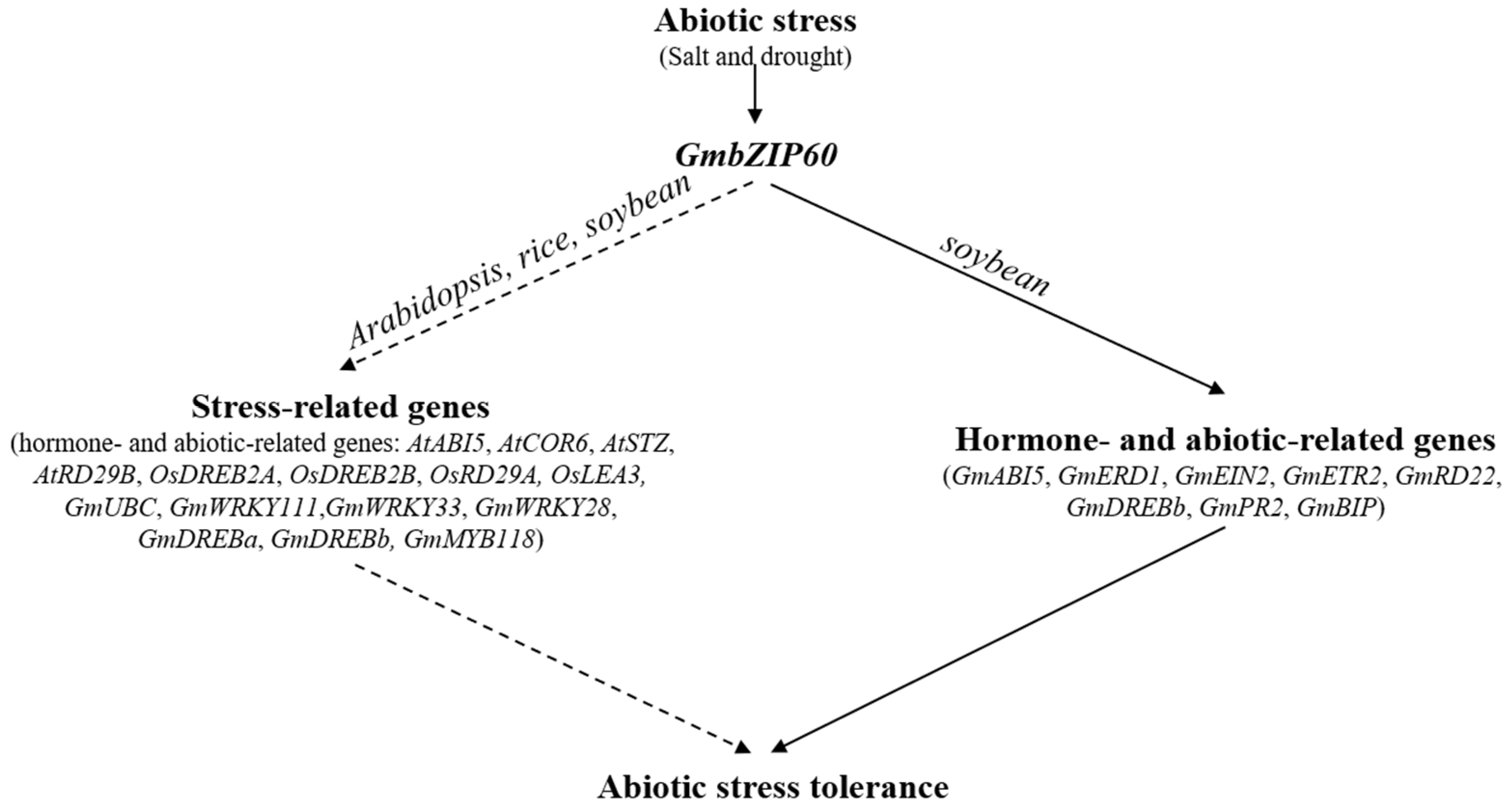
2.7. Identification of Target Genes for GmbZIP60
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. GmbZIP60 Gene Isolation, Vector Construction
4.3. Arabidopsis, Rice, and Soybean Transformation
4.4. Stress Tolerance Assays and Measurements of Physiological Indices
4.5. Quantitative Real-Time PCR
4.6. Chromatin Immunoprecipitation (ChIP) Analysis
4.7. Detection of Photosynthetic Activity Fluorescence Parameters (FPAD)
4.8. Detection of Leaf Water Content
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, S.; Zhao, X.; Hu, Y.; Liu, S.; Nan, H.; Li, X.; Fang, C.; Cao, D.; Shi, X.; Kong, L.; et al. Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 2017, 49, 1546–1718. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Fang, C.; Cheng, Q.; Su, T.; Kou, K.; Kong, L.; Zhang, C.; Li, H.; Hou, Z.; Zhang, Y.; et al. Genetic basis and adaptation trajectory of soybean from its temperate origin to tropics. Nat. Commun. 2021, 12, 5445. [Google Scholar] [PubMed]
- Min, C.W.; Gupta, R.; Kim, S.W.; Lee, S.E.; Kim, Y.C.; Bae, D.W.; Han, W.Y.; Lee, B.W.; Ko, J.M.; Agrawal, G.K.; et al. Comparative Biochemical and Proteomic Analyses of Soybean Seed Cultivars Differing in Protein and Oil Content. J. Agric. Food Chem. 2015, 63, 7134–7142. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Sun, L.; Jiang, S.; Ren, H.; Sun, R.; Wei, Z.; Hong, H.; Luan, X.; Wang, J.; Wang, X.; et al. Soybean genetic resources contributing to sustainable protein production. Theor. Appl. Genet. 2022, 135, 4095–4121. [Google Scholar]
- Kim, W.J.; Kang, B.H.; Kang, S.; Shin, S.; Chowdhury, S.; Jeong, S.C.; Choi, M.S.; Park, S.K.; Moon, J.K.; Ryu, J.; et al. A Genome-Wide Association Study of Protein, Oil, and Amino Acid Content in Wild Soybean (Glycine soja). Plants 2023, 12, 1665. [Google Scholar] [CrossRef]
- Gaffield, K.N.; Goodband, R.D.; DeRouchey, J.M.; Tokach, M.D.; Woodworth, J.A.-O.; Denny, G.; Gebhardt, J.T. A review of soybean processing byproducts and their use in swine and poultry diets. Transl. Anim. Sci. 2024, 8, txae063. [Google Scholar]
- Nicolás Marro, N.C. Gabriel Grilli,Carolina Alvarez,Diana Labuckas,Damián Maestri & Carlos Urcelay, Soybean yield, protein content and oil quality in response to interaction of arbuscular mycorrhizal fungi and native microbial populations from mono- and rotation-cropped soils. Appl. Soil Ecol. 2020, 152, 103575. [Google Scholar]
- Ray, D.K.; Gerber, J.S.; MacDonald, G.K.; West, P.C. Climate variation explains a third of global crop yield variability. Nat. Commun. 2015, 6, 5989. [Google Scholar] [CrossRef]
- Stolz, M.L.; McCormick, C. The bZIP Proteins of Oncogenic Viruses. Viruses 2020, 12, 757. [Google Scholar] [CrossRef]
- Gachon, F.; Olela, F.F.; Schaad, O.; Descombes, P.; Schibler, U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006, 4, 25–36. [Google Scholar]
- Li, M.; Yao, T.; Lin, W.; Hinckley, W.E.; Galli, M.; Muchero, W.; Gallavotti, A.; Chen, J.G.; Huang, S.C. Double DAP-seq uncovered synergistic DNA binding of interacting bZIP transcription factors. Nat. Commun. 2023, 14, 2600. [Google Scholar] [PubMed]
- Li, H.; Chen, J.; Zhao, Q.; Han, Y.; Li, L.; Sun, C.; Wang, K.; Wang, Y.; Zhao, M.; Chen, P.; et al. Basic leucine zipper (bZIP) transcription factor genes and their responses to drought stress in ginseng, Panax ginseng C.A. Meyer. BMC Genom. 2021, 22, 316. [Google Scholar]
- Han, H.; Wang, C.; Yang, X.; Wang, L.; Ye, J.; Xu, F.; Liao, Y.; Zhang, W. Role of bZIP transcription factors in the regulation of plant secondary metabolism. Planta 2023, 258, 13. [Google Scholar] [PubMed]
- Zhang, M.; Liu, Y.; Cai, H.; Guo, M.; Chai, M.; She, Z.; Ye, L.; Cheng, Y.; Wang, B.; Qin, Y. The bZIP Transcription Factor GmbZIP15 Negatively Regulates Salt- and Drought-Stress Responses in Soybean. Int. J. Mol. Sci. 2020, 21, 7778. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, T.F.; Ma, J.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Wei, W.L.; Xu, Z.S. The Soybean bZIP Transcription Factor Gene GmbZIP2 Confers Drought and Salt Resistances in Transgenic Plants. Int. J. Mol. Sci. 2020, 21, 670. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Q.Y.; Xu, P.; Wang, X.H.; Dai, S.J.; Liu, Z.N.; Xu, M.; Cao, X.; Cui, X.Y. GmTRAB1, a Basic Leucine Zipper Transcription Factor, Positively Regulates Drought Tolerance in Soybean (Glycine max. L). Plants 2024, 13, 3104. [Google Scholar] [CrossRef]
- Kataoka, R.; Takahashi, M.; Suzuki, N. Coordination between bZIP28 and HSFA2 in the regulation of heat response signals in Arabidopsis. Plant Signal Behav. 2017, 12, e1376159. [Google Scholar] [PubMed]
- Choi, J.; Lim, C.W.; Lee, S.C. Role of pepper bZIP transcription factor CaADBZ1 in abscisic acid signalling and drought stress response. Physiol. Plant 2025, 177, e70159. [Google Scholar]
- Zhang, M.; Liu, Y.; Li, Z.; She, Z.; Chai, M.; Aslam, M.; He, Q.; Huang, Y.; Chen, F.; Chen, H.; et al. The bZIP transcription factor GmbZIP15 facilitates resistance against Sclerotinia sclerotiorum and Phytophthora sojae infection in soybean. iScience 2021, 24, 102642. [Google Scholar]
- Zhang, L.; Fu, X.; Ye, J.; Chen, S.; Jin, J.; Liu, W.; Zhang, Z.; Zhou, L.; Chen, S.; Fang, W.; et al. CmbZIP19 inhibits lateral bud elongation via the brassinolide pathway in chrysanthemum. Plant J. 2025, 121, e70080. [Google Scholar]
- Gangappa, S.N.; Botto, J.F. The Multifaceted Roles of HY5 in Plant Growth and Development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Y.; Shi, H.; Guo, M.; Chai, M.; He, Q.; Yan, M.; Cao, D.; Zhao, L.; Cai, H.; et al. Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family. BMC Genom. 2018, 19, 159. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wang, S.; Dao, Y.; Wang, J.; Wang, K. Arabidopsis thaliana trehalose-6-phosphate phosphatase gene TPPI enhances drought tolerance by regulating stomatal apertures. J. Exp. Bot. 2020, 71, 4285–4297. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.M.; Pereira, J.S.; Maroco, J.; Rodrigues, M.L.; Ricardo, C.P.; Osório, M.L.; Carvalho, I.; Faria, T.; Pinheiro, C. How plants cope with water stress in the field. Photosynthesis and growth. Ann. Bot. 2002, 89, 907–916. [Google Scholar] [CrossRef]
- Pruthi, R.; Chaudhary, C.; Chapagain, S.; Abozaid, M.M.E.; Rana, P.; Kondi, R.K.R.; Fritsche-Neto, R.; Subudhi, P.K. Deciphering the genetic basis of salinity tolerance in a diverse panel of cultivated and wild soybean accessions by genome-wide association mapping. Theor. Appl. Genet. 2024, 137, 238. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, R.; Wang, H.; Ren, S.; Shi, L.; Huang, S.; Wei, Z.; Guo, B.; Jin, J.; Zhong, Y.; et al. OsWRKY28 positively regulates salinity tolerance by directly activating OsDREB1B expression in rice. Plant Cell Rep. 2023, 42, 223–234. [Google Scholar] [CrossRef]
- Li, X.P.; Tian, A.G.; Luo, G.Z.; Gong, Z.Z.; Zhang, J.S.; Chen, S.Y. Soybean DRE-binding transcription factors that are responsive to abiotic stresses. Theor. Appl. Genet. 2005, 110, 1355–1362. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Chen, M.; Chen, J.; Zhou, Y.B.; Xu, Z.S.; et al. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef]
- Ma, Z.; Jin, Y.M.; Wu, T.; Hu, L.; Zhang, Y.; Jiang, W.; Du, X. OsDREB2B, an AP2/ERF transcription factor, negatively regulates plant height by conferring GA metabolism in rice. Front. Plant Sci. 2022, 13, 1007811. [Google Scholar] [CrossRef]
- Msanne, J.; Lin, J.; Stone, J.M.; Awada, T. Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 2011, 234, 97–107. [Google Scholar] [CrossRef]
- Duan, J.; Cai, W. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS ONE 2012, 7, e45117. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, Y.; Wei, J.J.; Zhang, W.K.; Chen, S.Y.; Zhang, J.S. Regulation of seed traits in soybean. Abiotech 2023, 4, 372–385. [Google Scholar] [PubMed]
- Majidian, P.; Ghorbani, H.R.; Farajpour, M. Achieving agricultural sustainability through soybean production in Iran: Potential and challenges. Heliyon 2024, 10, e26389. [Google Scholar]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 45, 523–539. [Google Scholar] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar]
- Tsugama, D.; Liu, S.; Takano, T. The bZIP Protein VIP1 Is Involved in Touch Responses in Arabidopsis Roots. Plant Physiol. 2016, 171, 1355–1365. [Google Scholar]
- Lozano-Sotomayor, P.; Chávez Montes, R.A.; Silvestre-Vañó, M.; Herrera-Ubaldo, H.; Greco, R.; Pablo-Villa, J.; Galliani, B.M.; Diaz-Ramirez, D.; Weemen, M.; Boutilier, K.; et al. Altered expression of the bZIP transcription factor DRINK ME affects growth and reproductive development in Arabidopsis thaliana. Plant J. 2016, 88, 437–451. [Google Scholar]
- Tsugama, D.; Liu, S.; Takano, T. A bZIP protein, VIP1, is a regulator of osmosensory signaling in Arabidopsis. Plant Physiol. 2012, 159, 144–155. [Google Scholar] [PubMed]
- Tsugama, D.; Liu, S.; Takano, T. A bZIP protein, VIP1, interacts with Arabidopsis heterotrimeric G protein beta subunit, AGB1. Plant Physiol. Biochem. 2013, 71, 240–246. [Google Scholar]
- Bu, Y.; Yu, Y.; Song, T.; Zhang, D.; Shi, C.; Zhang, S.; Zhang, W.; Chen, D.; Xiang, J.; Zhang, X. The transcription factor TabZIP156 acts as a positive regulator in response to drought tolerance in Arabidopsis and wheat (Triticum aestivum L.). Plant Physiol. Biochem. 2024, 216, 109086. [Google Scholar]
- Chai, M.; Fan, R.; Huang, Y.; Jiang, X.; Wai, M.H.; Yang, Q.; Su, H.; Liu, K.; Ma, S.; Chen, Z.; et al. GmbZIP152, a Soybean bZIP Transcription Factor, Confers Multiple Biotic and Abiotic Stress Responses in Plant. Int. J. Mol. Sci. 2022, 23, 10935. [Google Scholar] [CrossRef]
- Wu, X.; Jia, Y.; Ma, Q.; Wang, T.; Xu, J.; Chen, H.; Wang, M.; Song, H.; Cao, S. The transcription factor bZIP44 cooperates with MYB10 and MYB72 to regulate the response of Arabidopsis thaliana to iron deficiency stress. New Phytol. 2024, 242, 2586–2603. [Google Scholar] [CrossRef]
- Dietrich, K.; Weltmeier, F.; Ehlert, A.; Weiste, C.; Stahl, M.; Harter, K.; Droge-Laser, W. Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 2011, 23, 381–395. [Google Scholar] [CrossRef]
- Hartmann, L.; Pedrotti, L.; Weiste, C.; Fekete, A.; Schierstaedt, J.; Gottler, J.; Kempa, S.; Krischke, M.; Dietrich, K.; Mueller, M.J.; et al. Crosstalk between Two bZIP Signaling Pathways Orchestrates Salt-Induced Metabolic Reprogramming in Arabidopsis Roots. Plant Cell 2015, 27, 2244–2260. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H.; Sato, K.; Berberich, T.; Miyazaki, A.; Ozaki, R.; Imai, R.; Kusano, T. LIP19, a basic region leucine zipper protein, is a Fos-like molecular switch in the cold signaling of rice plants. Plant Cell Physiol. 2005, 46, 1623–1634. [Google Scholar] [CrossRef] [PubMed]
- Weltmeier, F.; Ehlert, A.; Mayer, C.S.; Dietrich, K.; Wang, X.; Schütze, K.; Alonso, R.; Harter, K.; Vicente-Carbajosa, J.; Dröge-Laser, W. Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J. 2006, 25, 3133–3143. [Google Scholar] [CrossRef] [PubMed]
- Hepworth, S.R.; Zhang, Y.; McKim, S.; Li, X.; Haughn, G.W. BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 2005, 17, 1434–1448. [Google Scholar] [CrossRef]
- Ma, H.; Liu, C.; Li, Z.; Ran, Q.; Xie, G.; Wang, B.; Fang, S.; Chu, J.; Zhang, J. ZmbZIP4 Contributes to Stress Resistance in Maize by Regulating ABA Synthesis and Root Development. Plant Physiol. 2018, 178, 753–770. [Google Scholar] [CrossRef]
- Zong, W.; Yang, J.; Fu, J.; Xiong, L. Synergistic regulation of drought-responsive genes by transcription factor OsbZIP23 and histone modification in rice. J. Integr. Plant Biol. 2020, 62, 723–729. [Google Scholar] [CrossRef]
- Miyamoto, K.; Nishizawa, Y.; Minami, E.; Nojiri, H.; Yamane, H.; Okada, K. Overexpression of the bZIP transcription factor OsbZIP79 suppresses the production of diterpenoid phytoalexin in rice cells. J. Plant Physiol. 2015, 173, 19–27. [Google Scholar] [CrossRef]
- Brookbank, B.P.; Patel, J.; Gazzarrini, S.; Nambara, E. Role of Basal ABA in Plant Growth and Development. Genes 2021, 12, 1936. [Google Scholar] [CrossRef]
- Jiang, M.; Song, Y.; Yang, R.; Zheng, C.; Zheng, Y.; Zhang, H.; Li, S.; Tan, Y.; Huang, J.; Shu, Q.; et al. Melatonin activates the OsbZIP79-OsABI5 module that orchestrates nitrogen and ROS homeostasis to alleviate nitrogen-limitation stress in rice. Plant Commun. 2023, 4, 100674. [Google Scholar]
- Zhang, L.; Zhang, L.; Xia, C.; Zhao, G.; Liu, J.; Jia, J.; Kong, X. A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiol. Plant 2015, 153, 538–554. [Google Scholar] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [PubMed]
- Wu, H.; Ye, H.; Yao, R.; Zhang, T.; Xiong, L. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 2015, 232, 1–12. [Google Scholar]
- Singh, A.P.; Mani, B.; Giri, J. OsJAZ9 is involved in water-deficit stress tolerance by regulating leaf width and stomatal density in rice. Plant Physiol. Biochem. 2021, 162, 161–170. [Google Scholar] [CrossRef]
- Cao, W.H.; Liu, J.; Zhou, Q.Y.; Cao, Y.R.; Zheng, S.F.; Du, B.X.; Zhang, J.S.; Chen, S.Y. Expression of tobacco ethylene receptor NTHK1 alters plant responses to salt stress. Plant Cell Environ. 2006, 29, 1210–1219. [Google Scholar]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.J. Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in Plant Defense: Do We Understand What They Are Whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef] [PubMed]
- Zeng, P.; Vadnais, D.A.; Zhang, Z.; Polacco, J.C. Refined glufosinate selection in Agrobacterium-mediated transformation of soybean [Glycine max (L.) Merrill]. Plant Cell Rep. 2004, 22, 478–482. [Google Scholar]
- Paz, M.M.; Martinez, J.C.; Kalvig, A.B.; Fonger, T.M.; Wang, K. Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep. 2006, 25, 206–213. [Google Scholar]
- Do, V.G.; Kim, S.; Win, N.M.; Kwon, S.I.; Kweon, H.; Yang, S.; Park, J.; Do, G.; Lee, Y. Efficient Regeneration of Transgenic Rice from Embryogenic Callus via Agrobacterium-Mediated Transformation: A Case Study Using GFP and Apple MdFT1 Genes. Plants 2024, 13, 2803. [Google Scholar] [CrossRef]
- Dai, X.; Bai, Y.; Zhao, L.; Dou, X.; Liu, Y.; Wang, L.; Li, Y.; Li, W.; Hui, Y.; Huang, X.; et al. H2A.Z Represses Gene Expression by Modulating Promoter Nucleosome Structure and Enhancer Histone Modifications in Arabidopsis. Mol. Plant 2018, 11, 635. [Google Scholar] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chai, M.; Yang, F.; Cai, S.; Liu, T.; Xu, X.; Huang, Y.; Xi, X.; Yang, J.; Cao, Z.; Sun, L.; et al. Overexpression of the Transcription Factor GmbZIP60 Increases Salt and Drought Tolerance in Soybean (Glycine max). Int. J. Mol. Sci. 2025, 26, 3455. https://doi.org/10.3390/ijms26073455
Chai M, Yang F, Cai S, Liu T, Xu X, Huang Y, Xi X, Yang J, Cao Z, Sun L, et al. Overexpression of the Transcription Factor GmbZIP60 Increases Salt and Drought Tolerance in Soybean (Glycine max). International Journal of Molecular Sciences. 2025; 26(7):3455. https://doi.org/10.3390/ijms26073455
Chicago/Turabian StyleChai, Mengnan, Fan Yang, Shuping Cai, Tingyu Liu, Xiaoyuan Xu, Youmei Huang, Xinpeng Xi, Jiahong Yang, Zhuangyuan Cao, Ling Sun, and et al. 2025. "Overexpression of the Transcription Factor GmbZIP60 Increases Salt and Drought Tolerance in Soybean (Glycine max)" International Journal of Molecular Sciences 26, no. 7: 3455. https://doi.org/10.3390/ijms26073455
APA StyleChai, M., Yang, F., Cai, S., Liu, T., Xu, X., Huang, Y., Xi, X., Yang, J., Cao, Z., Sun, L., Dou, D., Fang, X., Yan, M., & Cai, H. (2025). Overexpression of the Transcription Factor GmbZIP60 Increases Salt and Drought Tolerance in Soybean (Glycine max). International Journal of Molecular Sciences, 26(7), 3455. https://doi.org/10.3390/ijms26073455





