Genome-Wide Identification and Expression Analysis of GASA Genes in Hevea brasiliensis Reveals Their Involvement in Response to Cold Stress
Abstract
1. Introduction
2. Results
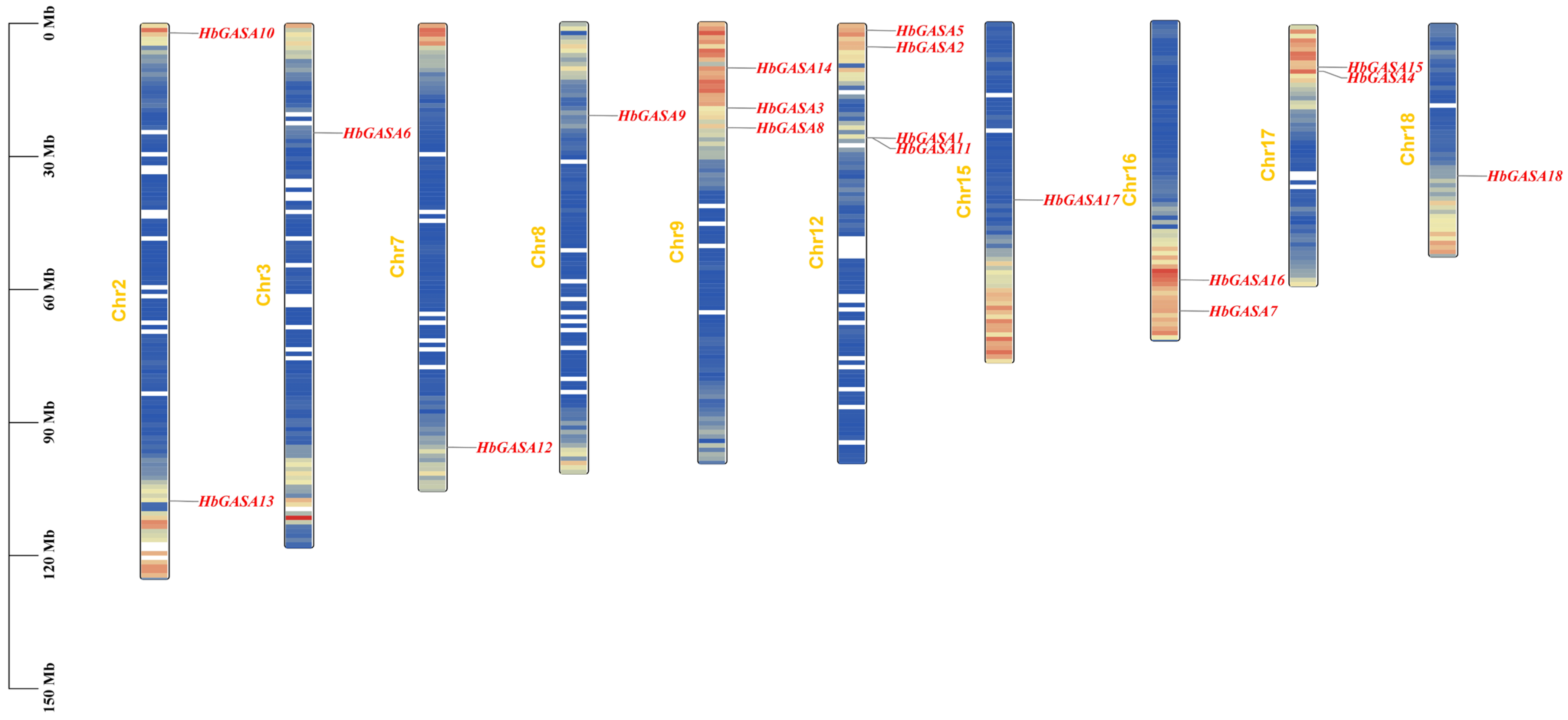
2.1. Identification and Characteristics of HbGASA Genes
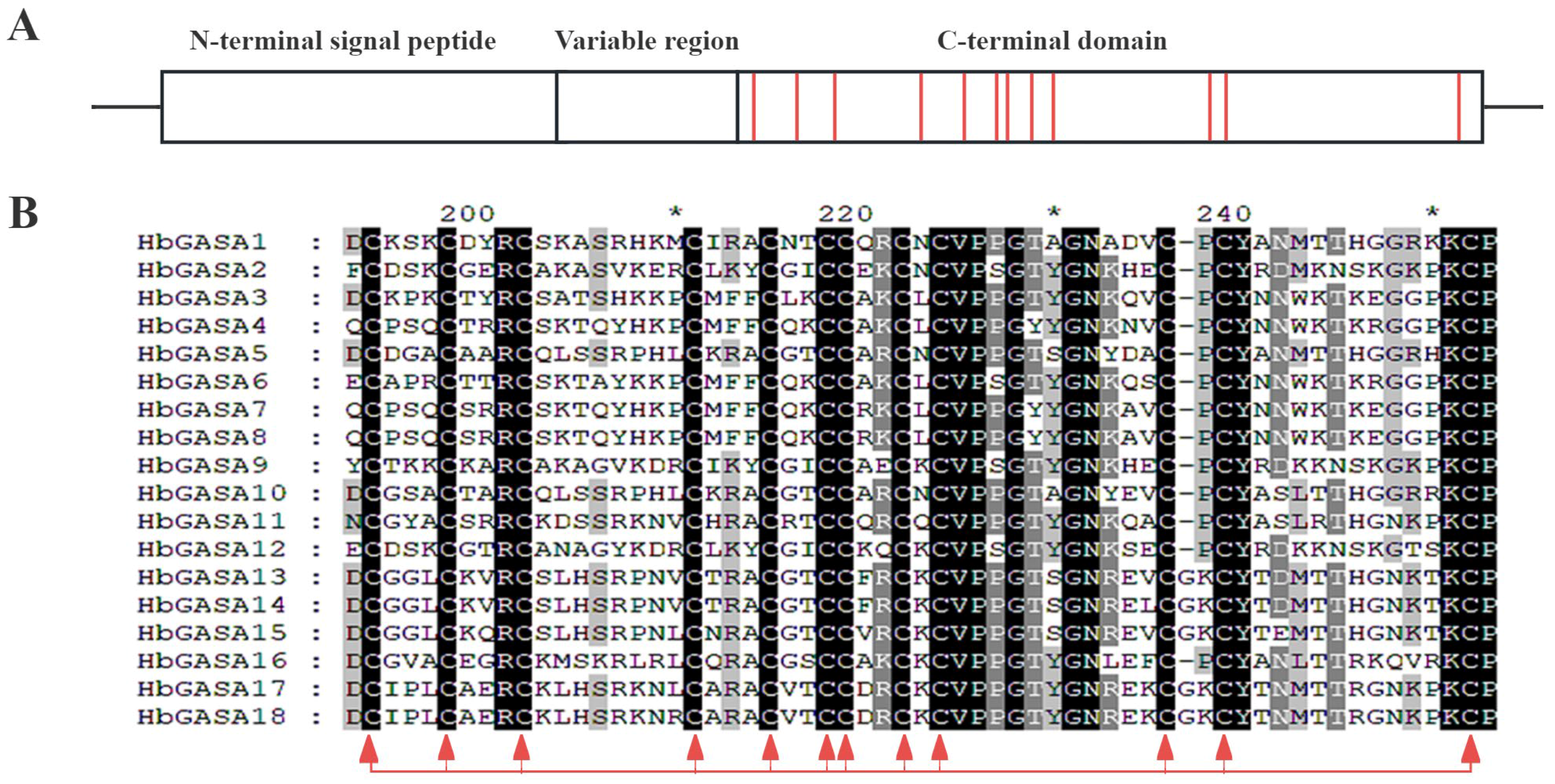
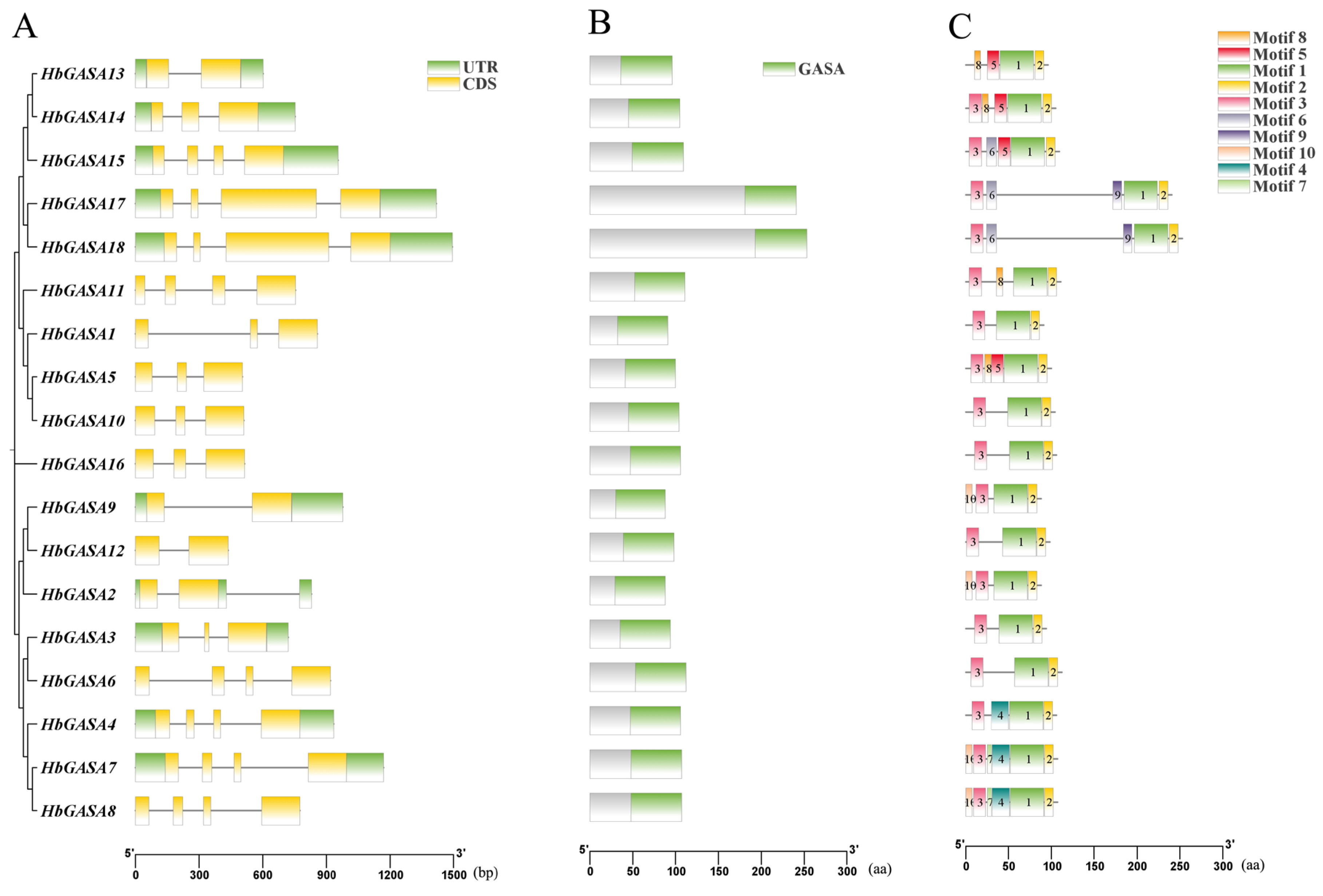
2.2. Protein and Gene Structure of HbGASA Genes
2.3. Promoter Region Analysis of HbGASA Genes
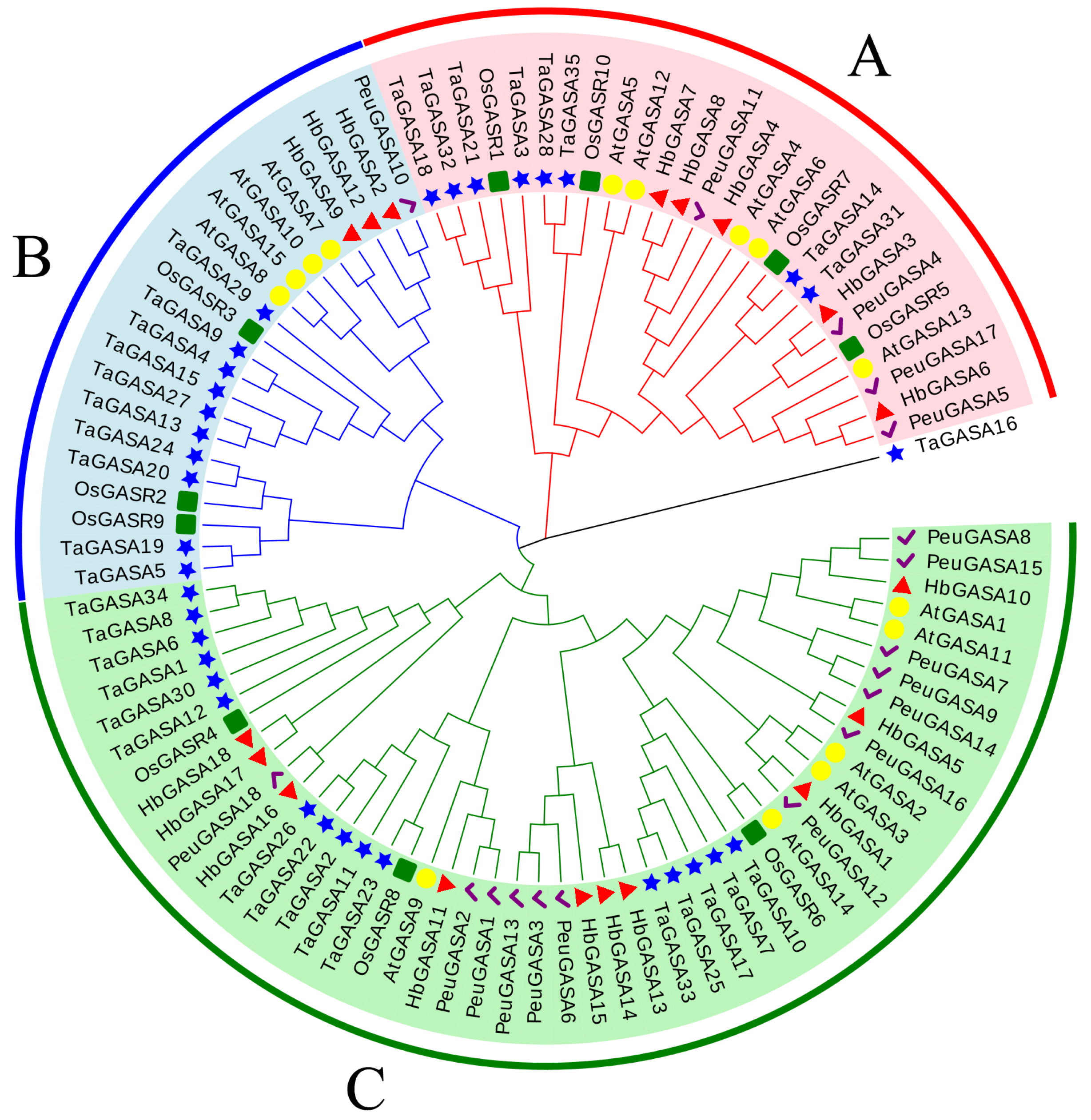
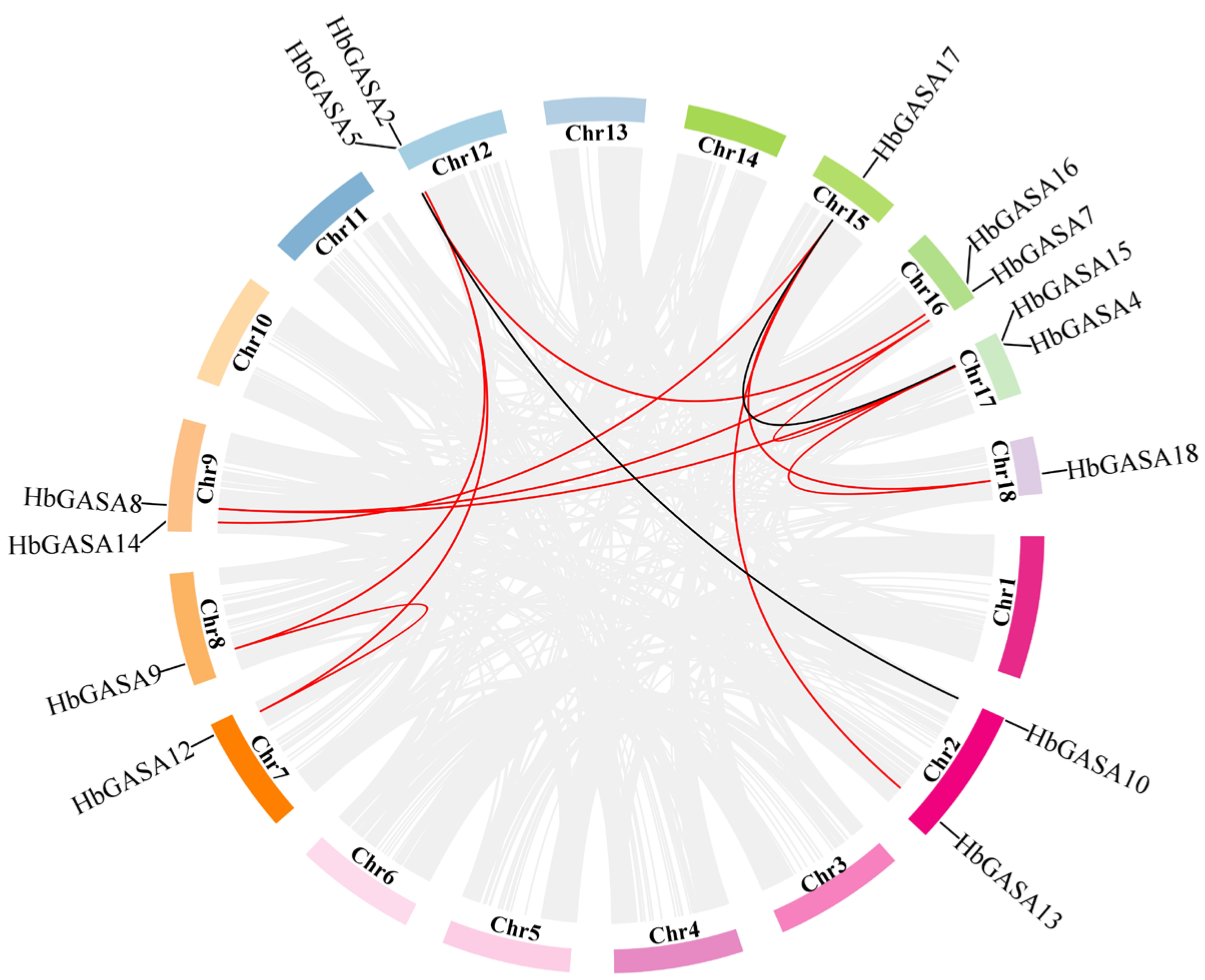
2.4. HbGASAs Duplication Analysis
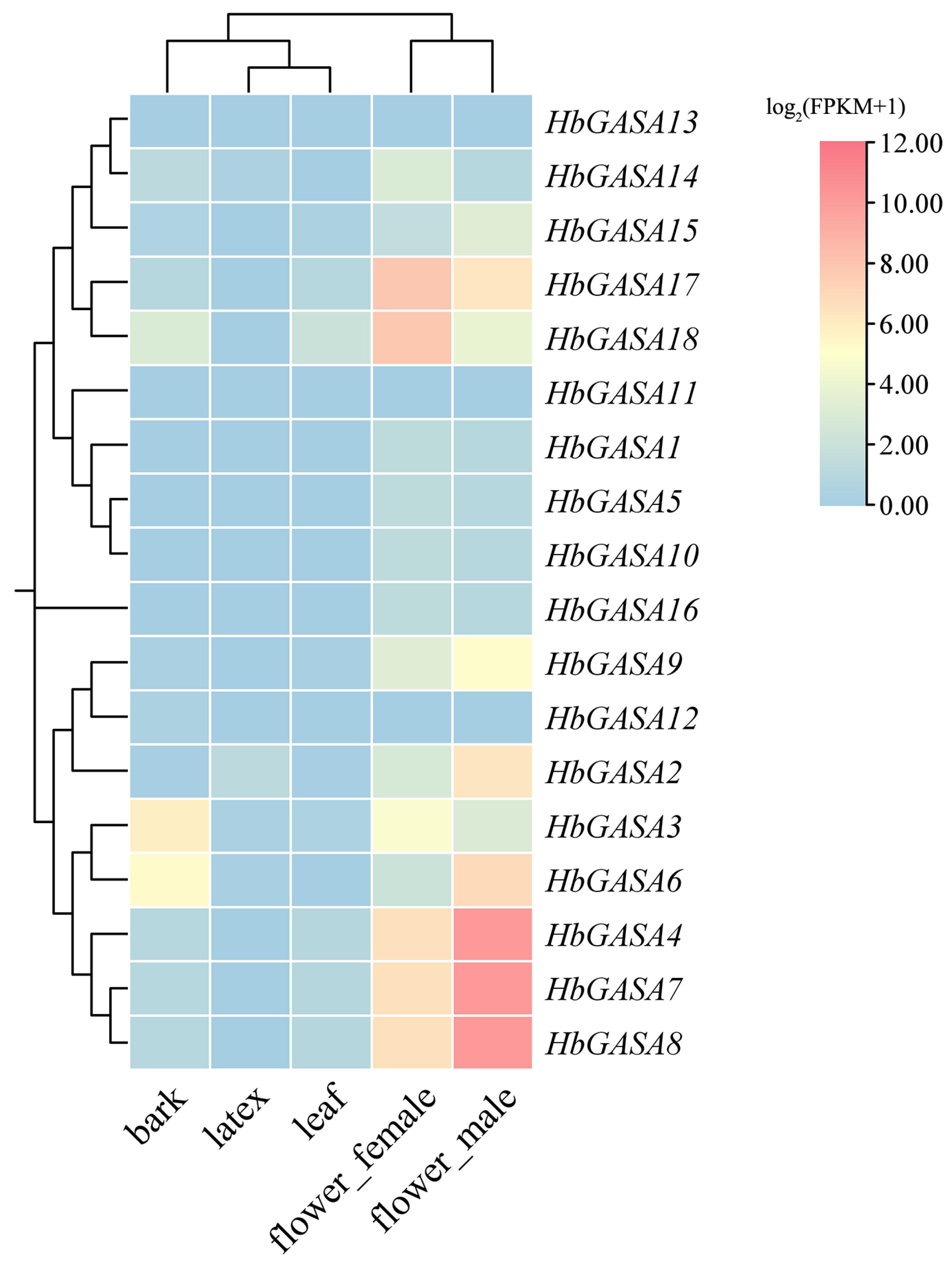
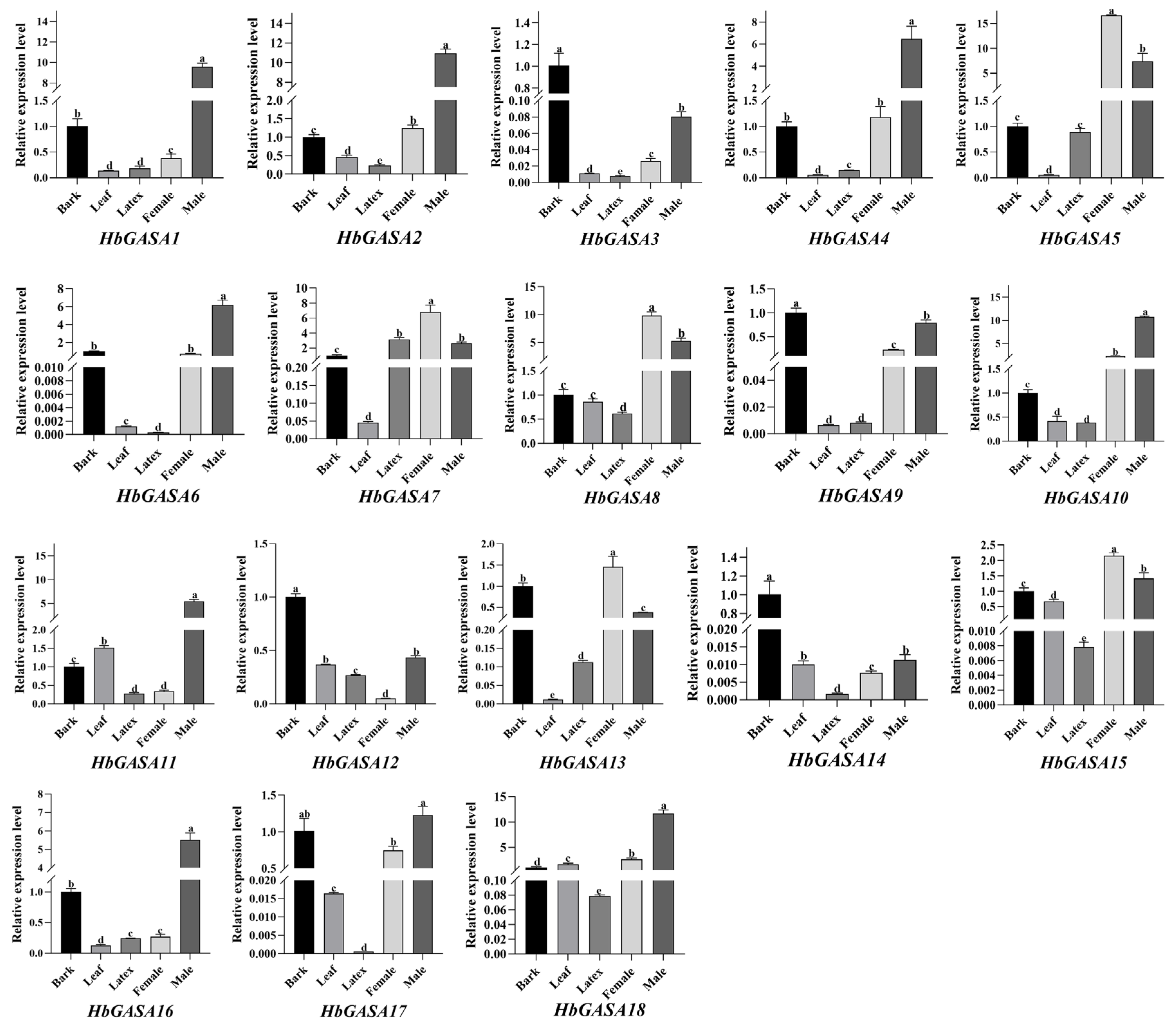
2.5. The Tissue-Specific Expression Patterns of HbGASA Genes
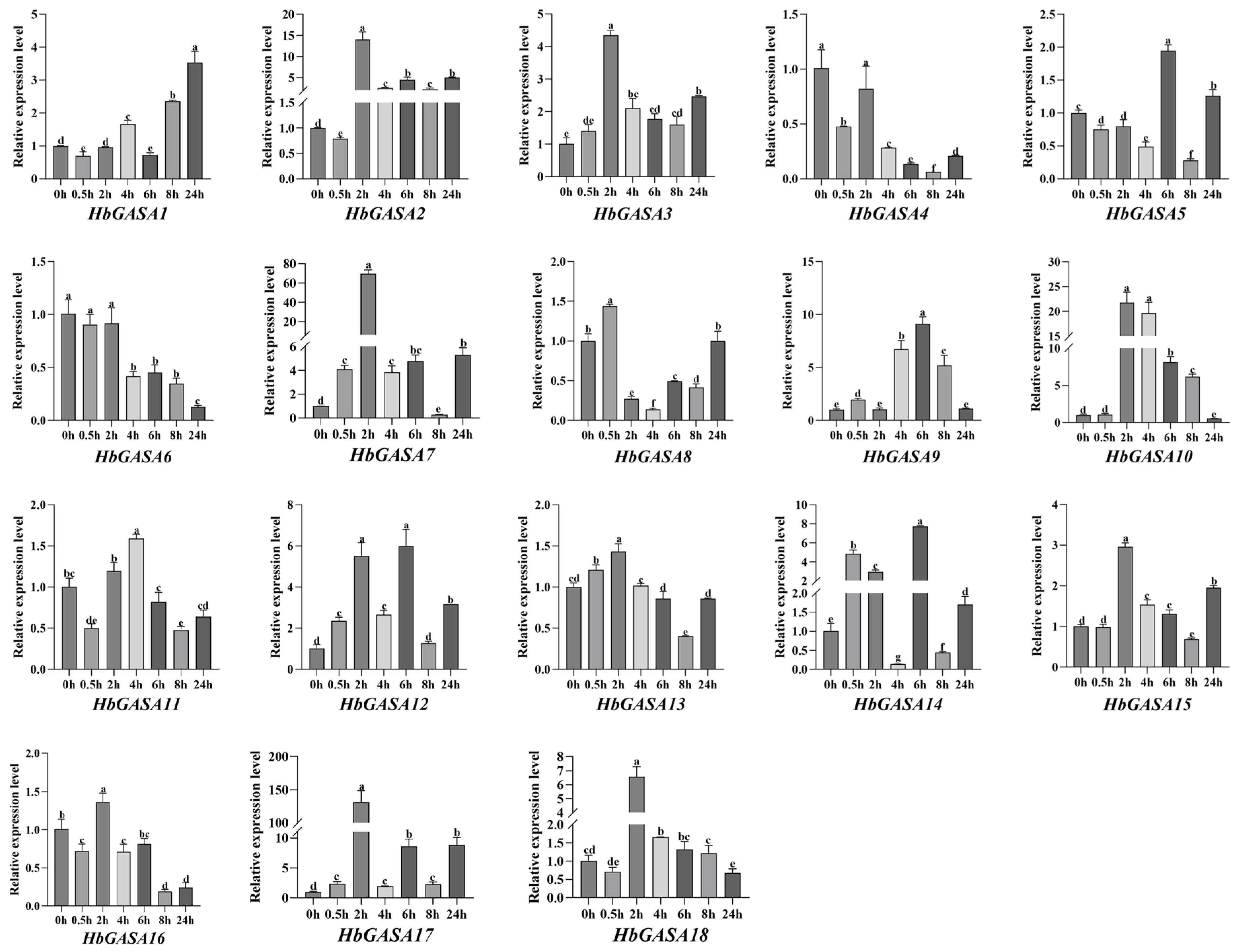
2.6. Expression Profiles of HbGASAs Under Cold Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Stress Treatment
4.2. Identification and Chromosomal Distribution of HbGASA Genes
4.3. Physicochemical Properties of HbGASA Proteins
4.4. Subcellular Localization Prediction and Signal Peptide Analysis of HbGASA Proteins
4.5. Analysis of Conserved Motifs and Gene Structures
4.6. Analysis of Promoters and Phylogeny of HbGASA Genes
4.7. Collinearity Analysis and Visualization of HbGASA Genes
4.8. Tissue-Specific Expression Profiles of HbGASA Genes
4.9. RNA Isolation and qRT-PCR Reaction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| MeJA | Methyl jasmonate |
| GA | Gibberellic acid |
| SA | Salicylic acid |
| GASA | Gibberellic Acid Stimulated in Arabidopsis |
| qRT-PCR | quantitative real time PCR |
References
- Bouteraa, M.T.; Ben Romdhane, W.; Baazaoui, N.; Alfaifi, M.Y.; Chouaibi, Y.; Ben Akacha, B.; Ben Hsouna, A.; Kacániová, M.; Cavar Zeljkovic, S.; Garzoli, S.; et al. GASA proteins: Review of their functions in plant environmental stress tolerance. Plants 2023, 12, 2045. [Google Scholar] [CrossRef]
- De la Fuente, J.I.; Amaya, I.; Castillejo, C.; Sánchez-Sevilla, J.F.; Quesada, M.A.; Botella, M.A.; Valpuestaet, V. The strawberry gene FaGAST affects plant growth through inhibition of cell elongation. J. Exp. Bot. 2006, 10, 2401–2411. [Google Scholar]
- Roxrud, I.; Lid, S.E.; Fletcher, J.C.; Schmidt, E.D.; Opsahl-Sorteberg, H.-G. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007, 48, 471–483. [Google Scholar]
- Fan, S.; Zhang, D.; Zhang, L.; Gao, C.; Xin, M.; Tahir, M.M.; Li, Y.; Ma, J.; Han, M. Comprehensive analysis of GASA family members in the Malus domestica genome: Identification, characterization, and their expressions in response to apple flower induction. BMC Genom. 2017, 18, 827. [Google Scholar]
- Ben-Nissan, G.; Lee, J.Y.; Borohov, A.; Weiss, D. GIP, a Petunia hybrida GA-induced cysteine-rich protein: A possible role in shoot elongation and flowering transition. Plant J. 2004, 37, 229–238. [Google Scholar]
- Wang, L.; Wang, Z.; Xu, Y.; Joo, S.-H.; Kim, S.-K.; Xue, Z.; Xu, Z.; Wang, Z.; Chong, K. OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J. 2009, 57, 498–510. [Google Scholar]
- Zhong, C.; Xu, H.; Ye, S.; Wang, S.; Li, L.; Zhang, S.; Wang, X. Gibberellic acid-stimulated Arabidopsis6 serves as an integrator of gibberellin, abscisic acid, and glucose signaling during seed germination in Arabidopsis. Plant Physiol. 2015, 169, 2288–2303. [Google Scholar]
- Zhang, S.; Yang, C.; Peng, J.; Sun, S.; Wang, X. GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana. Plant Mol. Biol. 2009, 69, 745–759. [Google Scholar]
- Zimmermann, R.; Sakai, H.; Hochholdinger, F. The gibberellic acid stimulated-like gene family in maize and its role in lateral root development. Plant Physiol. 2010, 152, 356–365. [Google Scholar]
- Bouteraa, M.T.; Romdhane, W.B.; Ben Hsouna, A.; Amor, F.; Ebel, C.; Ben Saad, R. Genome-wide characterization and expression profiling of GASA gene family in Triticum turgidum ssp. durum (desf.) husn. (Durum wheat) unveils its involvement in environmental stress responses. Phytochemistry 2023, 206, 113544. [Google Scholar]
- Cheng, X.; Wang, S.; Xu, D.; Liu, X.; Li, X.; Xiao, W.; Cao, J.; Jiang, H.; Min, X.; Wang, J.; et al. Identification and analysis of the GASR gene family in common wheat (Triticum aestivum L.) and characterization of TaGASR34, a gene associated with seed dormancy and germination. Front. Genet. 2019, 10, 980. [Google Scholar] [CrossRef]
- Herbel, V.; Sieber-Frank, J.; Wink, M. The antimicrobial peptide snakin-2 is up-regulated in the defense response of tomatoes (Solanum lycopersicum) as part of the jasmonate-dependent signaling pathway. J. Plant Physiol. 2017, 208, 1–6. [Google Scholar] [CrossRef]
- Ko, C.-B.; Woo, Y.-M.; Lee, D.J.; Lee, M.-C.; Kim, C.S. Enhanced tolerance to heat stress in transgenic plants expressing the GASA4 gene. Plant Physiol. Biochem. 2006, 44, 722–728. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, X. Overexpression of GASA5 increases the sensitivity of Arabidopsis to heat stress. J. Plant Physiol. 2011, 168, 2093–2101. [Google Scholar] [CrossRef]
- Wang, H.; Wei, T.; Wang, X.; Zhang, L.; Yang, M.; Chen, L.; Song, W.; Wang, C.; Chen, C. Transcriptome analyses from mutant Salvia miltiorrhiza reveals important roles for SmGASA4 during plant development. Int. J. Mol. Sci. 2018, 19, 2088. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Yu, H.; Zhong, C.; Zhang, X.; Peng, J.; Wang, X. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J. Exp. Bot. 2013, 64, 1637–1647. [Google Scholar] [CrossRef]
- Lee, S.-C.; Han, S.-K.; Kim, S.-R. Salt- and ABA-inducible OsGASR1 is involved in salt tolerance. J. Plant Biol. 2015, 58, 96–101. [Google Scholar] [CrossRef]
- Li, K.-L.; Bai, X.; Li, Y.; Cai, H.; Ji, W.; Tang, L.-L.; Wen, Y.-D.; Zhu, Y.-M. GsGASA1 mediated root growth inhibition in response to chronic cold stress is marked by the accumulation of DELLAs. J. Plant Physiol. 2011, 168, 2153–2160. [Google Scholar] [CrossRef]
- Priyadarshan, P.M.; Goncalves, P.S. Hevea gene pool for breeding. Genet. Resour. Crop Evol. 2003, 50, 101–114. [Google Scholar] [CrossRef]
- Priyadarshan, P.M. Refinements to Hevea rubber breeding. Tree Genet. Genomes 2017, 13, 20. [Google Scholar] [CrossRef]
- Priyadarshan, P.M.; Hoa, T.T.T.; Huasun, H.; de Gonçalves, P.S. Yielding potential of rubber (Hevea brasiliensis) in sub-optimal environments. J. Crop Improv. 2005, 14, 221–247. [Google Scholar]
- An, B.; Wang, Q.; Zhang, X.; Zhang, B.; Luo, H.; He, C. Comprehensive transcriptional and functional analyses of HbGASA genes reveal their roles in fungal pathogen resistance in Hevea brasiliensis. Tree Genet. Genomes 2018, 14, 41. [Google Scholar]
- Cheng, H.; Song, X.; Hu, Y.; Wu, T.; Yang, Q.; An, Z.; Feng, S.; Deng, Z.; Wu, W.; Zeng, X.; et al. Chromosome-level wild Hevea brasiliensis genome provides new tools for genomic-assisted breeding and valuable loci to elevate rubber yield. Plant Biotechnol. J. 2023, 21, 1058–1072. [Google Scholar]
- Muhammad, I.; Li, W.Q.; Jing, X.Q.; Zhou, M.R.; Shalmani, A.; Ali, M.; Wei, X.Y.; Sharif, R.; Liu, W.T.; Chen, K.M. A systematic in silico prediction of gibberellicacid stimulated GASA family members: A novel small peptide contributes to floral architecture and transcriptomic changes induced by external stimuli in rice. J. Plant Physiol. 2019, 234–235, 117–132. [Google Scholar] [CrossRef]
- Ahmad, B.; Yao, J.; Zhang, S.; Li, X.; Zhang, X.; Yadav, V.; Wang, X. Genome-wide characterization and expression profiling of GASA genes during different stages of seed development in Grapevine (Vitis vinifera L.) predict their involvement in seed development. Int. J. Mol. Sci. 2020, 21, 1088. [Google Scholar] [CrossRef]
- Qiao, K.; Ma, C.; Lv, J.; Zhang, C.; Ma, Q.; Fan, S. Identification, characterization, and expression profiles of the GASA genes in Cotton. J. Cotton Res. 2021, 4, 7. [Google Scholar]
- Yin, D.; Li, G.; Qu, Y. Genome-wide identification and bioinformatics analysis of the GASA gene family in Ricinus communis. Mol. Plant Breed. 2023, 1–14. (In Chinese) [Google Scholar]
- Wu, K.; Qu, Y.; Rong, H.; Han, X.; Tian, Y.; Xu, L. Identification and expression analysis of the Populus trichocarpa GASA-gene family. Int. J. Mol. Sci. 2022, 23, 1507. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, X. Expression pattern of GASA, downstream genes of DELLA, in Arabidopsis. Chin. Sci. Bull. 2008, 53, 3839–3846. [Google Scholar]
- Lü, L.; Chen, X.; Zhang, Y.; Liu, Q.; Wang, L.; Ma, L.; Li, H. Bioinformatics identification of GASA gene family expression profiles in Wheat. J. Crops. 2018, 34, 58–67. [Google Scholar]
- Xu, G.X.; Guo, C.C.; Shan, H.Y.; Kong, H.Z. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar]
- Han, S.; Jiao, Z.; Niu, M.-X.; Yu, X.; Huang, M.; Liu, C.; Wang, H.-L.; Zhou, Y.; Mao, W.; Wang, X.; et al. Genome-wide comprehensive analysis of the GASA gene family in Populus. Int. J. Mol. Sci. 2021, 22, 12336. [Google Scholar] [CrossRef]
- Nahirnak, V.; Rivarola, M.; de Urreta, M.G.; Paniego, N.; Hopp, H.E.; Almasia, N.I.; Vazquez-Rovere, C. Genome-wide analysis of the Snakin/GASA gene family in Solanum Tuberosum cv. Kennebec. Am. J. Potato Res. 2016, 93, 172–188. [Google Scholar]
- Qu, J.; Kang, S.G.; Hah, C.; Jang, J.C. Molecular and cellular characterization of GA-Stimulated Transcripts GASA4 and GASA6 in Arabidopsis thaliana. Plant Sci. 2016, 246, 550. [Google Scholar]
- Li, M.; Wang, X.; Cao, Y.; Liu, X.; Lin, Y.; Ou, Y.; Zhang, H.; Liu, J. Strength comparison between cold-inducible promoters of Arabidopsis cor15a and cor15b genes in potato and tabacco. Plant Physiol. Biochem. 2013, 71, 77–86. [Google Scholar]
- Xue, J.Y.; Zhao, T.; Liu, Y.; Zhang, Y.X.; Zhang, G.Q.; Chen, H.; Zhou, G.C.; Zhang, S.Z. Genome-wide analysis of the nucleotide binding site leucine-rich repeat genes of four orchids revealed extremely low numbers of disease resistance genes. Front. Genet. 2020, 10, 1286. [Google Scholar]
| Gene Name | Gene ID | Protein (aa) | MW (Da) | pI | GRAVY | Instability Index | Aliphatic Index | Asp + Glu | Arg + Lys | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|
| HbGASA1 | LOC110646399 | 91 | 10,007.97 | 9.05 | −0.071 | 35.77 | 74.07 | 5 | 14 | Extracellular |
| HbGASA2 | LOC110639973 | 88 | 9680.57 | 8.92 | −0.006 | 27.25 | 57.61 | 6 | 14 | Extracellular |
| HbGASA3 | LOC110671420 | 94 | 10,413.46 | 9.12 | −0.059 | 38.58 | 58.19 | 4 | 14 | Extracellular |
| HbGASA4 | LOC110645670 | 106 | 11,639.83 | 9.27 | −0.176 | 37.78 | 61.60 | 2 | 14 | Extracellular |
| HbGASA5 | LOC110655875 | 100 | 10,764.64 | 7.46 | 0.184 | 64.30 | 82.90 | 7 | 8 | Extracellular |
| HbGASA6 | LOC110637234 | 112 | 12,424.58 | 9.45 | −0.446 | 54.82 | 46.25 | 3 | 16 | Extracellular |
| HbGASA7 | LOC110665251 | 107 | 12,053.17 | 9.38 | −0.407 | 45.44 | 45.61 | 2 | 15 | Extracellular |
| HbGASA8 | LOC110651389 | 107 | 12,116.24 | 9.42 | −0.427 | 53.83 | 49.25 | 2 | 15 | Extracellular |
| HbGASA9 | LOC110667031 | 88 | 9709.73 | 9.33 | −0.194 | 35.92 | 60.00 | 5 | 18 | Chloroplast |
| HbGASA10 | LOC110673766 | 104 | 11,066.93 | 8.92 | −0.015 | 45.58 | 78.85 | 4 | 11 | Extracellular |
| HbGASA11 | LOC110646398 | 111 | 12,325.53 | 9.46 | −0.154 | 56.89 | 73.78 | 4 | 18 | Extracellular |
| HbGASA12 | LOC110647740 | 98 | 10,406.24 | 9.16 | −0.179 | 45.23 | 61.84 | 5 | 15 | Extracellular |
| HbGASA13 | LOC110643317 | 96 | 10,572.41 | 8.9 | −0.328 | 50.72 | 63.85 | 8 | 15 | Chloroplast |
| HbGASA14 | LOC110632043 | 105 | 11,442.52 | 8.75 | 0.078 | 59.26 | 83.52 | 8 | 14 | Extracellular |
| HbGASA15 | LOC110660492 | 109 | 11,904.03 | 8.75 | −0.083 | 46.10 | 79.54 | 9 | 15 | Extracellular |
| HbGASA16 | LOC110655855 | 106 | 11,499.55 | 9.01 | −0.013 | 49.44 | 79.25 | 6 | 14 | Extracellular |
| HbGASA17 | LOC110667249 | 241 | 25,225.32 | 9.79 | −0.226 | 62.00 | 66.64 | 9 | 32 | Extracellular |
| HbGASA18 | LOC110659225 | 253 | 26,546.86 | 9.98 | −0.355 | 66.68 | 61.58 | 6 | 33 | Extracellular |
| Primer Name | Forward Primer (5ʹ-3ʹ) | Reverse Primer (5ʹ-3ʹ) |
|---|---|---|
| HbYLS8 | CCTCGTCGTCATCCGATTC | CAGGCACCTCAGTGATGTC |
| HbGASA1 | AGCAAGGCATCAAGGCACAA | TTGGCATAGCAAGGGCAAAC |
| HbGASA2 | GTGACTCCAAGTGTGGGGAG | ATAAGTTCCAGAGGGCACGC |
| HbGASA3 | GCCTCAGGTGTACTTCCCTC | AGCACGAAACTTAGCACCCT |
| HbGASA4 | ACCCAACACCACCTTGACAG | GTCTTGCTGCATCTCCTCGT |
| HbGASA5 | TGCTTCCCTCCCACAACAAA | GCACAACACGTACCACATGC |
| HbGASA6 | CAGCTTACAAGAAGCCGTGC | TTGTTGTAGCAAGGGCAGGA |
| HbGASA7 | GTTATGGCATCGCATGGTCA | ATTGGGTCTTGCTGCACCTC |
| HbGASA8 | GGCTAAATTTGTTGCTGCCTTCCT | GCCTTTGGTTGTCATAGTGGTG |
| HbGASA9 | CAAGTCCCCTTACTGCACGA | AGAGGGCACACACTTGCATT |
| HbGASA10 | ACCTTGCTCCAACCAATCCC | CAAGTCCCACATGCCCTCTT |
| HbGASA11 | GCTCCTTCAGGTTTGTTTGGG | TCCTTGCATCTCCTTGAGCAT |
| HbGASA12 | TCGTTCAGTTCACCAGGGCT | ATCCTGCATTCGCACACCTC |
| HbGASA13 | TTGTGGAGGGTTGTGCAAGG | CACGCACTTGCACCTAAAGC |
| HbGASA14 | TGAAGAGCAAAGCACCGAGTT | TGCAAACTACACCTCACCTTG |
| HbGASA15 | GGCAGTCCGTTTGCTTCTTG | GAGCCTCCTGTTTCCACCTC |
| HbGASA16 | ATGCGAGGGAAGGTGCAAAA | GGACAGAACTCAAGGTTGCC |
| HbGASA17 | CCAAGGCTGTGTTGTTTCTGG | AGGAGTCTTGGCTGGTGGTA |
| HbGASA18 | CCCAAATACTAGTCGCCCCA | GGAGTTGGTGCCTTGACTGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cong, Y.; Hu, Y.; Deng, Z.; Wu, W.; Wu, T.; Zhao, Y.; An, Z. Genome-Wide Identification and Expression Analysis of GASA Genes in Hevea brasiliensis Reveals Their Involvement in Response to Cold Stress. Int. J. Mol. Sci. 2025, 26, 3454. https://doi.org/10.3390/ijms26073454
Cong Y, Hu Y, Deng Z, Wu W, Wu T, Zhao Y, An Z. Genome-Wide Identification and Expression Analysis of GASA Genes in Hevea brasiliensis Reveals Their Involvement in Response to Cold Stress. International Journal of Molecular Sciences. 2025; 26(7):3454. https://doi.org/10.3390/ijms26073454
Chicago/Turabian StyleCong, Yuying, Yanshi Hu, Zhi Deng, Wenguan Wu, Tingkai Wu, Yanhong Zhao, and Zewei An. 2025. "Genome-Wide Identification and Expression Analysis of GASA Genes in Hevea brasiliensis Reveals Their Involvement in Response to Cold Stress" International Journal of Molecular Sciences 26, no. 7: 3454. https://doi.org/10.3390/ijms26073454
APA StyleCong, Y., Hu, Y., Deng, Z., Wu, W., Wu, T., Zhao, Y., & An, Z. (2025). Genome-Wide Identification and Expression Analysis of GASA Genes in Hevea brasiliensis Reveals Their Involvement in Response to Cold Stress. International Journal of Molecular Sciences, 26(7), 3454. https://doi.org/10.3390/ijms26073454






