The Relationship Between Bacterial Flora in Saliva and Esophageal Mucus and Endoscopic Severity in Patients with Eosinophilic Esophagitis
Abstract
1. Introduction
2. Results
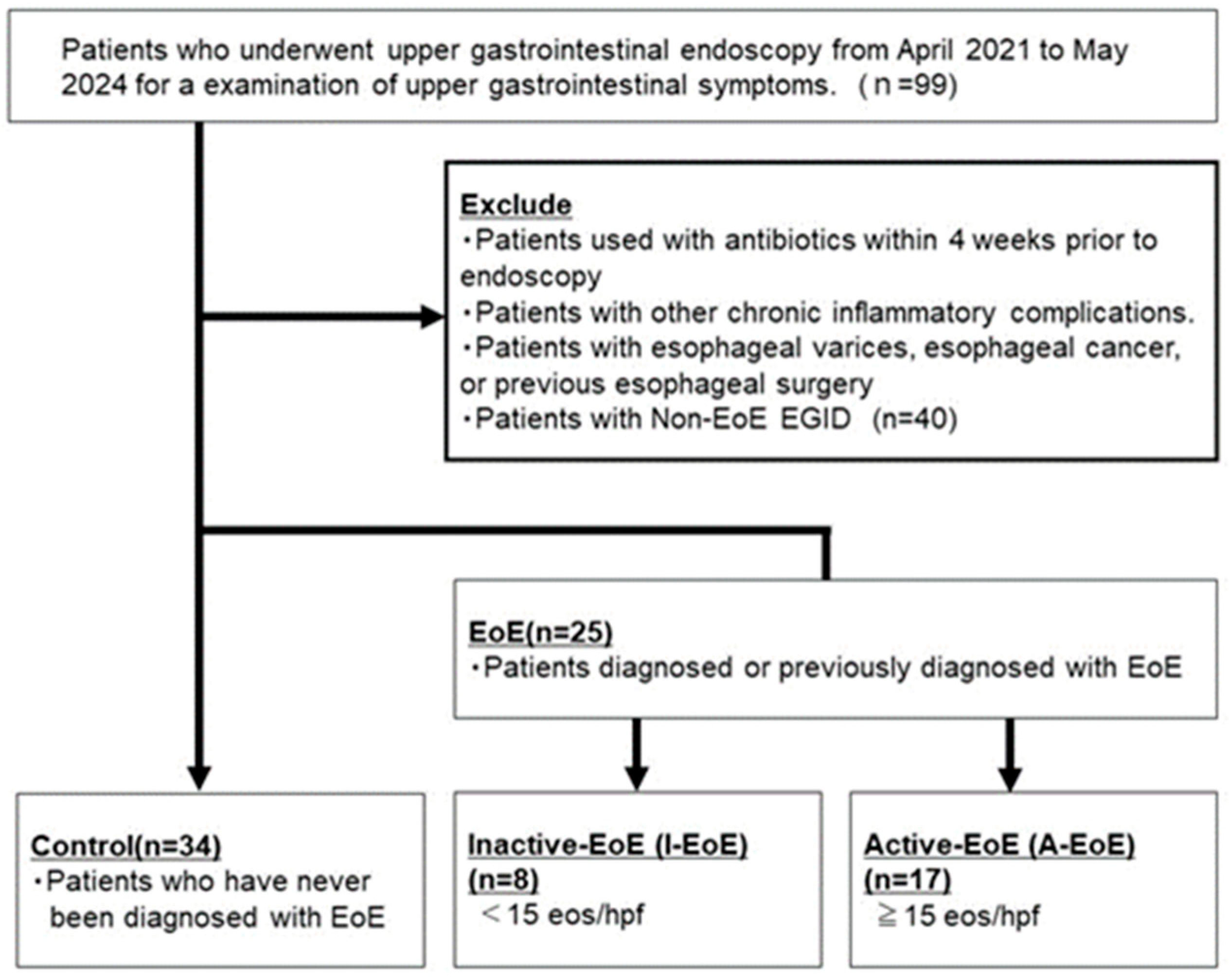
2.1. Patient Characteristics
2.2. Comparative Analyses of the Alpha Diversity Indices Among the Three Groups Using Saliva and Esophageal Mucus
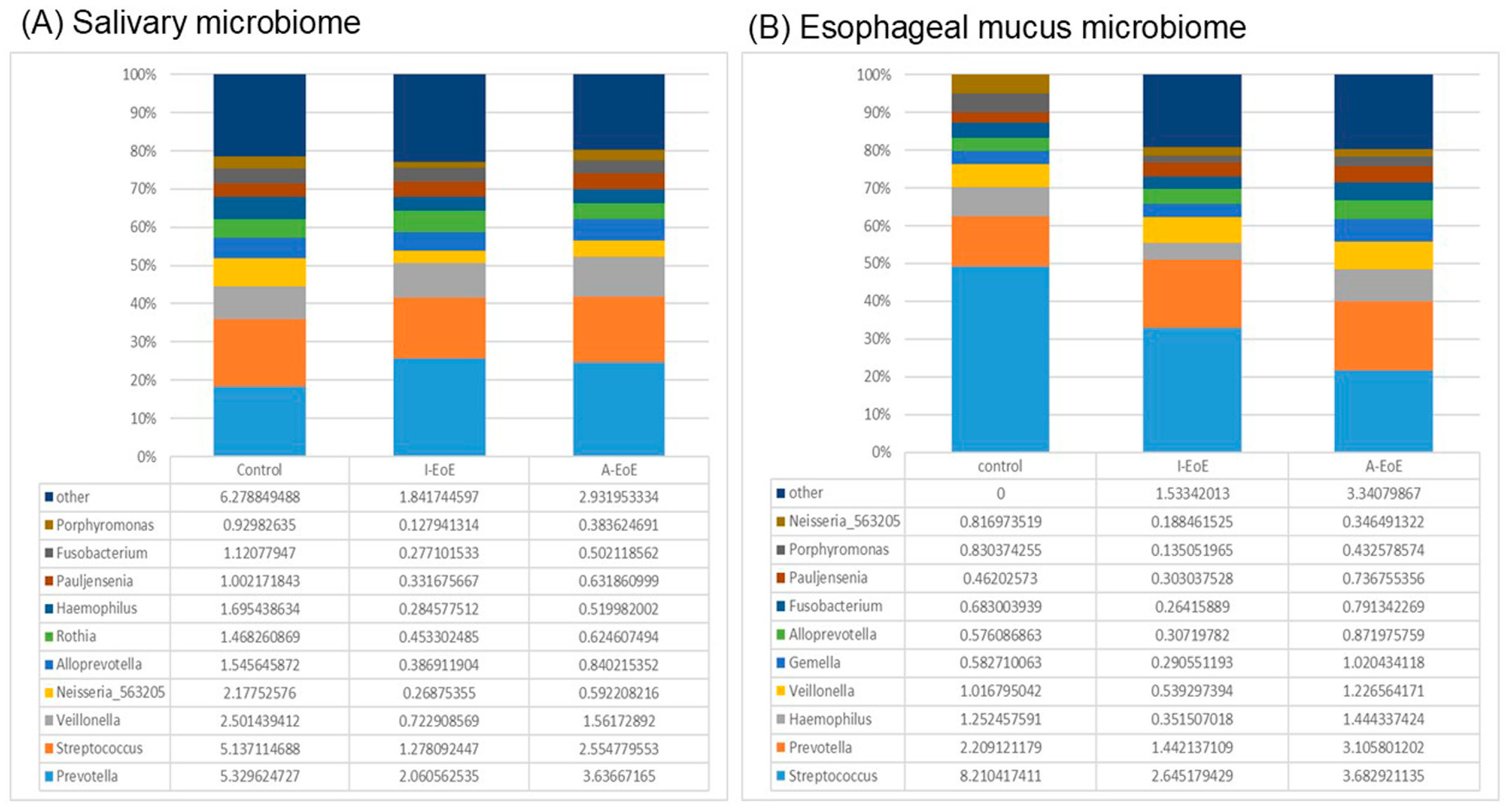
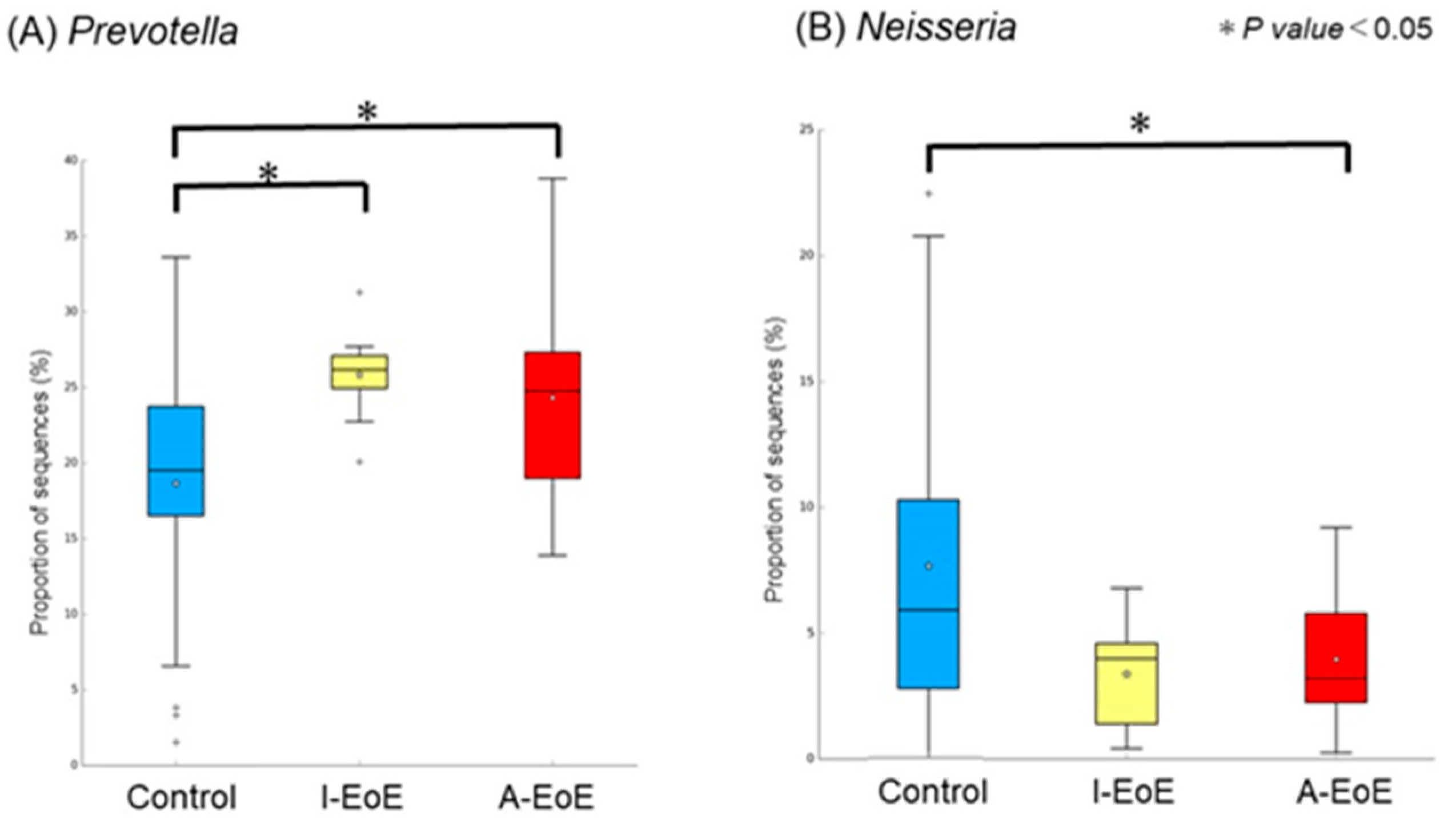
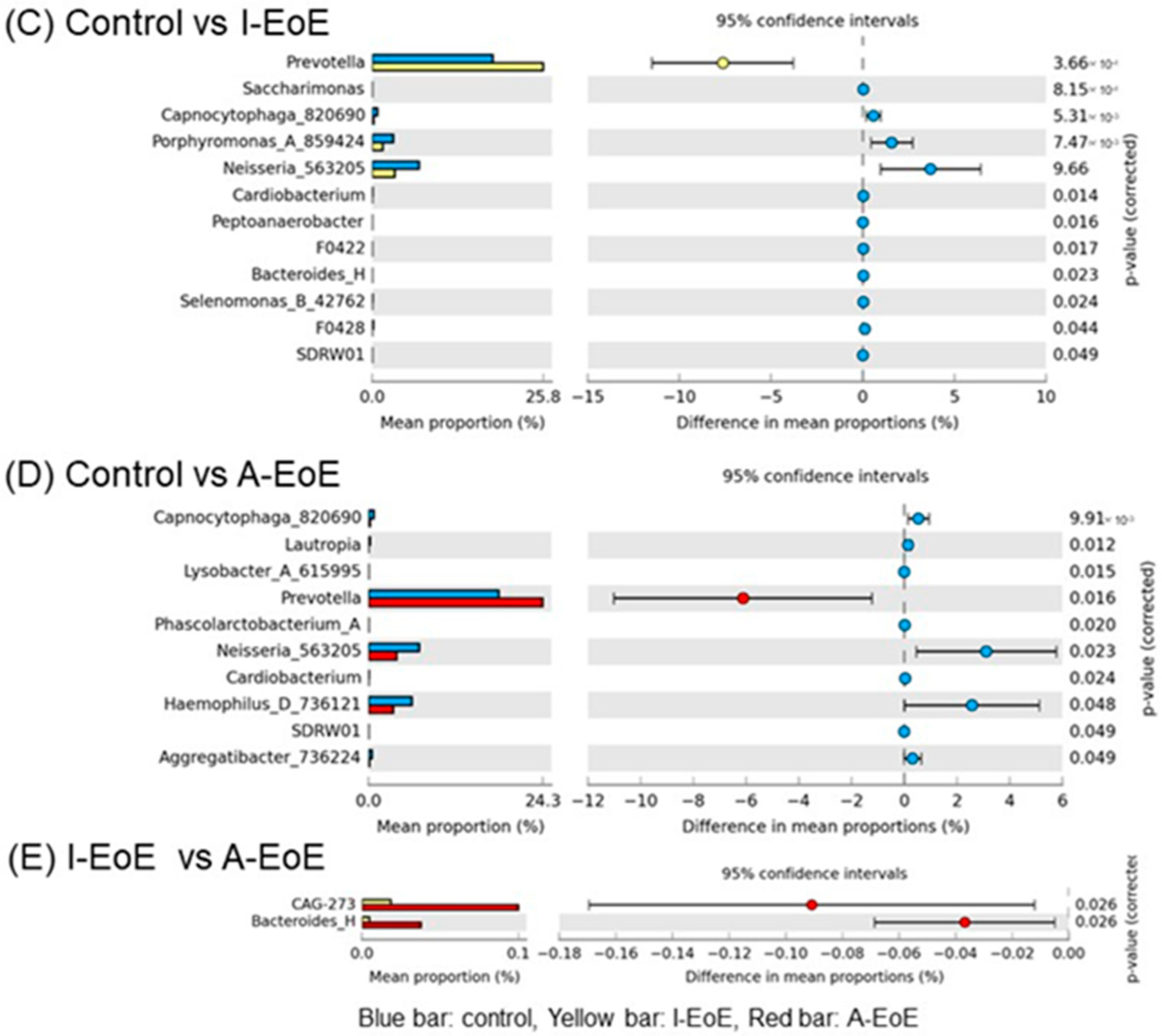
2.3. Comparative Analyses of Saliva Taxonomic Composition
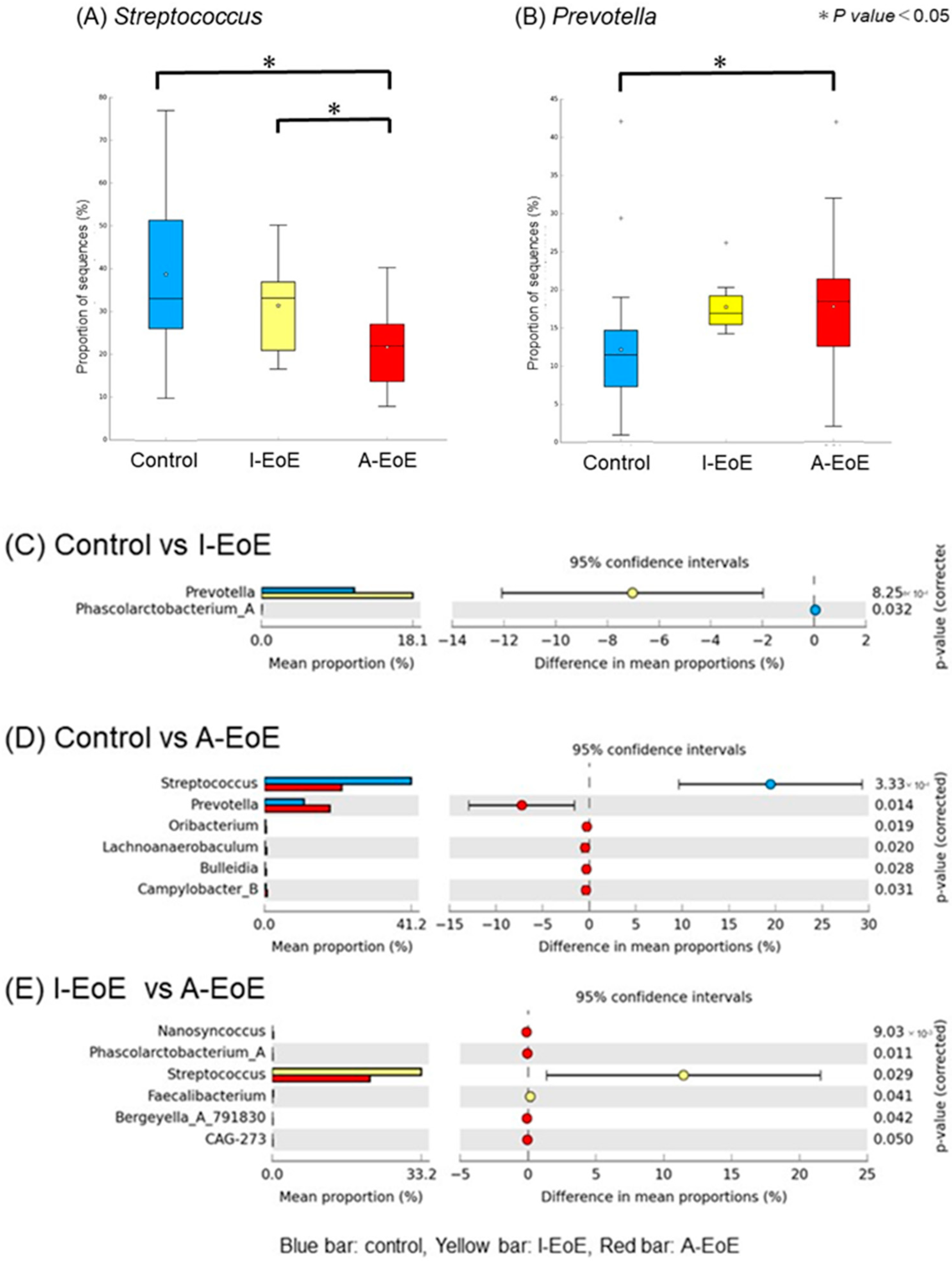
2.4. Comparative Taxonomic Analysis of the Esophageal Mucosa
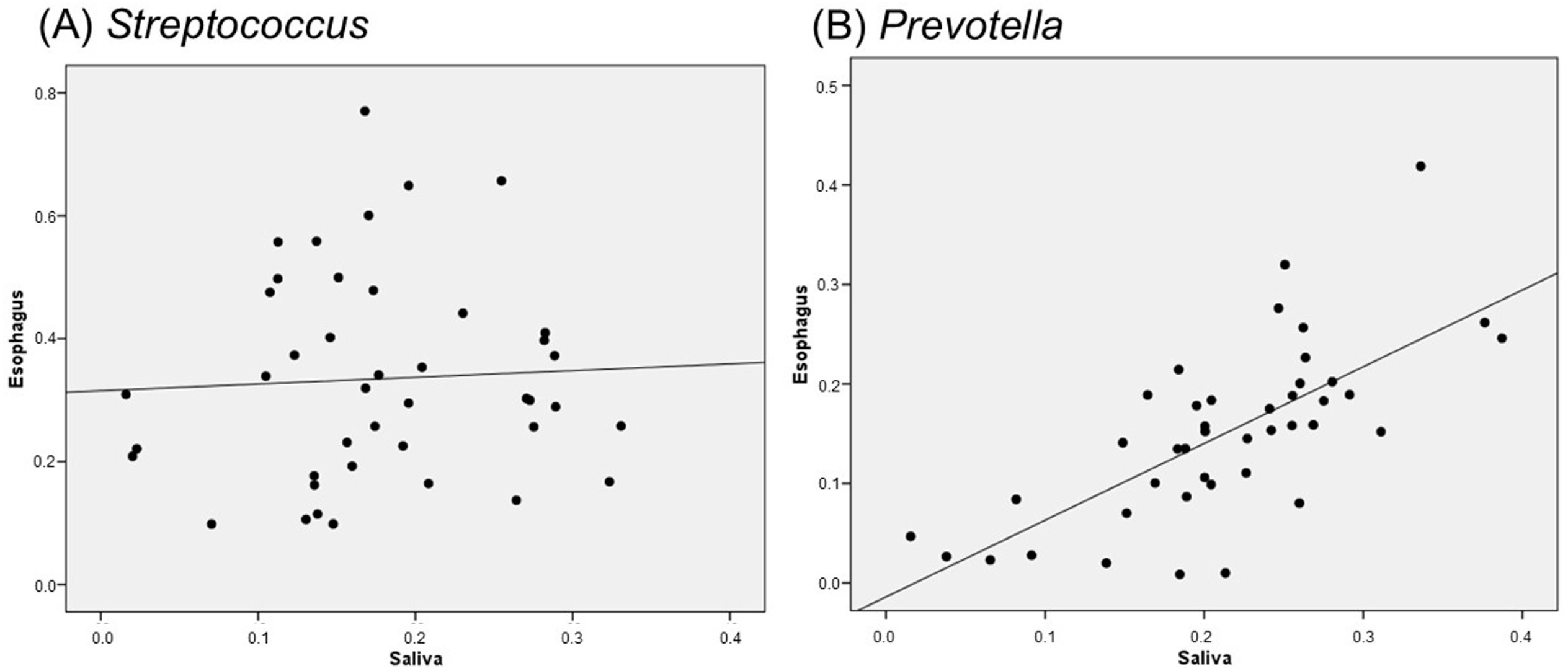
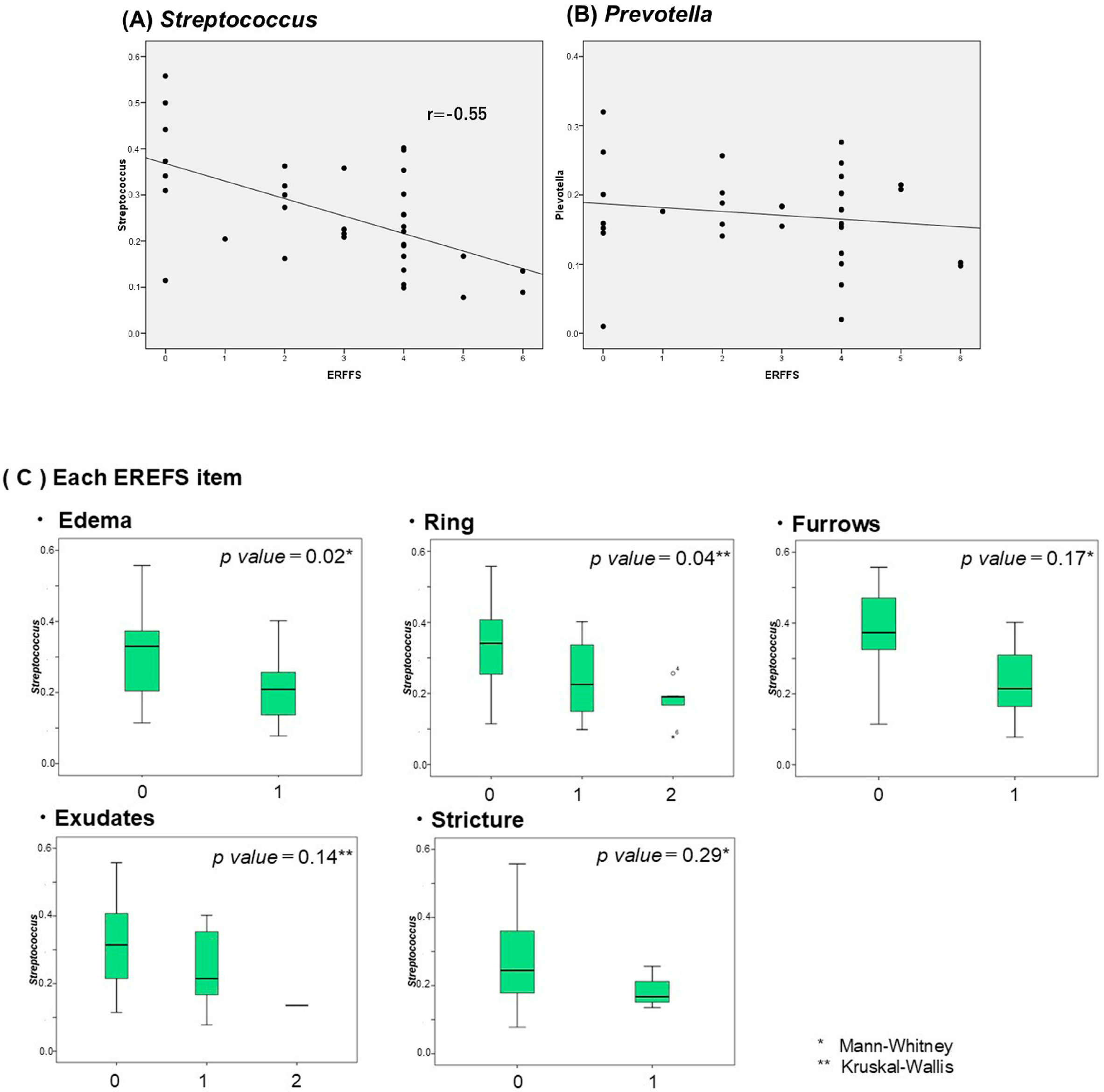
2.5. Correlation Between Salivary and Esophageal Flora
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Patients and Sample Collection
4.3. RNA Extraction
4.4. Reverse Transcription Quantitative Polymerase Chain Reaction
4.5. DNA Extraction, PCR Amplification, Library Preparation, and 16S rRNA Sequencing
4.6. Bioinformatics Analysis
4.7. Alpha Diversity
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EoE | Eosinophilic esophagitis |
| A-EoE | Active eosinophilic esophagitis |
| I-EoE | Inactive eosinophilic esophagitis |
| EREFS | Eosinophilic esophagitis endoscopic reference score |
| IL | Interleukin |
| EGD | Esophago-gastroduodenoscopy |
| PPI | Proton pump inhibitor |
| GERD | Gastroesophageal reflex disease |
| hpf | high-power field |
| IBD | inflammatory bowel disease |
| TSLP | Thymic stromal lymphopoietin |
| CCL | CC chemokine ligand |
| CXCL | CXC motif chemokine ligand |
References
- Barchi, B.; Vespa, E.; Passaretti, S.; Dell’Anna, G.; Fasulo, E.; Yacoub, M.; Albarello, L.; Sinagra, E.; Massimino, L.; Ungaro, F.; et al. The Dual Lens of Endoscopy and Histology in the Diagnosis and Management of Eosinophilic Gastrointestinal Disorders-A Comprehensive Review. Diagnostics 2024, 22, 858. [Google Scholar] [CrossRef]
- Furuta, G.T.; Katzka, D.A. Eosinophilic Esophagitis. N. Engl. J. Med. 2015, 373, 1640–1648. [Google Scholar] [PubMed]
- Dellon, E.S.; Liacouras, C.A.; Molina-Infante, J.; Furuta, G.T.; Spergel, J.M.; Zevit, N.; Spechler, S.J.; Attwood, S.E.; Straumann, A.; Aceves, S.S.; et al. Updated International Consensus Diagnostic Criteria for Eosinophilic Esophagitis: Proceedings of the AGREE Conference. Gastroenterology 2018, 155, 1022–1033.e10. [Google Scholar]
- Lucendo, A.J.; Molina-Infante, J.; Arias, Á.; von Arnim, U.; Bredenoord, A.J.; Bussmann, C.; Amil Dias, J.; Bove, M.; González-Cervera, J.; Larsson, H.; et al. Guidelines on Eosinophilic Esophagitis: Evidence-Based Statements and Recommendations for Diagnosis and Management in Children and Adults. United Eur. Gastroenterol. J. 2017, 5, 335–358. [Google Scholar]
- Hirano, I.; Chan, E.S.; Rank, M.A.; Sharaf, R.N.; Stollman, N.H.; Stukus, D.R.; Wang, K.; Greenhawt, M.; Falck-Ytter, Y.T.; AGA Institute Clinical Guidelines Committee; et al. AGA Institute and the Joint Task Force on Allergy-Immunology Practice Parameters Clinical Guidelines for the Management of Eosinophilic Esophagitis. Gastroenterology 2020, 158, 1776–1786. [Google Scholar]
- Dobbins, J.W.; Sheahan, D.G.; Behar, J. Eosinophilic Gastroenteritis with Esophageal Involvement. Gastroenterology 1977, 72, 1312–1316. [Google Scholar]
- Jensen, E.T.; Kuhl, J.T.; Martin, L.J.; Langefeld, C.D.; Dellon, E.S.; Rothenberg, M.E. Early-Life Environmental Exposures Interact with Genetic Susceptibility Variants in Pediatric Patients with Eosinophilic Esophagitis. J. Allergy Clin. Immunol. 2018, 141, 632–637.e5. [Google Scholar] [CrossRef] [PubMed]
- Hahn, J.W.; Kim, S.; Yon, D.K. Global Incidence and Prevalence of Eosinophilic Esophagitis, 1976–2022, and Projections to 2037: A Post-Hoc Analysis. Clin. Gastroenterol. Hepatol. 2023, 21, S1542–S3565. [Google Scholar]
- Angerami Almeida, K.; de Queiroz Andrade, E.; Burns, G.; Hoedt, E.C.; Mattes, J.; Keely, S.; Collison, A. The Microbiota in Eosinophilic Esophagitis: A Systematic Review. J. Gastroenterol. Hepatol. 2022, 37, 1673–1684. [Google Scholar]
- Kottyan, L.C.; Rothenberg, M.E. Genetics of Eosinophilic Esophagitis. Mucosal Immunol. 2017, 10, 580–588. [Google Scholar] [CrossRef]
- Sherrill, J.D.; Blanchard, C. Genetics of Eosinophilic Esophagitis. Dig. Dis. 2014, 32, 22–29. [Google Scholar] [PubMed]
- Simon, D.; Cianferoni, A.; Spergel, J.M.; Aceves, S.; Holbreich, M.; Venter, C.; Rothenberg, M.E.; Terreehorst, I.; Muraro, A.; Lucendo, A.J.; et al. Eosinophilic Esophagitis Is Characterized by a Non-IgE-Mediated Food Hypersensitivity. Allergy 2016, 71, 611–620. [Google Scholar] [PubMed]
- Rochman, M.; Travers, J.; Miracle, C.E.; Bedard, M.C.; Wen, T.; Azouz, N.P.; Caldwell, J.M.; Kc, K.; Sherrill, J.D.; Davis, B.P.; et al. Profound Loss of Esophageal Tissue Differentiation in Patients with Eosinophilic Esophagitis. J. Allergy Clin. Immunol. 2017, 140, 738–749.e3. [Google Scholar]
- McGowan, E.C.; Singh, R.; Katzka, D.A. Barrier Dysfunction in Eosinophilic Esophagitis. Curr. Gastroenterol. Rep. 2023, 25, 380–389. [Google Scholar]
- Odiase, E.; Zhang, X.; Chang, Y.; Nelson, M.; Balaji, U.; Gu, J.; Zhang, Q.; Pan, Z.; Spechler, S.J.; Souza, R.F. In Esophageal Squamous Cells from Eosinophilic Esophagitis Patients, Th2 Cytokines Increase Eotaxin-3 Secretion through Effects on Intracellular Calcium and a Non-Gastric Proton Pump. Gastroenterology 2021, 160, 2072–2088.e6. [Google Scholar] [PubMed]
- Arias, Á.; Vicario, M.; Bernardo, D.; Olalla, J.M.; Fortea, M.; Montalban-Arques, A.; Martínez-Fernández, P.; González-Castro, A.M.; Mota-Huertas, T.; Arias-González, L.; et al. Toll-like Receptors-Mediated Pathways Activate Inflammatory Responses in the Esophageal Mucosa of Adult Eosinophilic Esophagitis. Clin. Transl. Gastroenterol. 2018, 9, 147. [Google Scholar]
- Graves, D.T.; Corrêa, J.D.; Silva, T.A. The Oral Microbiota Is Modified by Systemic Diseases. J. Dent. Res. 2019, 98, 148–156. [Google Scholar]
- Facchin, S.; Calgaro, M.; Pandolfo, M.; Caldart, F.; Ghisa, M.; Greco, E.; Sattin, E.; Valle, G.; Dellon, E.S.; Vitulo, N.; et al. Salivary Microbiota Composition May Discriminate between Patients with Eosinophilic Oesophagitis (EoE) and Non-EoE Subjects. Aliment. Pharmacol. Ther. 2022, 56, 450–462. [Google Scholar] [PubMed]
- Hiremath, G.; Shilts, M.H.; Boone, H.H.; Correa, H.; Acra, S.; Tovchigrechko, A.; Rajagopala, S.V.; Das, S.R. The Salivary Microbiome Is Altered in Children with Eosinophilic Esophagitis and Correlates with Disease Activity. Clin. Transl. Gastroenterol. 2019, 10, e00039. [Google Scholar]
- Yano, Y.; Etemadi, A.; Abnet, C.C. Microbiome and Cancers of the Esophagus: A Review. Microorganisms 2021, 9, 1764. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, N.; Wang, Z. Eosinophilic Esophagitis and Esophageal Microbiota. Front. Cell Infect. Microbiol. 2023, 13, 1206343. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.K.; Fang, R.; Wagner, B.D.; Choe, H.N.; Kelly, C.J.; Schroeder, S.; Moore, W.; Stevens, M.J.; Yeckes, A.; Amsden, K.; et al. Esophageal Microbiome in Eosinophilic Esophagitis. PLoS ONE 2015, 10, e0128346. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Takeda, K. Host-Microbiota Interactions in Rheumatoid Arthritis. Exp. Mol. Med. 2019, 51, 1–6. [Google Scholar] [CrossRef]
- Mangalam, A.K.; Murray, J. Microbial Monotherapy with Prevotella Histicola for Patients with Multiple Sclerosis. Expert Rev. Neurother. 2019, 19, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Abdelbary, M.M.H.; Hatting, M.; Bott, A.; Dahlhausen, A.; Keller, D.; Trautwein, C.; Conrads, G. The Oral-Gut Axis: Salivary and Fecal Microbiome Dysbiosis in Patients with Inflammatory Bowel Disease. Front. Cell. Infect. Microbiol. 2022, 12, 1010853. [Google Scholar] [CrossRef]
- Larsen, J.M. The Immune Response to Prevotella Bacteria in Chronic Inflammatory Disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef]
- Blais, L.L.; Montgomery, T.L.; Amiel, E.; Deming, P.B.; Krementsov, D.N. Probiotic and commensal gut microbial therapies in multiple sclerosis and its animal models: Comprehensive review. Gut Microbes 2021, 13, 194289. [Google Scholar] [CrossRef]
- Cani, P.D. Human gut microbiome: Hope, threats and promises. Gut 2018, 67, 1716–1725. [Google Scholar] [CrossRef]
- Gao, Y.; Nanan, R.; Micia, L.; Tan, J.; Sominsky, L.; Quinn, T.P.; O’Hely, M.; Ponsonby, A.; Tang, M.L.K.; Collier, F.; et al. The maternal gut microbiome during pregnancy and offspring allergy and asthma. J. Allergy Clin. Immunol. 2021, 148, 669–678. [Google Scholar] [CrossRef]
- Ahmed, H.S. The impact of Prevotella on Neurobiology in Aging: Deciphering Dendritic Cell Activity and Inflammatory Dynamics. Mol. Neurobiol. 2024, 61, 9240–9251. [Google Scholar] [CrossRef]
- Nabe, T. Steroid-Resistant Asthema and Neutrophils. Biol. Pharm. Bull. 2020, 43, 31–35. [Google Scholar]
- Luo, W.; Hu, J.; Xu, W.; Dong, J. Distinct spatial and temporal for Th1, Th2, and Th17 cells in asthema. Front. Immunol. 2022, 12, 974066. [Google Scholar]
- Baring, C.; Bezema, T.; Calder, P.C.; Charloux, A.; Frossard, N.; Garssen, J.; Haworth, O.; Dilevskaya, K.; Levi-Schaffer, F.; Lonsdorfer, E.; et al. Activation of Resolution Pathways to Prevent and Fight Chronic Inflammation: Lessons from Asthma and Inflammatory Bowel Disease. Front. Immunol. 2019, 23, 1699. [Google Scholar]
- Li, H.; Zhang, Z.; Zhang, H.; Guo, Y.; Yao, Z. Update on the Pathogenesis and Therapy of Atopic Dermatitis. Clin. Rev. Allergy Immunol. 2021, 61, 324–338. [Google Scholar] [CrossRef]
- Sharma, G.; Garg, N.; Hasan, S.; Saffarini, D.; Shirodkar, S. Fumarate and Nitrite Reduction by Prevotella Nigrescens and Prevotella Buccae Isolated from Chronic Periodontitis Patients. Microb. Pathog. 2023, 176, 106022. [Google Scholar] [PubMed]
- Lianto, P.; Zhang, Y.; Che, H. Signals from the Various Immune Cells in Promoting Food Allergy-Induced Eosinophilic Esophagitis like Disease. Asia Pac. Allergy 2019, 9, e28. [Google Scholar]
- Holvoet, S.; Doucet-Ladevèze, R.; Perrot, M.; Barretto, C.; Nutten, S.; Blanchard, C. Beneficial Effect of Lactococcus Lactis NCC 2287 in a Murine Model of Eosinophilic Esophagitis. Allergy 2016, 71, 1753–1761. [Google Scholar]
- Tagg, J.R. Prevention of Streptococcal Pharyngitis by Anti-Streptococcus Pyogenes Bacteriocin-like Inhibitory Substances (BLIS) Produced by Streptococcus Salivarius. Indian J. Med. Res. 2004, 119, 13–16. [Google Scholar]
- Wescombe, P.A.; Dyet, K.H.; Dierksen, K.P.; Power, D.A.; Jack, R.W.; Burton, J.P.; Inglis, M.A.; Wescombe, A.L.; Tagg, J.R. Salivaricin G32, a Homolog of the Prototype Streptococcus Pyogenes Nisin-like Lantibiotic SA-FF22, Produced by the Commensal Species Streptococcus Salivarius. Int. J. Microbiol. 2012, 2012, 738503. [Google Scholar] [CrossRef]
- Beak, D.; Lee, S. Anti-inflammatory efficacy of human-derived Streptococcus salivarius on Peridontopathogen-induced inflammation. J. Microbiol. Biotechnol. 2023, 33, 998–1005. [Google Scholar]
- Torre, F.; Sota, J.; Insalaco, A.; Conti, G.; Giudice, E.; Lubrano, R.; Breda, L.; Maggio, M.C.; Civino, A.; Mastrorilli, V.; et al. Preliminary data revealing efficacy of Sterptococcus salivarius (SSK12) in Periodic Fever, Apthous stomatitis, Pharygitis, and cervical Adenitis (PFAPA) syndrome: A multicenter study from the AIDA Network PFAPA syndrome registry. Front. Med. 2023, 10, 1105605. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Qiu, J.; Zhang, T.; Wu, X.; Zhou, J.; Park, S. Quantitative Changes of Veillonella, Streptococcus, and Neisseria in the Oral Cavity of Patients with Recurrent Aphthous Stomatitis: A Systematic Review and Meta-Analysis. Arch. Oral Biol. 2021, 129, 105198. [Google Scholar] [CrossRef] [PubMed]
- Di Spirito, F.; Pisano, M.; Di Palo, M.P.; Franci, G.; Rupe, A.; Fiorino, A.; Rengo, C. Peri-Implantitis-Associated Microbiota before and after Peri-Implantitis Treatment, the Biofilm “Competitive Balancing” Effect: A Systematic Review of Randomized Controlled Trials. Microorganisms 2024, 12, 1965. [Google Scholar] [CrossRef]
- Han, Y.; Wang, B.; Gao, H.; He, C.; Hua, R.; Liang, C.; Xin, S.; Wang, Y.; Xu, J. Insight into the Relationship between Oral Microbiota and the Inflammatory Bowel Disease. Microorganisms 2022, 10, 1868. [Google Scholar] [CrossRef] [PubMed]
- Laserna-Mendieta, E.J.; FitzGerald, J.A.; Arias-Gonzalez, L.; Ollala, J.M.; Bernardo, D.; Claesson, M.J.; Lucendo, A.J. Esophageal microbiome in active eosinophilic esophagitis and changes induced by different therapies. Sci. Rep. 2021, 11, 7113. [Google Scholar] [CrossRef]
- Furuta, G.T.; Fillon, S.A.; Williamson, K.M.; Robertson, C.E.; Stevens, M.J.; Aceves, S.S.; Arva, N.C.; Chehade, M.; Collins, M.H.; Davis, C.M.; et al. Mucosal microbiota associated with eosinophilic esophagitis and eosinophilic gastritis. J. Pediatr. Gastroenterol. Nutr. 2023, 76, 347–354. [Google Scholar]
- Hirano, I.; Moy, N.; Heckman, M.G.; Thomas, C.S.; Gonsalves, N.; Achem, S.R. Endoscopic Assessment of the Oesophageal Features of Eosinophilic Oesophagitis: Validation of a Novel Classification and Grading System. Gut 2013, 62, 489–495. [Google Scholar] [CrossRef]
- Dellon, E.S.; Muir, A.B.; Katzka, D.A.; Shah, S.C.; Sauer, B.G.; Aceves, S.S.; Furuta, G.T.; Gonsalves, N.; Hirano, I. ACG Clinical Guideline: Diagnosis and Management of Eosinophilic Esophagitis. Am. J. Gastroenterol. 2025, 120, 31–59. [Google Scholar] [CrossRef]
- Gall, A.; Fero, J.; McCoy, C.; Claywell, B.C.; Sanchez, C.A.; Blount, P.L.; Li, X.; Vaughan, T.L.; Matsen, F.A.; Reid, B.J.; et al. Bacterial Composition of the Human Upper Gastrointestinal Tract Microbiome Is Dynamic and Associated with Genomic Instability in a Barrett’s Esophagus Cohort. PLoS ONE 2015, 10, e0129055. [Google Scholar] [CrossRef]
- Dellon, E.S.; Cotton, C.C.; Gebhart, J.H.; Higgins, L.L.; Beitia, R.; Woosley, J.T.; Shaheen, N.J. Accuracy of the Eosinophilic Esophagitis Endoscopic Reference Score in Diagnosis and Determining Response to Treatment. Clin. Gastroenterol. Hepatol. 2016, 14, 31–39. [Google Scholar] [CrossRef]
- Juge, N. Relationship between Mucosa-Associated Gut Microbiota and Human Diseases. Biochem. Soc. Trans. 2022, 50, 1225–1236. [Google Scholar] [PubMed]
- Shimizu, H.; Arai, K.; Tang, J.; Hosoi, K.; Funayama, R. 5-Aminosalicylate Intolerance Causing Exacerbation in Pediatric Ulcerative Colitis. Pediatr. Int. 2017, 59, 583–587. [Google Scholar] [PubMed]
- Hayashi, A.; Mikami, Y.; Miyamoto, K.; Kamada, N.; Sato, T.; Mizuno, S.; Naganuma, M.; Teratani, T.; Aoki, R.; Fukuda, S.; et al. Intestinal Dysbiosis and Biotin Deprivation Induce Alopecia Through Overgrowth of Lactobacillus Murinus in Mice. Cell Rep. 2017, 20, 1513–1524. [Google Scholar] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar]

| Control (n = 28) | EoE | p-Value 3 Groups | p-Value | ||
|---|---|---|---|---|---|
| Inactive (n = 8) | Active (n = 17) | 2 Groups Cont vs. EoE | |||
| Gender (M) | 11 (39%) | 5 (62%) | 10 (59%) | 0.3 | 0.52 |
| Age (mean ± SD; yr) | 49 [29–62] | 52 [37–66] | 50 [42–57] | 0.9 | 0.23 |
| Allergic comorbidities (n, %) | |||||
| Asthma | 0 | 3 (38%) | 5 (29%) | 0.04 | <0.01 |
| Eczema | 1 (0%) | 6 (75%) | 3 (18%) | <0.01 | <0.01 |
| Allergic rhinitis | 6 (21%) | 2 (25%) | 12 (71%) | 0.01 | 0.05 |
| Food allergies | 6 (21%) | 4 (40%) | 11 (65%) | 0.02 | <0.01 |
| Clinical symptoms (n, %) | |||||
| Dysphagia | 4 (14%) | 6 (75%) | 13 (76%) | <0.01 | <0.01 |
| Abdominal pain | 9 (32%) | 2 (25%) | 3 (18%) | 0.32 | 0.2 |
| Nausea, vomiting | 1 (0%) | 0 | 5 (29%) | 0.05 | 0.08 |
| Endoscopy findings (n, %) | |||||
| Edema | - | 0 [0–0] | 1 [1–1] | <0.01 | |
| Rings | - | 0 [0–1] | 1 [1–2] | <0.01 | |
| Furrows | - | 0 [0–1] | 1 [1–1] | <0.01 | |
| Exudates | - | 0 [0–1] | 1 [1–1] | <0.01 | |
| Structure | - | 0 [0–0] | 0 [0–0] | 0.28 | |
| EREFS | - | 2 [2–0] | 4 [4–4] | 0.02 | |
| The number of eosinophilic infiltration (median IQR: eos/hpf) | - | 0 [0–20] | 65 [24–85] | <0.01 | |
| Medication (n, %) | |||||
| Proton pump inhibitors | 6 (21%) | 7 (88%) | 4 (24%) | 0.59 | 0.27 |
| Topical steroids | 0 (0%) | 2 (25%) | 2 (12%) | <0.01 | 0.04 |
| Antiallergic medicine | 0 (0%) | 6 (75%) | 7 (41%) | <0.01 | <0.01 |
| H. pylori infection (uninfected/ exposed/currently Infected) | 22/5/1 | 5/3/0 | 14/3/0 | 0.85 | 0.6 |
| Cytokines expression in esophageal mucosa | |||||
| IL-4 | 12.5 (2.3–111.3) | 240.2 (159.4–333.7) | 8.4 (5.6–12.4) | 0.03 | 0.53 |
| IL-13 | 1.0 (0.53–17.1) | 19.6 (7.9–29.4) | 1.5 (0.74–2.4) | 0.02 | 0.26 |
| IL-33 | 1.7 (1.0–12.6) | 26.4 (4.8–84.9) | 3.2 (2.9–3.3) | 0.02 | 0.25 |
| TGF-beta1 | 2.0 (1.1–6.1) | 3.5 (2.0–5.2) | 4.0 (2.4–9.7) | 0.65 | 0.19 |
| TSLP | 2.9 (1.8–13.5) | 8.9 (5.5–15.5) | 3.4 (2.3–4.6) | 0.58 | 0.80 |
| CCL26 | 1.4 (0.97–6.7) | 42.0 (21.2–113.4) | 0.38 (0.11–0.49) | 0.01 | 0.11 |
| CXCL16 | 1.0 (0.72–1.9) | 6.0 (2.5–25.2) | 1.2 (0.69–2.4) | 0.03 | 0.76 |
| Target Gene | Sequence | |
|---|---|---|
| IL-4 | FP | ACATCTTTGCTGCCTCCAA |
| RP | AGGCAGCGAGTGTCCTTCT | |
| IL-13 | FP | GATTCTGCCCGCACAAGGT |
| RP | CGATTTTGGTGTCTCGGACAT | |
| IL-33 | FP | GGCTGAGAATTACCATACAAGG |
| RP | AGTGTTTTTCAGATGGGATGA | |
| TGFβ1 | FP | CGCGTGCTAATGGTGGAAA |
| RP | TGTGTGTACTCTGCTTGAACTTG | |
| CCL26 | FP | TCCCTCCTGAGTCTCCACCTT |
| RP | CTGTATTGGAAGCAGCAGGTCTT | |
| CXCL16 | FP | CCTATGTGCTGTGCAAGAGGAG |
| RP | CTGGGCAACATAGAGTCCGTCT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sasahira, M.; Matsumoto, H.; Go, T.T.; Yo, S.; Monden, S.; Ninomiya, T.; Oosawa, M.; Handa, O.; Umegaki, E.; Inoue, R.; et al. The Relationship Between Bacterial Flora in Saliva and Esophageal Mucus and Endoscopic Severity in Patients with Eosinophilic Esophagitis. Int. J. Mol. Sci. 2025, 26, 3026. https://doi.org/10.3390/ijms26073026
Sasahira M, Matsumoto H, Go TT, Yo S, Monden S, Ninomiya T, Oosawa M, Handa O, Umegaki E, Inoue R, et al. The Relationship Between Bacterial Flora in Saliva and Esophageal Mucus and Endoscopic Severity in Patients with Eosinophilic Esophagitis. International Journal of Molecular Sciences. 2025; 26(7):3026. https://doi.org/10.3390/ijms26073026
Chicago/Turabian StyleSasahira, Momoyo, Hiroshi Matsumoto, Tei Tei Go, Shogen Yo, Shuzo Monden, Takahirao Ninomiya, Motoyasu Oosawa, Osamu Handa, Eiji Umegaki, Ryo Inoue, and et al. 2025. "The Relationship Between Bacterial Flora in Saliva and Esophageal Mucus and Endoscopic Severity in Patients with Eosinophilic Esophagitis" International Journal of Molecular Sciences 26, no. 7: 3026. https://doi.org/10.3390/ijms26073026
APA StyleSasahira, M., Matsumoto, H., Go, T. T., Yo, S., Monden, S., Ninomiya, T., Oosawa, M., Handa, O., Umegaki, E., Inoue, R., & Shiotani, A. (2025). The Relationship Between Bacterial Flora in Saliva and Esophageal Mucus and Endoscopic Severity in Patients with Eosinophilic Esophagitis. International Journal of Molecular Sciences, 26(7), 3026. https://doi.org/10.3390/ijms26073026






