On the Significance of the Terminal Location of Prion-Forming Regions of Yeast Proteins
Abstract
1. Introduction
2. Results
2.1. N-Terminal Extensions to Sup35 Unpredictably Alter Prion Phenotype
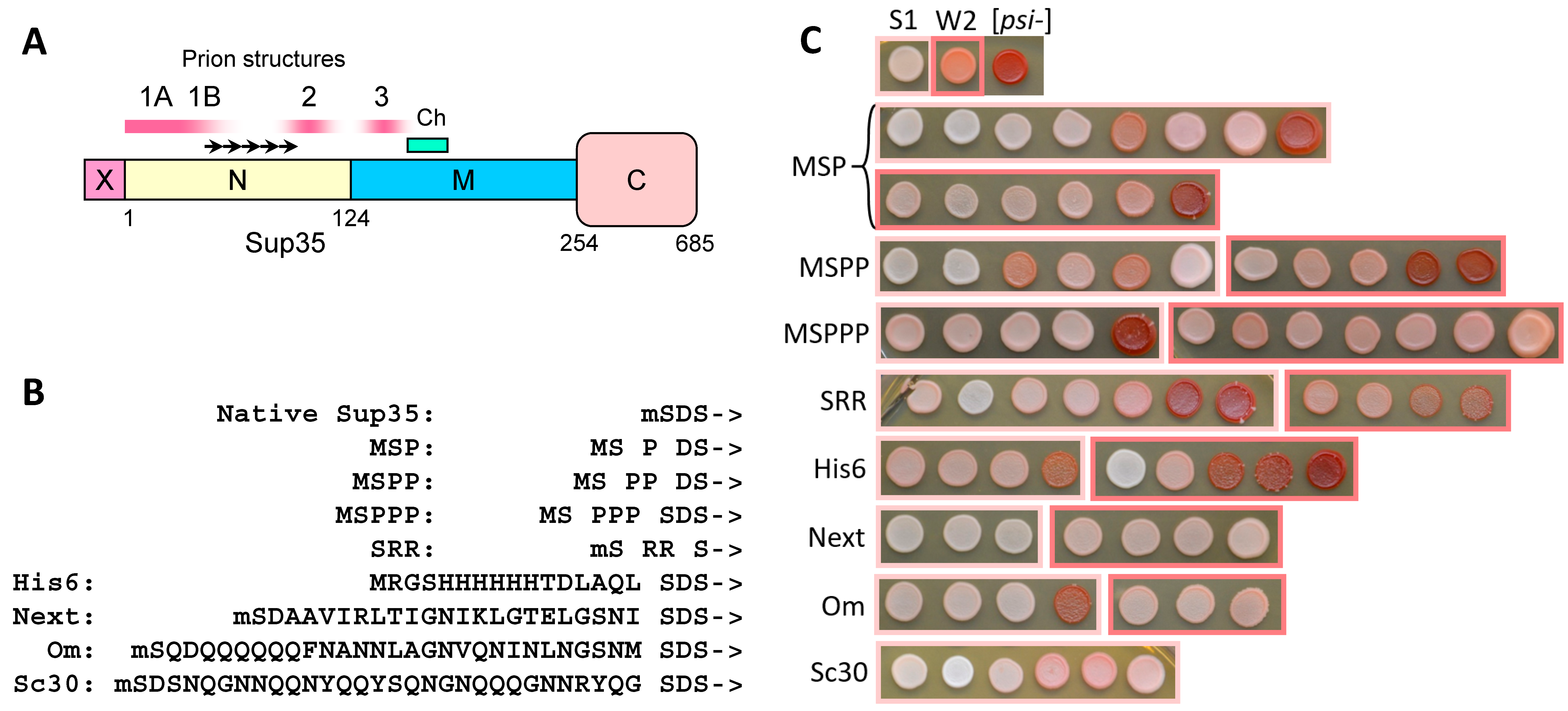
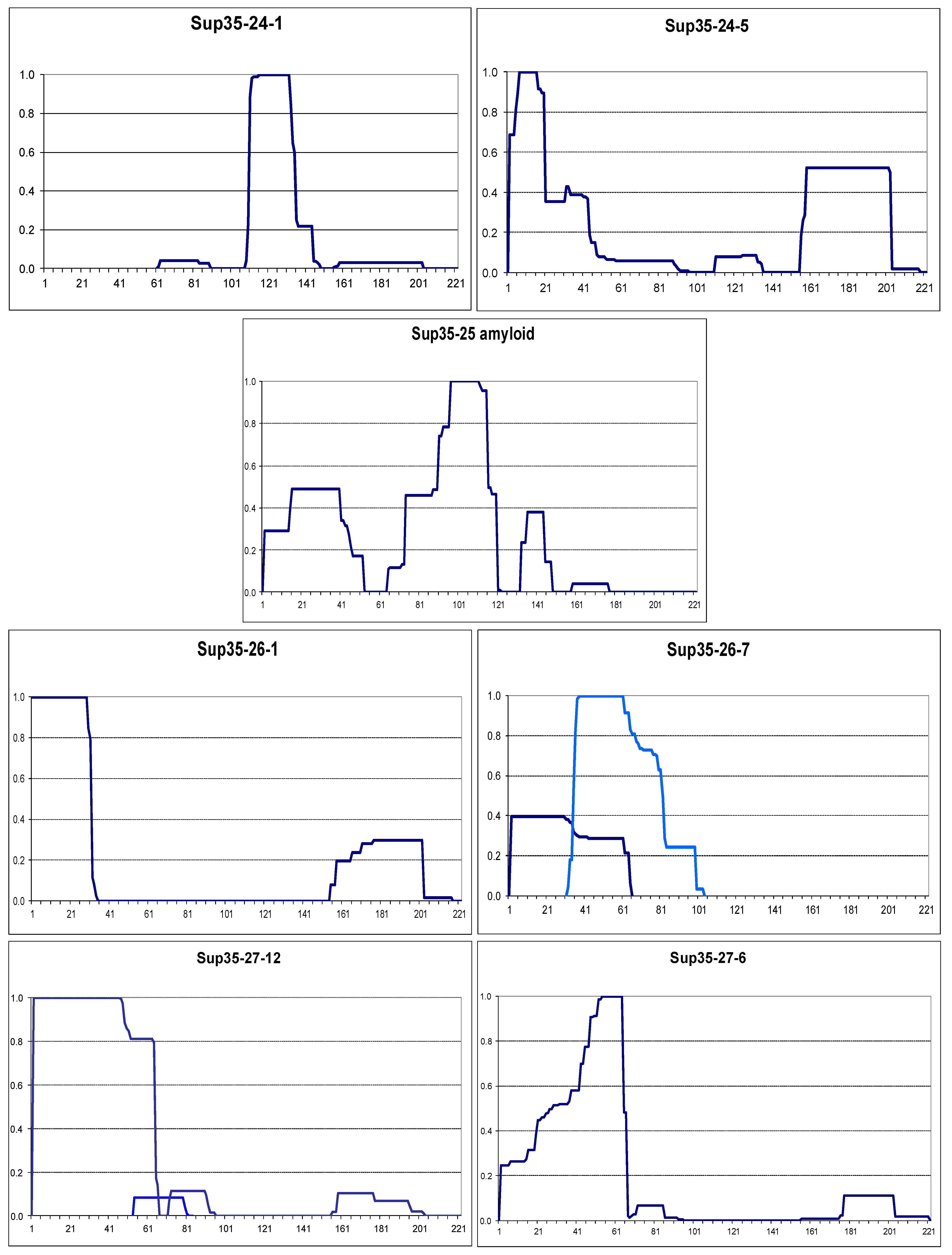
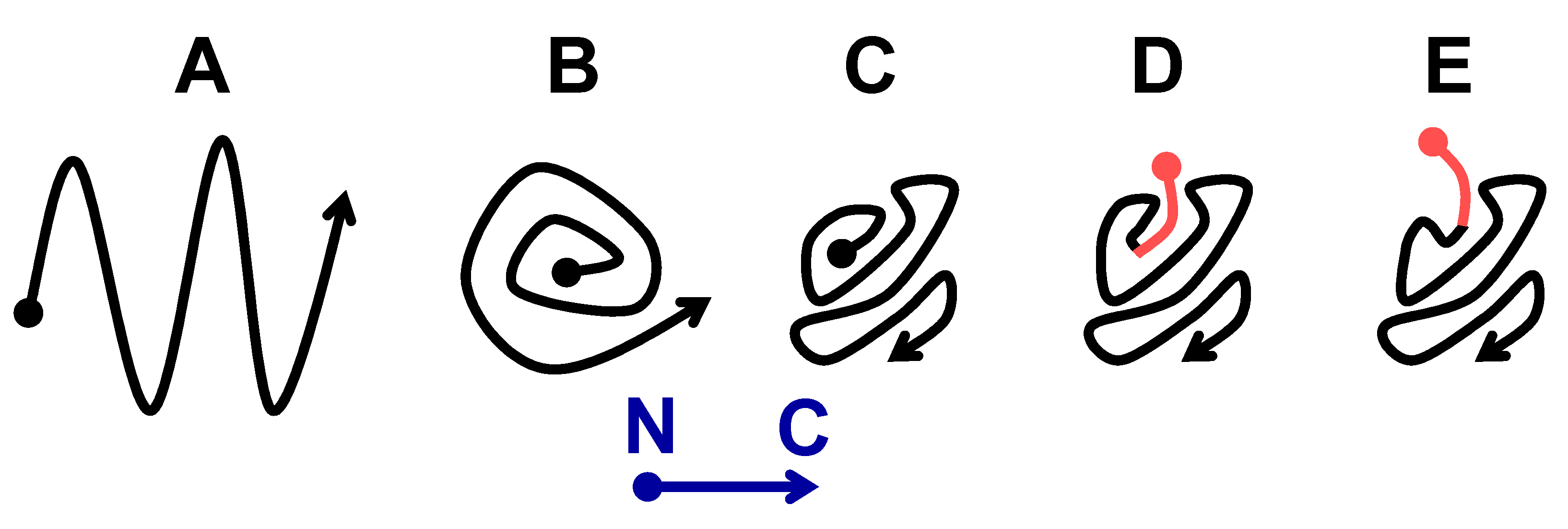
2.2. The Larger N-Terminal Extensions Cause Major Alteration in Amyloid Structure
2.2.1. General Notes
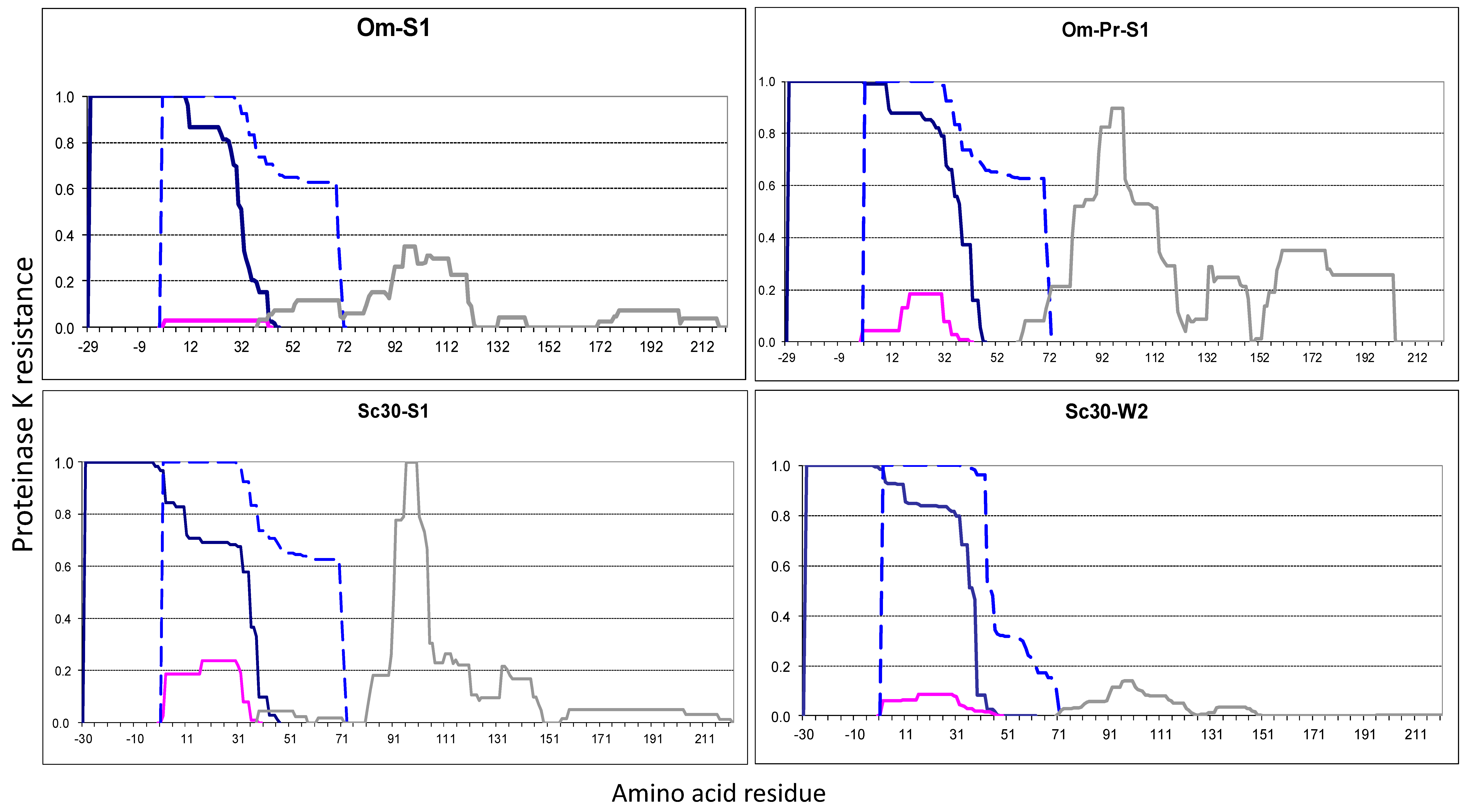
2.2.2. The Om and Sc30 Extensions Dramatically Alter the Original Prion Fold
2.3. N-Terminal Random Poorly Amyloidogenic Extensions Can Acquire Amyloid Fold
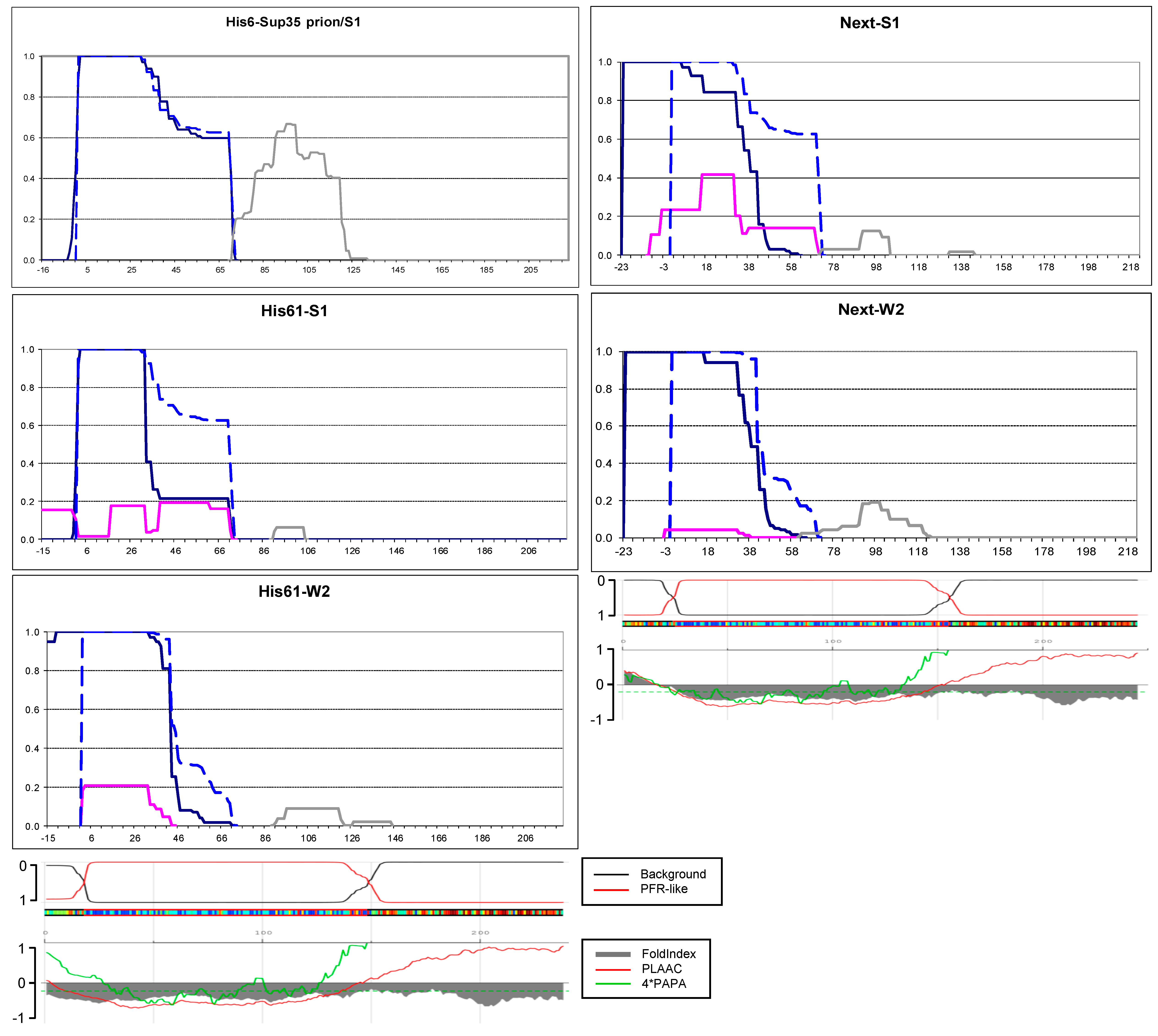
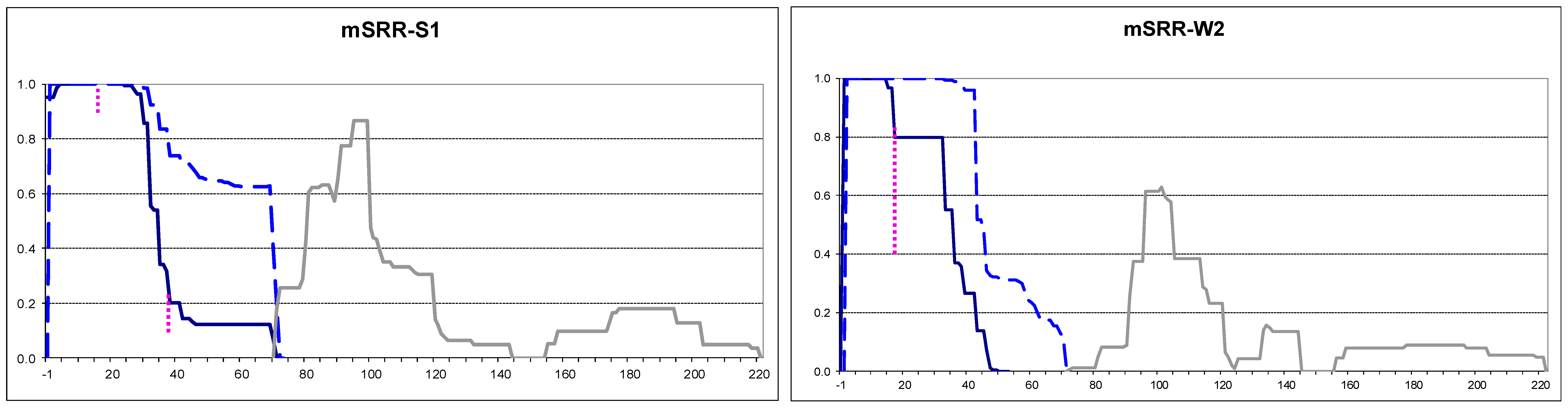
2.4. Small Extensions Can Alter Prion Structure
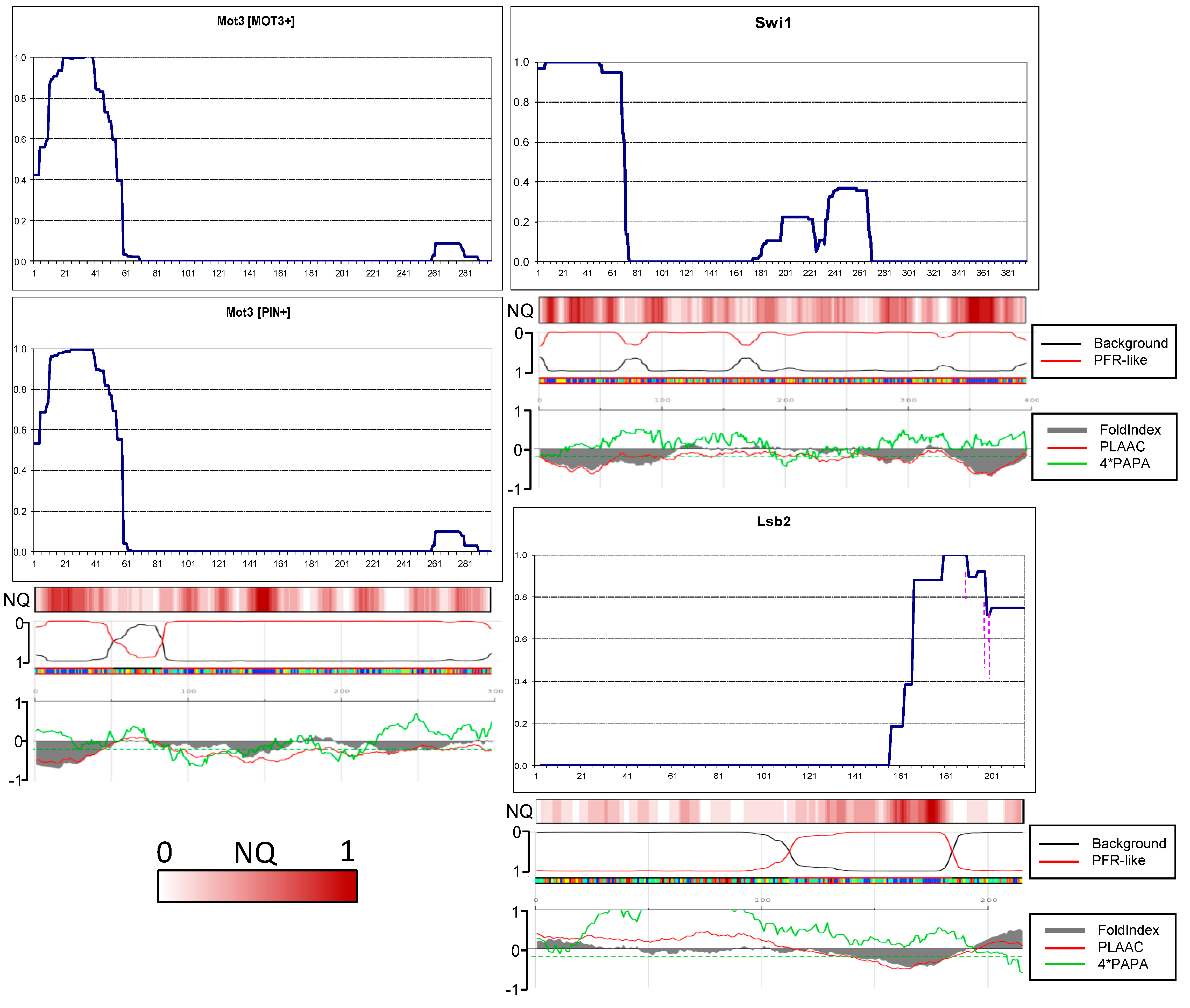
2.5. Terminal Location Is Insufficient for Prion Structure Formation by QN-Rich Sequences Despite Increasing Its Probability
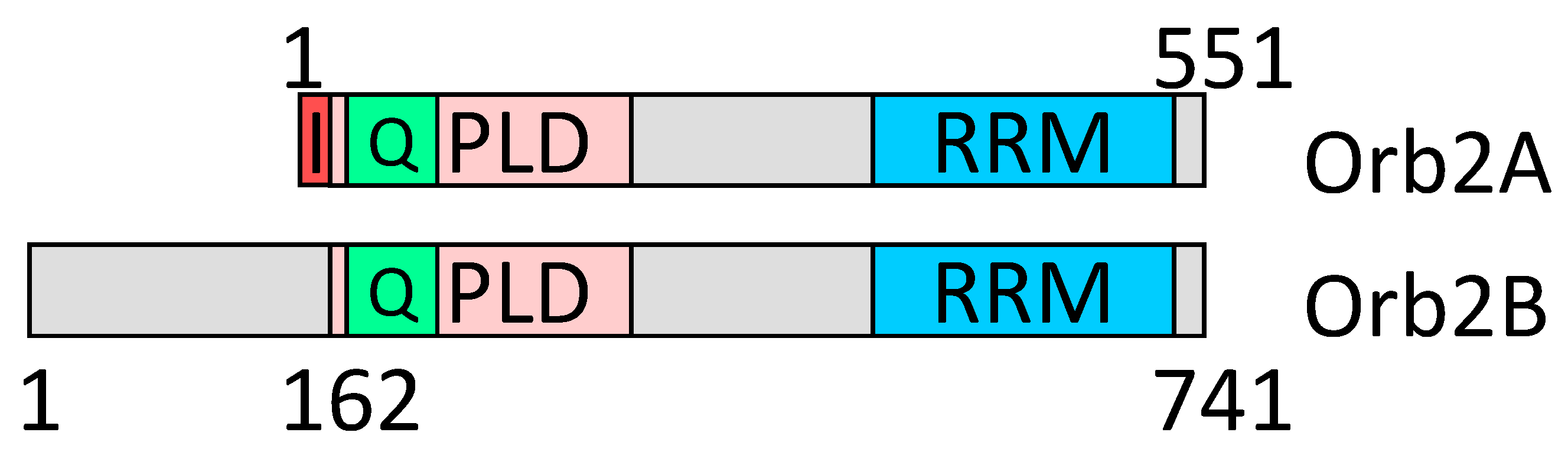
2.6. Amyloid Structures of Prion and Candidate Prion Proteins
3. Discussion
4. Materials and Methods
4.1. Yeast Strains and Media
4.2. Plasmids
4.3. Obtaining of Prions and Genetic Procedures
4.4. Western Blotting
4.5. PK Digestion, Mass Spectrometry and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PK | Proteinase K |
| MALDI-TOF | Matrix-assisted Laser Desorption/Ionization Time of Flight |
References
- Halfmann, R.; Wright, J.R.; Alberti, S.; Lindquist, S.; Rexach, M. Prion formation by a yeast GLFG nucleoporin. Prion 2012, 6, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Zhouravleva, G.A.; Bondarev, S.A.; Trubitsina, N.P. How Big Is the Yeast Prion Universe? Int. J. Mol. Sci. 2023, 24, 11651. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.C.S.; Lindquist, S. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev. 2009, 23, 2320–2332. [Google Scholar] [CrossRef] [PubMed]
- Chakrabortee, S.; Byers, J.S.; Jones, S.; Garcia, D.M.; Bhullar, B.; Chang, A.; She, R.; Lee, L.; Fremin, B.; Lindquist, S.; et al. Intrinsically Disordered Proteins Drive Emergence and Inheritance of Biological Traits. Cell 2016, 167, 369–381.e12. [Google Scholar] [CrossRef]
- Roberts, B.T.; Wickner, R.B. Heritable activity: A prion that propagates by covalent autoactivation. Genes Dev. 2003, 17, 2083–2087. [Google Scholar] [CrossRef]
- Suzuki, G.; Shimazu, N.; Tanaka, M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 2012, 336, 355–359. [Google Scholar] [CrossRef]
- Harrison, P.M. Intrinsically Disordered Compositional Bias in Proteins: Sequence Traits, Region Clustering, and Generation of Hypothetical Functional Associations. Bioinform. Biol. Insights 2024, 18, 11779322241287484. [Google Scholar] [CrossRef]
- Fomicheva, A.; Ross, E.D. From Prions to Stress Granules: Defining the Compositional Features of Prion-Like Domains That Promote Different Types of Assemblies. Int. J. Mol. Sci. 2021, 22, 1251. [Google Scholar] [CrossRef]
- Wickner, R.B. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science 1994, 264, 566–569. [Google Scholar] [CrossRef]
- Dergalev, A.A.; Alexandrov, A.I.; Ivannikov, R.I.; Ter-Avanesyan, M.D.; Kushnirov, V.V. Yeast Sup35 Prion Structure: Two Types, Four Parts, Many Variants. Int. J. Mol. Sci. 2019, 20, 2633. [Google Scholar] [CrossRef]
- Ohhashi, Y.; Yamaguchi, Y.; Kurahashi, H.; Kamatari, Y.O.; Sugiyama, S.; Uluca, B.; Piechatzek, T.; Komi, Y.; Shida, T.; Müller, H.; et al. Molecular basis for diversification of yeast prion strain conformation. Proc. Natl. Acad. Sci. USA 2018, 115, 2389–2394. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.-H.H.; Komi, Y.; Nakagawa, Y.; Kamatari, Y.O.; Nomura, T.; Kimura, H.; Shida, T.; Burke, J.; Tamai, S.; Ishida, Y.; et al. Exposed Hsp70-binding site impacts yeast Sup35 prion disaggregation and propagation. Proc. Natl. Acad. Sci. USA 2024, 121, e2318162121. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-W.; King, C.-Y. A complete catalog of wild-type Sup35 prion variants and their protein-only propagation. Curr. Genet. 2020, 66, 97–122. [Google Scholar] [CrossRef] [PubMed]
- Dagkesamanskaia, A.R.; Kushnirov, V.V.; Paushkin, S.V.; Ter-Avanesian, M.D. Fusion of glutathione S-transferase with the N-terminus of yeast Sup35p protein inhibits its prion-like properties. Genetika 1997, 33, 610–615. [Google Scholar]
- Galliamov, A.A.; Malukhina, A.D.; Kushnirov, V.V. Mapping of Prion Structures in the Yeast Rnq1. Int. J. Mol. Sci. 2024, 25, 3397. [Google Scholar] [CrossRef]
- Dergalev, A.A.; Urakov, V.N.; Agaphonov, M.O.; Alexandrov, A.I.; Kushnirov, V. V Dangerous Stops: Nonsense Mutations Can Dramatically Increase Frequency of Prion Conversion. Int. J. Mol. Sci. 2021, 22, 1542. [Google Scholar] [CrossRef]
- Tanaka, H.; Murata, K.; Hashimoto, W.; Kawai, S. Hsp104-dependent ability to assimilate mannitol and sorbitol conferred by a truncated Cyc8 with a C-terminal polyglutamine in Saccharomyces cerevisiae. PLoS ONE 2020, 15, e0242054. [Google Scholar] [CrossRef]
- Alberti, S.; Halfmann, R.; King, O.; Kapila, A.; Lindquist, S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009, 137, 146–158. [Google Scholar] [CrossRef]
- Crow, E.T.; Du, Z.; Li, L. A Small, Glutamine-Free Domain Propagates the [SWI+] Prion in Budding Yeast. Mol. Cell. Biol. 2011, 31, 3436–3444. [Google Scholar] [CrossRef]
- Chang, H.-Y.; Lin, J.-Y.; Lee, H.-C.; Wang, H.-L.; King, C.-Y. Strain-specific sequences required for yeast [PSI+] prion propagation. Proc. Natl. Acad. Sci. USA 2008, 105, 13345–13350. [Google Scholar] [CrossRef]
- Depace, A.H.; Santoso, A.; Hillner, P.; Weissman, J.S. A Critical Role for Amino-Terminal Glutamine/Asparagine Repeats in the Formation and Propagation of a Yeast Prion. Cell 1998, 93, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Goncharoff, D.K.; Cabral, R.; Applebey, S.V.; Pagadala, M.; Du, Z.; Li, L. Defining Key Residues of the Swi1 Prion Domain in Prion Formation and Maintenance. Mol. Cell. Biol. 2021, 41, e0004421. [Google Scholar] [CrossRef] [PubMed]
- Ross, E.D.; Baxa, U.; Wickner, R.B. Scrambled Prion Domains Form Prions and Amyloid. Mol. Cell. Biol. 2004, 24, 7206–7213. [Google Scholar] [CrossRef] [PubMed]
- Kushnirov, V.V.; Kochneva-Pervukhova, N.V.; Chechenova, M.B.; Frolova, N.S.; Ter-Avanesyan, M.D. Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J. 2000, 19, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Chernoff, Y.O.; Galkin, A.P.; Lewitin, E.; Chernova, T.A.; Newnam, G.P.; Belenkiy, S.M. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol. Microbiol. 2000, 35, 865–876. [Google Scholar] [CrossRef]
- Krishnan, R.; Lindquist, S.L. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 2005, 435, 765–772. [Google Scholar] [CrossRef]
- Ross, E.D.; Edskes, H.K.; Terry, M.J.; Wickner, R.B. Primary sequence independence for prion formation. Proc. Natl. Acad. Sci. USA 2005, 102, 12825–12830. [Google Scholar] [CrossRef]
- Grishin, A.V.; Rothenberg, M.; Downs, M.A.; Blumer, K.J. Mot3, a Zn Finger Transcription Factor That Modulates Gene Expression and Attenuates Mating Pheromone Signaling in Saccharomyces cerevisiae. Genetics 1998, 149, 879–892. [Google Scholar] [CrossRef]
- Peterson, C.L.; Herskowitz, I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 1992, 68, 573–583. [Google Scholar] [CrossRef]
- Spiess, M.; de Craene, J.-O.; Michelot, A.; Rinaldi, B.; Huber, A.; Drubin, D.G.; Winsor, B.; Friant, S. Lsb1 is a negative regulator of las17 dependent actin polymerization involved in endocytosis. PLoS ONE 2013, 8, e61147. [Google Scholar] [CrossRef]
- Chernova, T.A.; Kiktev, D.A.; Romanyuk, A.V.; Shanks, J.R.; Laur, O.; Ali, M.; Ghosh, A.; Kim, D.; Yang, Z.; Mang, M.; et al. Yeast Short-Lived Actin-Associated Protein Forms a Metastable Prion in Response to Thermal Stress. Cell Rep. 2017, 18, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.S.; Belmont, B.J.; Sante, J.M.; Rexach, M.F. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 2007, 129, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Otto, T.A.; Bergsma, T.; Dekker, M.; Mouton, S.N.; Gallardo, P.; Wolters, J.C.; Steen, A.; Onck, P.R.; Veenhoff, L.M. Nucleoporin Nsp1 surveils the phase state of FG-Nups. Cell Rep. 2024, 43, 114793. [Google Scholar] [CrossRef] [PubMed]
- Bidlingmaier, S.; Weiss, E.L.; Seidel, C.; Drubin, D.G.; Snyder, M. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 2449–2462. [Google Scholar] [CrossRef]
- Jonischkies, K.; Del Angel, M.; Demiray, Y.E.; Loaiza Zambrano, A.; Stork, O. The NDR family of kinases: Essential regulators of aging. Front. Mol. Neurosci. 2024, 17, 1371086. [Google Scholar] [CrossRef]
- Giolito, M.L.; Bigliani, G.; Meinero, R.; Taubas, J.V. Palmitoylation of CYSTM (CYSPD) proteins in yeast. J. Biol. Chem. 2024, 300, 105609. [Google Scholar] [CrossRef]
- Kajava, A.V.; Baxa, U.; Wickner, R.B.; Steven, A.C. A model for Ure2p prion filaments and other amyloids: The parallel superpleated beta-structure. Proc. Natl. Acad. Sci. USA 2004, 101, 7885–7890. [Google Scholar] [CrossRef]
- Shewmaker, F.; Kryndushkin, D.; Chen, B.; Tycko, R.; Wickner, R.B. Two prion variants of Sup35p have in-register parallel β-sheet structures, independent of hydration. Biochemistry 2009, 48, 5074–5082. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Hervas, R.; Rau, M.J.; Park, Y.; Zhang, W.; Murzin, A.G.; Fitzpatrick, J.A.J.; Scheres, S.H.W.; Si, K. Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila. Science 2020, 367, 1230–1234. [Google Scholar] [CrossRef]
- Majumdar, A.; Cesario, W.C.C.; White-Grindley, E.; Jiang, H.; Ren, F.; Khan, M.R.; Li, L.; Choi, E.M.-L.; Kannan, K.; Guo, F.; et al. Critical Role of Amyloid-like Oligomers of Drosophila Orb2 in the Persistence of Memory. Cell 2012, 148, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Alieva, M.K.; Nikishina, S.B.; Kireev, I.I.; Golyshev, S.A.; Tyurin-Kuzmin, P.A.; Ivanova, L.V.; Alexandrov, A.I.; Kushnirov, V.V.; Dergalev, A.A. A liquid-to-solid phase transition of biomolecular condensates drives in vivo formation of yeast amyloids and prions. bioRxiv 2023. [Google Scholar] [CrossRef]
- Kushnirov, V.V.; Kryndushkin, D.S.; Boguta, M.; Smirnov, V.N.; Ter-Avanesyan, M.D. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol. 2000, 10, 1443–1446. [Google Scholar] [CrossRef] [PubMed]

| Residue | # | S1 | W2 | [PIN+] |
|---|---|---|---|---|
| M | −4 | 0 | 0 | 0 |
| S | −3 | 0.94 | 0.96 | 0.29 |
| P | −2 | 1 | 1 | 0.66 |
| P | −1 | 1 | 1 | 0.94 |
| P | 1 | 1 | 1 | 1 |
| S | 2 | 1 | 1 | 1 |
| D | 3 | 1 | 1 | 1 |
| S | 4 | 1 | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galliamov, A.A.; Urakov, V.N.; Dergalev, A.A.; Kushnirov, V.V. On the Significance of the Terminal Location of Prion-Forming Regions of Yeast Proteins. Int. J. Mol. Sci. 2025, 26, 1637. https://doi.org/10.3390/ijms26041637
Galliamov AA, Urakov VN, Dergalev AA, Kushnirov VV. On the Significance of the Terminal Location of Prion-Forming Regions of Yeast Proteins. International Journal of Molecular Sciences. 2025; 26(4):1637. https://doi.org/10.3390/ijms26041637
Chicago/Turabian StyleGalliamov, Arthur A., Valery N. Urakov, Alexander A. Dergalev, and Vitaly V. Kushnirov. 2025. "On the Significance of the Terminal Location of Prion-Forming Regions of Yeast Proteins" International Journal of Molecular Sciences 26, no. 4: 1637. https://doi.org/10.3390/ijms26041637
APA StyleGalliamov, A. A., Urakov, V. N., Dergalev, A. A., & Kushnirov, V. V. (2025). On the Significance of the Terminal Location of Prion-Forming Regions of Yeast Proteins. International Journal of Molecular Sciences, 26(4), 1637. https://doi.org/10.3390/ijms26041637







