Characterization of Tetrathionate Hydrolase from Acidothermophilic Sulfur-Oxidizing Archaeon Metallosphaera cuprina Ar-4
Abstract
1. Introduction
2. Results
2.1. Gene Sequence Analysis and Structure Prediction of Tetrathionate Hydrolase of M. cuprina
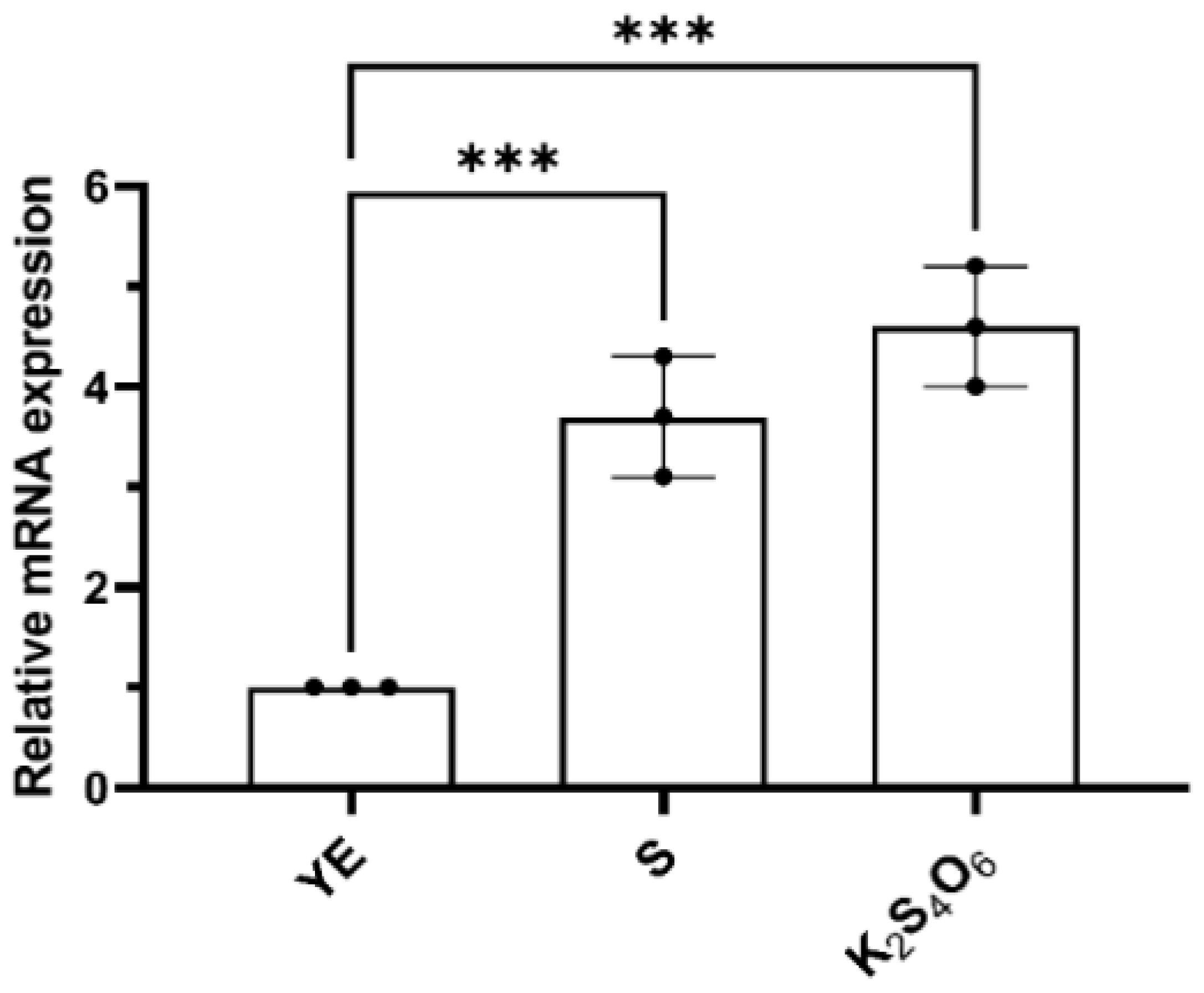
2.2. The TTH Gene Is Expressed Both in Tetrathionate and S0 Grown Cells of M. cuprina
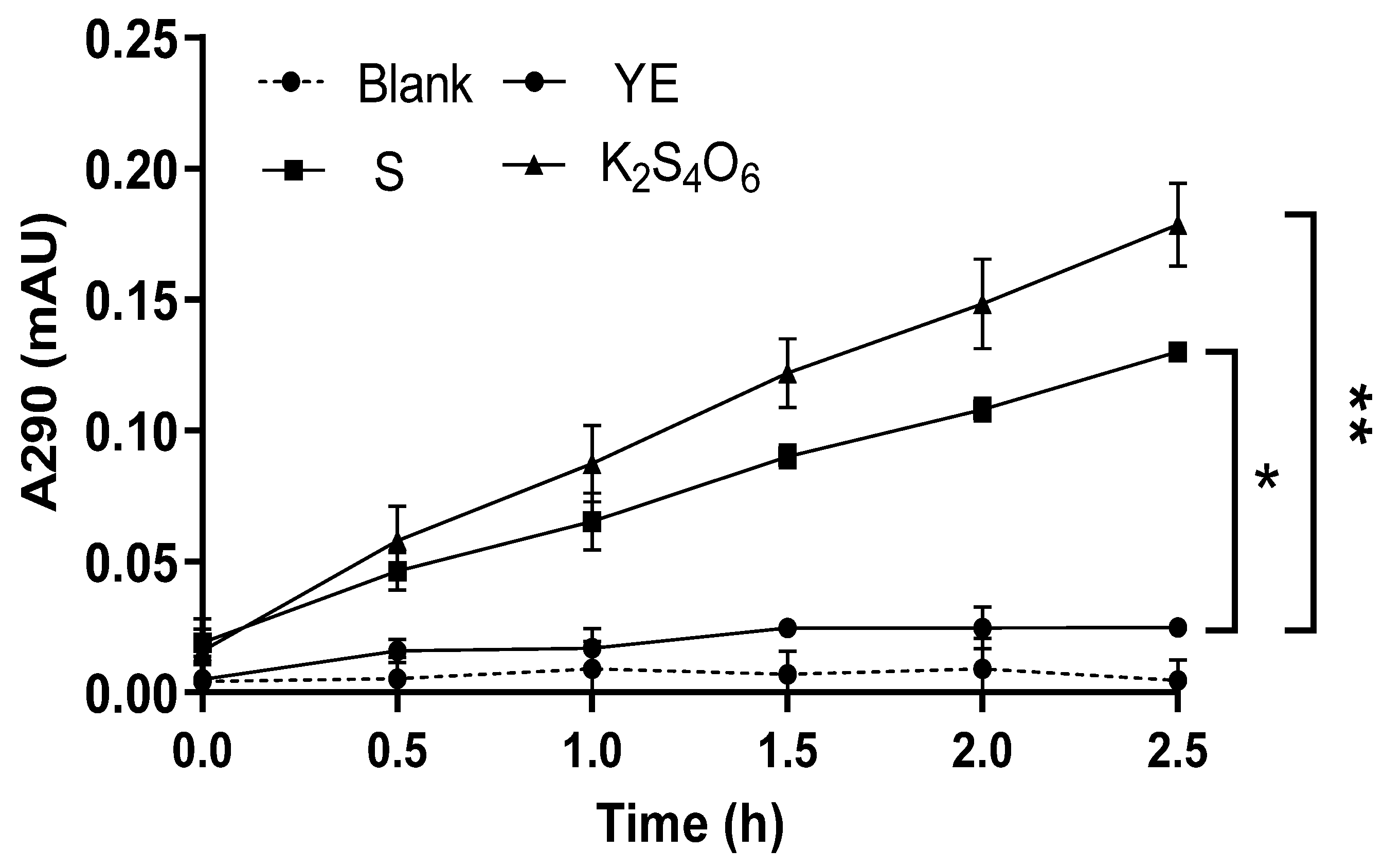
2.3. Preliminary Study on TTH Activity of M. cuprina
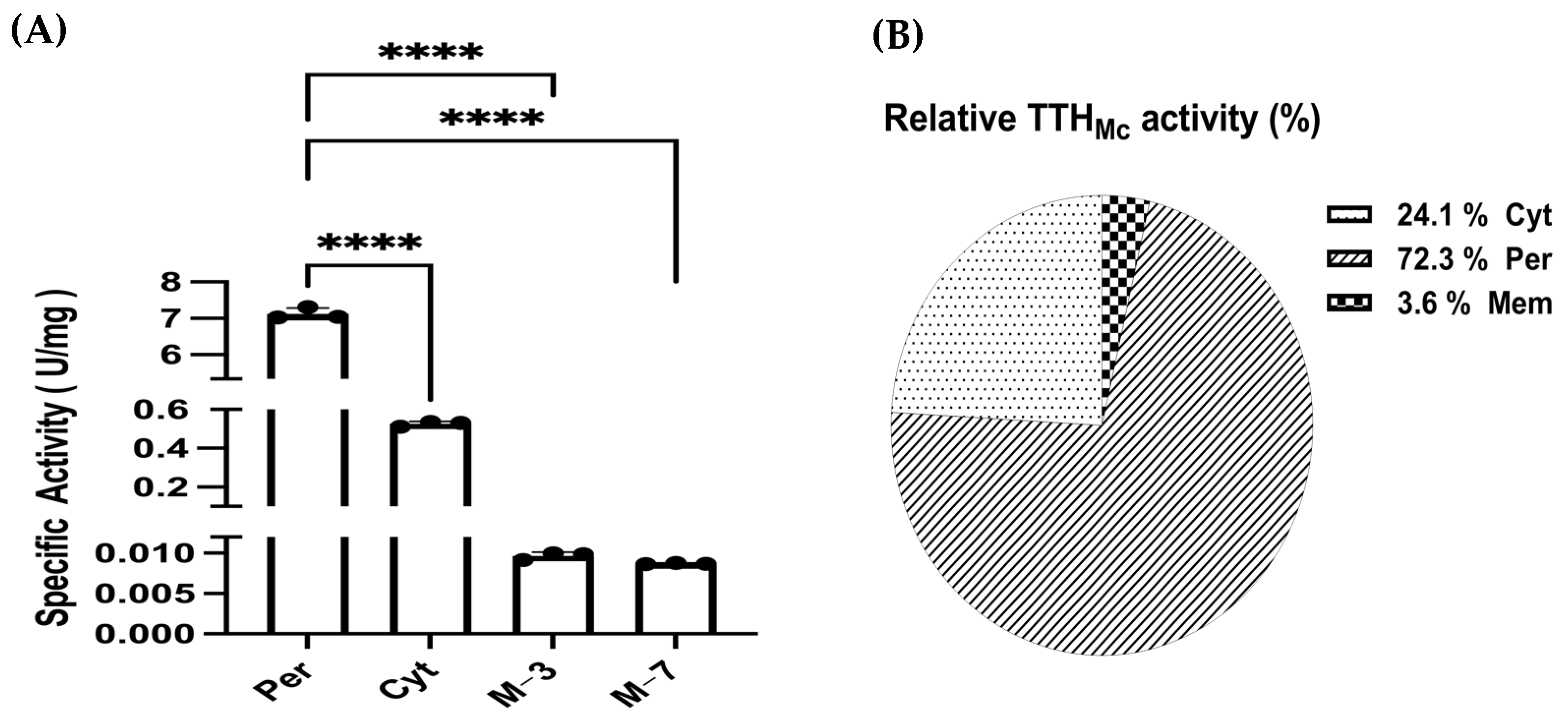
2.4. Tetrathionate Hydrolase Primarily Located in Cytoplasm and Periplasm
2.5. Purification of the Tetrathionate Hydrolase from Whole Cell
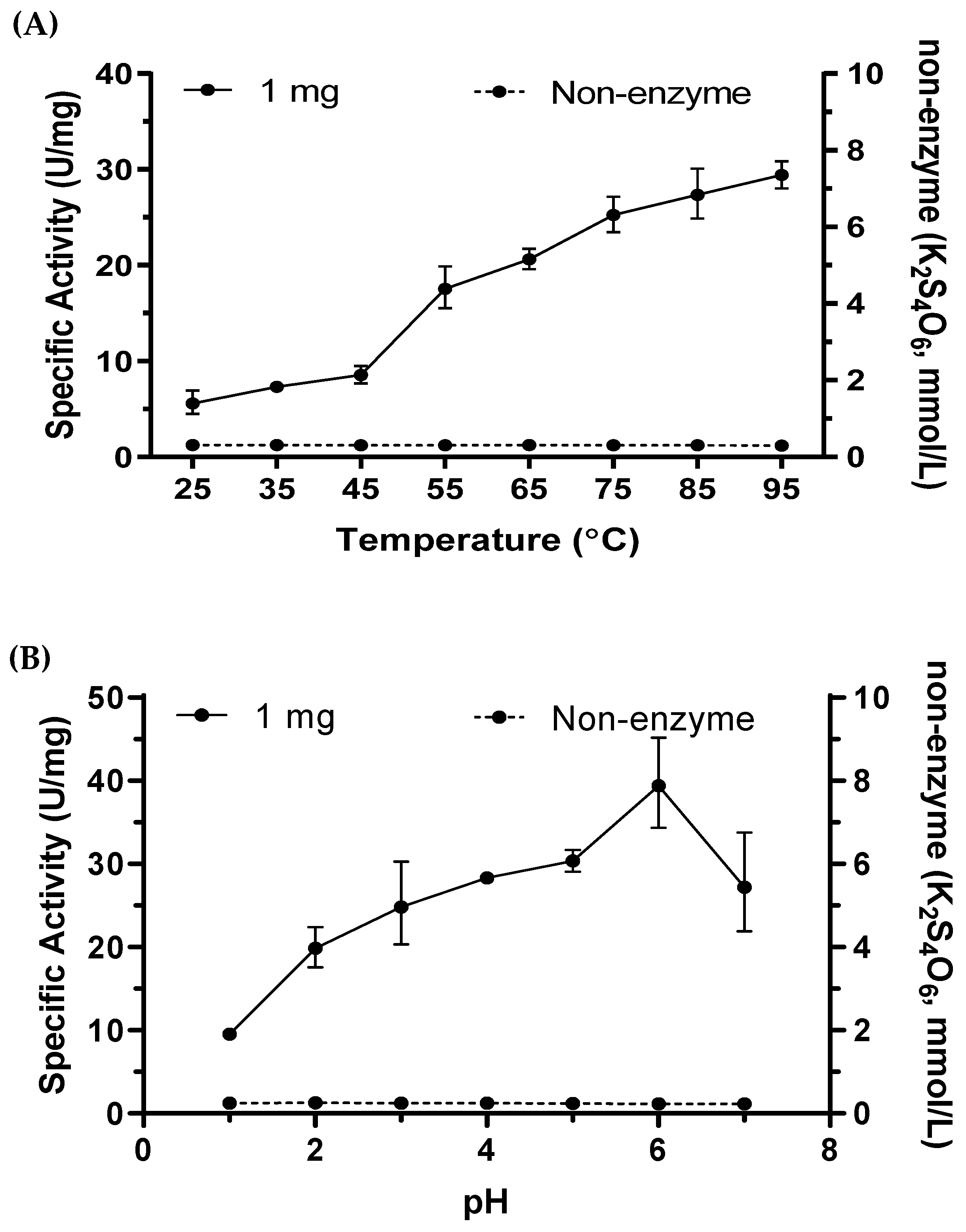
2.6. Tetrathionate Hydrolase of M. cuprina Is an Acid Resisting Thermophilic Enzyme
2.7. Hydrolysis Products of Tetrathionate by Tetrathionate Hydrolase
3. Discussion
3.1. TTH in Archaea and Bacteria Species
3.2. TTH Is Found in the Cytoplasm and Periplasm of K2S4O6 and S0-Grown Cells
3.3. Biochemical Properties of Tetrathionate Hydrolase
3.4. Evolutionary Insights of TTHs Functionality
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Sequence Analysis
4.3. RNA Extraction from Cells and Transcription Analysis for mcup_1281
4.4. Tetrathionate Hydrolases Purification from Whole Cells
4.5. Cell Fractionation Separation
4.6. Enzyme Characteristic Assay
4.6.1. Continuous Method of Enzyme Activity Assay
4.6.2. Discontinuous Method of Enzyme Activity Assay
4.7. Effects of Bivalent Ions on Enzyme Activity
4.8. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shi, J.C.; Huang, T.L.; Wen, G.; Liu, F.; Qiu, X.P.; Wang, B.S. The Variation Characteristic of Sulfides and VOSc in a Source Water Reservoir and Its Control Using a Water-Lifting Aerator. Int. J. Environ. Res. Public Health 2016, 13, 427. [Google Scholar] [CrossRef] [PubMed]
- Vavourakis, C.D.; Mehrshad, M.; Balkema, C.; van Hall, R.; Andrei, A.S.; Ghai, R.; Sorokin, D.Y.; Muyzer, G. Metagenomes and metatranscriptomes shed new light on the microbial-mediated sulfur cycle in a Siberian soda lake. BMC Biol. 2019, 17, 69. [Google Scholar] [CrossRef]
- Urbieta, M.S.; Rascovan, N.; Vazquez, M.P.; Donati, E. Genome analysis of the thermoacidophilic archaeon Acidianus copahuensis focusing on the metabolisms associated to biomining activities. BMC Genom. 2017, 18, 445. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Z.; Wei, G.; Yang, F.; Liu, X. In Silico Genome-Wide Analysis Reveals the Potential Links Between Core Genome of Acidithiobacillus thiooxidans and Its Autotrophic Lifestyle. Front. Microbiol. 2018, 9, 1255. [Google Scholar] [CrossRef]
- Parey, K.; Demmer, U.; Warkentin, E.; Wynen, A.; Ermler, U.; Dahl, C. Structural, biochemical and genetic characterization of dissimilatory ATP sulfurylase from Allochromatium vinosum. PLoS ONE 2013, 8, e74707. [Google Scholar] [CrossRef]
- Liu, L.J.; You, X.Y.; Guo, X.; Liu, S.J.; Jiang, C.Y. Metallosphaera cuprina sp. nov., an acidothermophilic, metal-mobilizing archaeon. Int. J. Syst. Evol. Microbiol. 2011, 61, 2395–2400. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.J.; Stockdreher, Y.; Koch, T.; Sun, S.T.; Fan, Z.; Josten, M.; Sahl, H.-G.; Wang, Q.; Luo, Y.-M.; Liu, S.-J.; et al. Thiosulfate transfer mediated by DsrE/TusA homologs from acidothermophilic sulfur-oxidizing archaeon Metallosphaera cuprina. J. Biol. Chem. 2014, 289, 26949–26959. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.J.; Liu, L.J.; Liu, C.; Yang, Z.F.; Liu, S.J.; Jiang, C.Y. Metallosphaera tengchongensis sp. nov., an acidothermophilic archaeon isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2015, 65, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B. Biomining—Biotechnologies for extracting and recovering metals from ores and waste materials. Curr. Opin. Biotechnol. 2014, 30, 24–31. [Google Scholar] [CrossRef]
- Batty, J.D.; Rorke, G.V. Development and commercial demonstration of the BioCOP™ thermophile process. Hydrometallurgy 2006, 83, 83–89. [Google Scholar] [CrossRef]
- Liu, L.J.; Jiang, Z.; Wang, P.; Qin, Y.L.; Xu, W.; Wang, Y.; Liu, S.-J.; Jiang, C.-Y. Physiology, Taxonomy, and Sulfur Metabolism of the Sulfolobales, an Order of Thermoacidophilic Archaea. Front. Microbiol. 2021, 12, 768283. [Google Scholar] [CrossRef] [PubMed]
- Auernik, K.S.; Kelly, R.M. Identification of components of electron transport chains in the extremely thermoacidophilic crenarchaeon Metallosphaera sedula through iron and sulfur compound oxidation transcriptomes. Appl. Environ. Microbiol. 2008, 74, 7723–7732. [Google Scholar] [CrossRef] [PubMed]
- Segerer, A.; Neuner, A.; Kristjansson, J.K.; Stetter, K.O. Acidianus infernus gen. nov., sp. nov., and Acidianus brierleyi Comb. nov.: Facultatively Aerobic, Extremely Acidophilic Thermophilic Sulfur-Metabolizing Archaebacteria. Int. J. Syst. Evol. Microbiol. 1986, 36, 559–564. [Google Scholar] [CrossRef]
- Brock, T.D.; Brock, K.M.; Belly, R.T.; Weiss, R.L. Sulfolobus: A new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 1972, 84, 54–68. [Google Scholar] [CrossRef]
- Protze, J.; Müller, F.; Lauber, K.; Naß, B.; Mentele, R.; Lottspeich, F.; Kletzin, A. An Extracellular Tetrathionate Hydrolase from the Thermoacidophilic Archaeon Acidianus ambivalens with an Activity Optimum at pH 1. Front. Microbiol. 2011, 2, 68. [Google Scholar] [CrossRef] [PubMed]
- Quatrini, R.; Appia-Ayme, C.; Denis, Y.; Jedlicki, E.; Holmes, D.S.; Bonnefoy, V. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genom. 2009, 10, 394. [Google Scholar] [CrossRef] [PubMed]
- Rohwerder, T.; Sand, W. The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 2003, 149, 1699–1710. [Google Scholar] [CrossRef]
- Nguyen, P.M.; Do, P.T.; Pham, Y.B.; Doan, T.O.; Nguyen, X.C.; Lee, W.K.; Nguyen, D.D.; Vadiveloo, A.; Um, M.-J.; Ngo, H.H. Roles, mechanism of action, and potential applications of sulfur-oxidizing bacteria for environmental bioremediation. Sci. Total Environ. 2022, 852, 158203. [Google Scholar] [CrossRef] [PubMed]
- Kanao, T.; Hase, N.; Nakayama, H.; Yoshida, K.; Nishiura, K.; Kosaka, M.; Kamimura, K.; Hirano, Y.; Tamada, T. Reaction mechanism of tetrathionate hydrolysis based on the crystal structure of tetrathionate hydrolase from Acidithiobacillus ferrooxidans. Protein Sci. 2021, 30, 328–338. [Google Scholar] [CrossRef]
- Chen, L.; Ren, Y.; Lin, J.; Liu, X.; Pang, X.; Lin, J. Acidithiobacillus caldus sulfur oxidation model based on transcriptome analysis between the wild type and sulfur oxygenase reductase defective mutant. PLoS ONE 2012, 7, e39470. [Google Scholar] [CrossRef]
- Gumulya, Y.; Boxall, N.J.; Khaleque, H.N.; Santala, V.; Carlson, R.P.; Kaksonen, A.H. In a quest for engineering acidophiles for biomining applications: Challenges and opportunities. Genes 2018, 9, 116. [Google Scholar] [CrossRef]
- Bugaytsova, Z.; Lindström, E.B. Localization, purification and properties of a tetrathionate hydrolase from Acidithiobacillus caldus. Eur. J. Biochem. 2004, 271, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.P.; Kelly, D. Cellular Location and Partial Purification of the ‘Thiosulphate-oxidizing Enzyme’ and ‘Trithionate Hydrolyase’ from Thiobacillus tepidarius. J. Gen. Microbiol. 1998, 134, 877–885. [Google Scholar] [CrossRef][Green Version]
- Krupovic, M.; Peixeiro, N.; Bettstetter, M.; Rachel, R.; Prangishvili, D. Archaeal Tetrathionate Hydrolase Goes Viral: Secretion of a Sulfur Metabolism Enzyme in the Form of Virus-Like Particles. Appl. Environ. Microbiol. 2012, 78, 5463–5465. [Google Scholar] [CrossRef] [PubMed]
- You, X.-Y.; Liu, C.; Wang, S.-Y.; Jiang, C.-Y.; Shah, S.A.; Prangishvili, D.; She, Q.; Liu, S.-J.; Garrett, R.A. Genomic analysis of Acidianus hospitalis W1 a host for studying crenarchaeal virus and plasmid life cycles. Extremophiles 2011, 15, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Kanao, T.; Onishi, M.; Kajitani, Y.; Hashimoto, Y.; Toge, T.; Kikukawa, H.; Kamimura, K. Characterization of tetrathionate hydrolase from the marine acidophilic sulfur-oxidizing bacterium, Acidithiobacillus thiooxidans strain SH. Biosci. Biotechnol. Biochem. 2018, 82, 152–160. [Google Scholar] [CrossRef]
- Kanao, T.; Kamimura, K.; Sugio, T. Identification of a gene encoding a tetrathionate hydrolase in Acidithiobacillus ferrooxidans. J. Biotechnol. 2007, 132, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Rzhepishevska, O.I.; Valdés, J.; Marcinkeviciene, L.; Gallardo, C.A.; Meskys, R.; Bonnefoy, V.; Holmes, D.S.; Dopson, M. Regulation of a novel Acidithiobacillus caldus gene cluster involved in metabolism of reduced inorganic sulfur compounds. Appl. Environ. Microbiol. 2007, 73, 7367–7372. [Google Scholar] [CrossRef] [PubMed]
- de Jong, G.A.H.; Hazeu, W.; Bos, P.; Kuenen, J.G. Polythionate degradation by tetrathionate hydrolase of Thiobacillus ferrooxidans. Microbiology 1997, 143, 499–504. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miura, Y.; Kawaoi, A. Determination of thiosulfate, thiocyanate and polythionates in a mixture by ion-pair chromatography with ultraviolet absorbance detection. J. Chromatogr. A 2000, 884, 81–87. [Google Scholar] [CrossRef]
- Kanao, T.; Matsumoto, C.; Shiraga, K.; Yoshida, K.; Takada, J.; Kamimura, K. Recombinant tetrathionate hydrolase from Acidithiobacillus ferrooxidans requires exposure to acidic conditions for proper folding. FEMS Microbiol. Lett. 2010, 309, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.B.; Li, Y.Q.; Lin, J.Q.; Pang, X.; Liu, X.M.; Liu, B.Q.; Wang, R.; Zhang, C.-J.; Wu, Y.; Lin, J.-Q.; et al. The Two-Component System RsrS-RsrR Regulates the Tetrathionate Intermediate Pathway for Thiosulfate Oxidation in Acidithiobacillus caldus. Front. Microbiol. 2016, 7, 1755. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Lin, J.Q.; Liu, X.M.; Pang, X.; Zhang, C.J.; Yang, C.L.; Gao, X.-Y.; Lin, C.-M.; Li, Y.-Q.; Li, Y.; et al. Sulfur Oxidation in the Acidophilic Autotrophic Acidithiobacillus spp. Front. Microbiol. 2018, 9, 3290. [Google Scholar] [CrossRef]
- Sugio, T.; Taha, T.M.; Takeuchi, F. Ferrous iron production mediated by tetrathionate hydrolase in tetrathionate-, sulfur-, and iron-grown Acidithiobacillus ferrooxidans ATCC 23270 cells. Biosci. Biotechnol. Biochem. 2009, 73, 1381–1386. [Google Scholar] [CrossRef][Green Version]
- Cao, X.; Hu, X.; Zhang, X.; Gao, S.; Ding, C.; Feng, Y.; Bao, W. Identification of metal ion binding sites based on amino acid sequences. PLoS ONE 2017, 12, e0183756. [Google Scholar] [CrossRef]
- Atkinson, H.J.; Morris, J.H.; Ferrin, T.E.; Babbitt, P.C. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE 2009, 4, e4345. [Google Scholar] [CrossRef] [PubMed]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef]
- Kalckar, H.M. The periplasmic galactose binding protein of Escherichia coli. Science 1971, 174, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Richter, O.M.; Schäfer, G. Purification and enzymic characterization of the cytoplasmic pyrophosphatase from the thermoacidophilic archaebacterium Thermoplasma acidophilum. Eur. J. Biochem. 1992, 209, 343–349. [Google Scholar] [CrossRef]
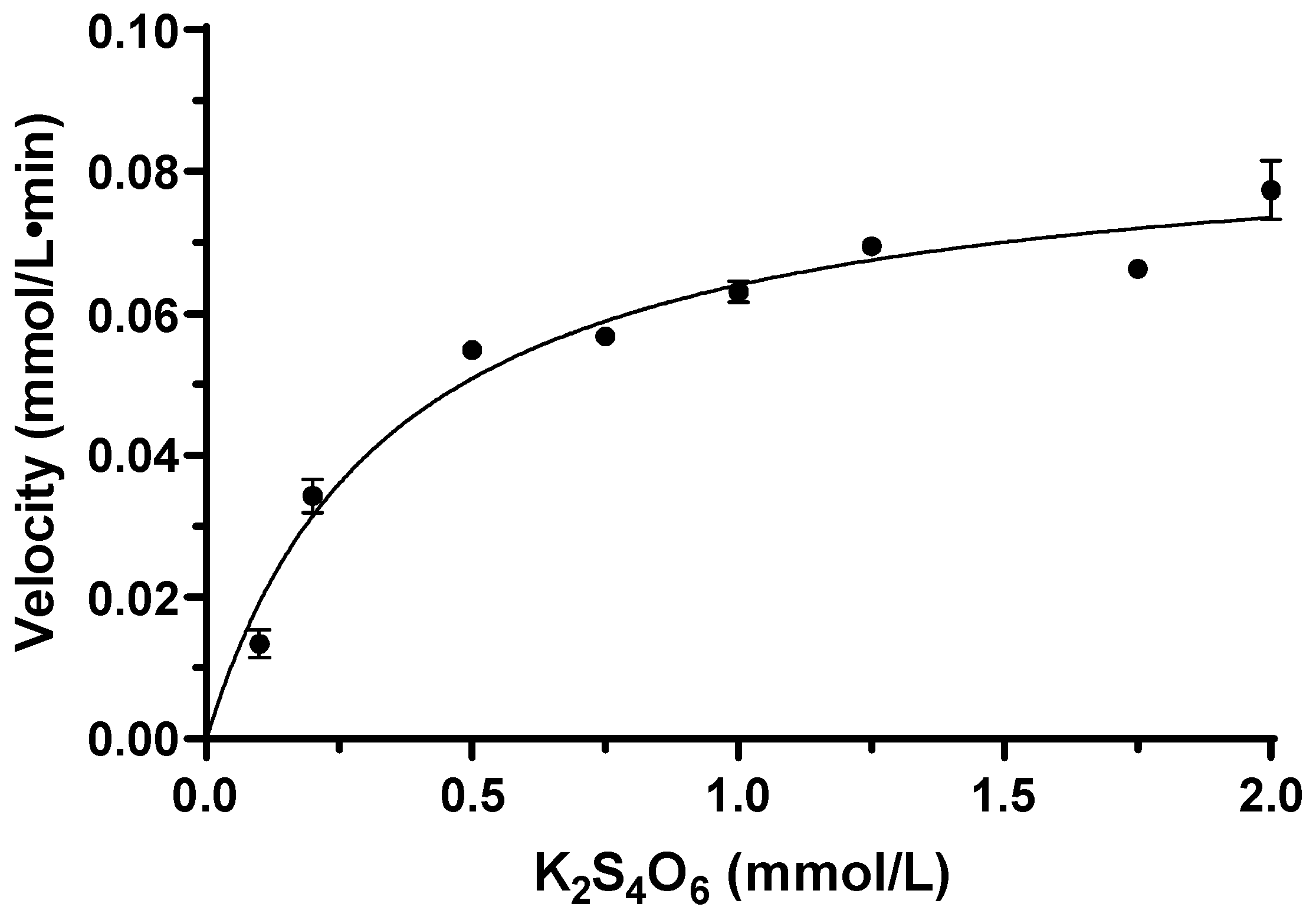
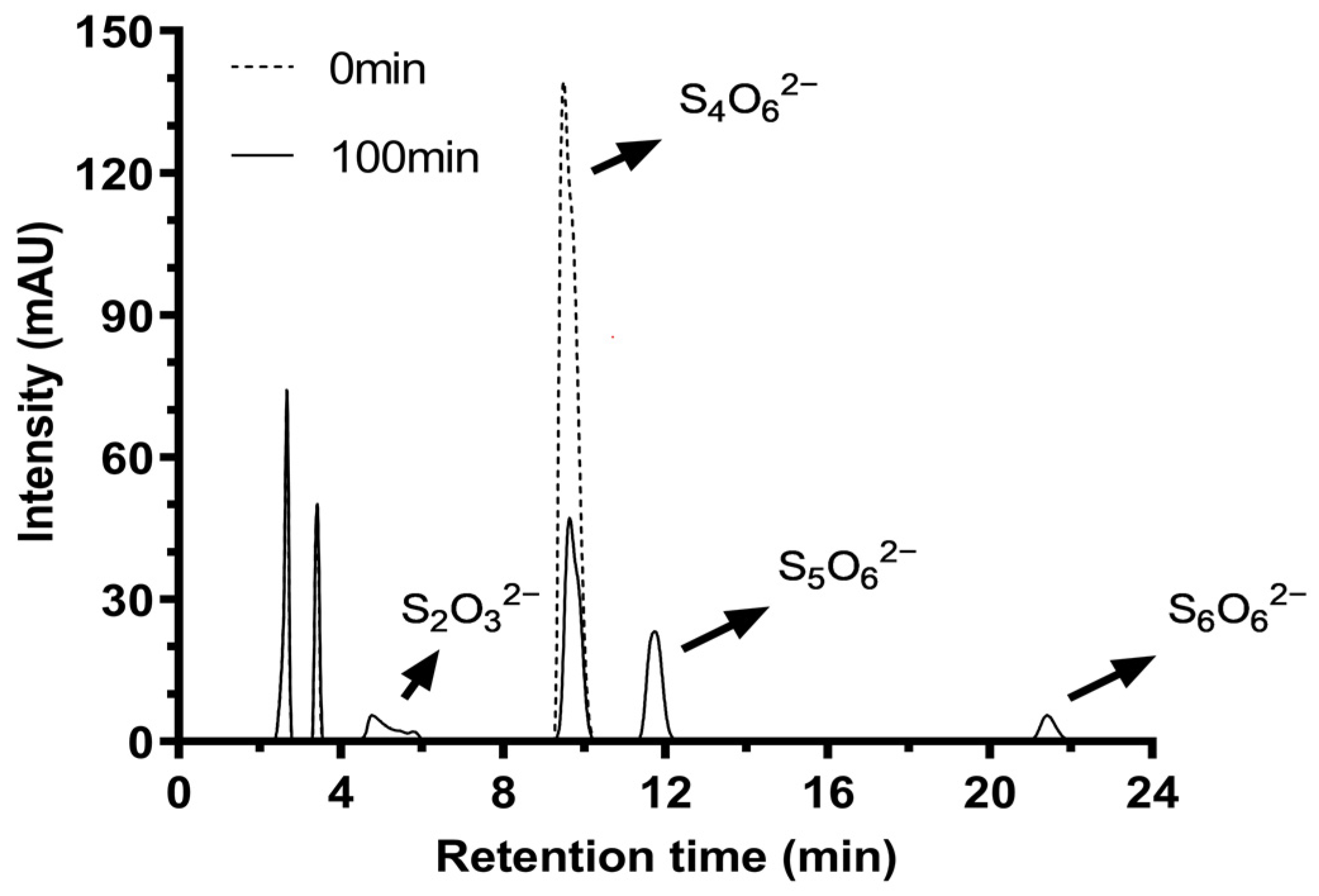
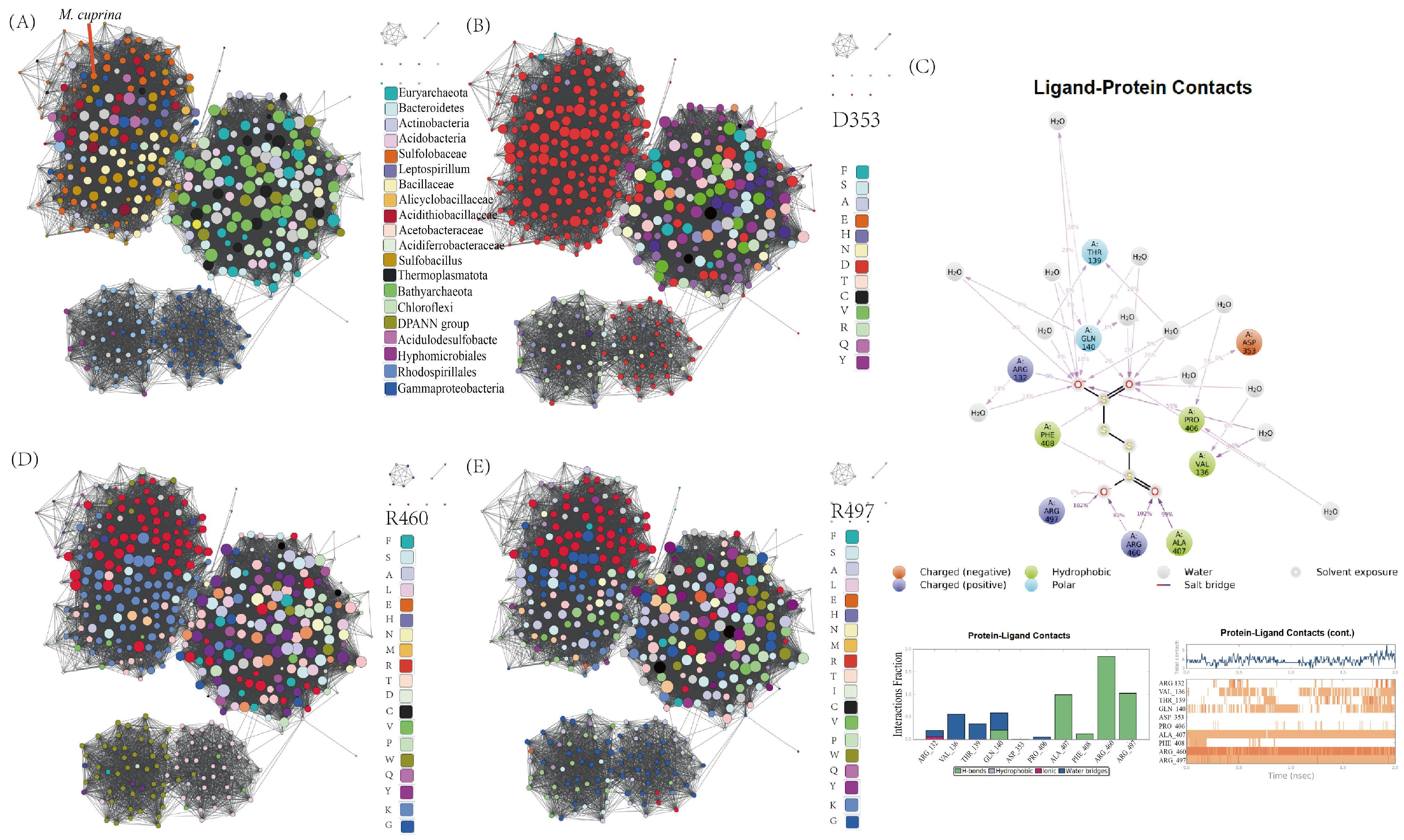
| Fraction | Total Protein (mg) a | Total Activity (U) b | Specific Activity (U/mg) c | Yield (%) | Fold |
|---|---|---|---|---|---|
| Cell extract | 79 ± 5 | 57 ± 1 | 0.7 ± 0.2 | 100.0 | 1.0 |
| DEAE | 36 ± 1 | 50 ± 6 | 1.4 ± 0.1 | 88.5 | 2.0 |
| HIC | 1.2 ± 0.04 | 2.6 ± 0.2 | 2.2 ± 0.1 | 4.6 | 3.1 |
| Size exclusion | 0.33 ± 0.01 | 1.2 ± 0.2 | 3.8 ± 0.1 | 2.2 | 5.4 |
| Q-sepharose | 0.058 ± 0.001 | 0.7 ± 0.04 | 12 ± 1 | 1.2 | 17 |
| Metal Ions | Concentration (mol/L) | Specific Activity (U/mg) | Residual Activity (%) |
|---|---|---|---|
| Control | - | 6.57 ± 0.12 | - |
| Mg2+ | 0.01 | 7.95 ± 0.04 | 121 |
| Zn2+ | 0.01 | 6.55 ± 0.12 | 94.8 |
| Fe2+ | 0.01 | 6.23 ± 0.09 | 94.5 |
| Ni2+ | 0.01 | 6.10 ± 0.00 | 92.8 |
| Mn2+ | 0.01 | 6.23 ± 0.09 | 87.7 |
| Ca2+ | 0.01 | 3.93 ± 0.05 | 59.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Li, L.-Z.; Liu, L.-J.; Qin, Y.-L.; Li, X.-T.; Yin, H.-Q.; Li, D.-F.; Liu, S.-J.; Jiang, C.-Y. Characterization of Tetrathionate Hydrolase from Acidothermophilic Sulfur-Oxidizing Archaeon Metallosphaera cuprina Ar-4. Int. J. Mol. Sci. 2025, 26, 1338. https://doi.org/10.3390/ijms26031338
Wang P, Li L-Z, Liu L-J, Qin Y-L, Li X-T, Yin H-Q, Li D-F, Liu S-J, Jiang C-Y. Characterization of Tetrathionate Hydrolase from Acidothermophilic Sulfur-Oxidizing Archaeon Metallosphaera cuprina Ar-4. International Journal of Molecular Sciences. 2025; 26(3):1338. https://doi.org/10.3390/ijms26031338
Chicago/Turabian StyleWang, Pei, Liang-Zhi Li, Li-Jun Liu, Ya-Ling Qin, Xiu-Tong Li, Hua-Qun Yin, De-Feng Li, Shuang-Jiang Liu, and Cheng-Ying Jiang. 2025. "Characterization of Tetrathionate Hydrolase from Acidothermophilic Sulfur-Oxidizing Archaeon Metallosphaera cuprina Ar-4" International Journal of Molecular Sciences 26, no. 3: 1338. https://doi.org/10.3390/ijms26031338
APA StyleWang, P., Li, L.-Z., Liu, L.-J., Qin, Y.-L., Li, X.-T., Yin, H.-Q., Li, D.-F., Liu, S.-J., & Jiang, C.-Y. (2025). Characterization of Tetrathionate Hydrolase from Acidothermophilic Sulfur-Oxidizing Archaeon Metallosphaera cuprina Ar-4. International Journal of Molecular Sciences, 26(3), 1338. https://doi.org/10.3390/ijms26031338







