Combinatorial Effects of Cisplatin and PARP Inhibitor Olaparib on Survival, Intestinal Integrity, and Microbiome Modulation in Murine Model
Abstract
1. Introduction
2. Results
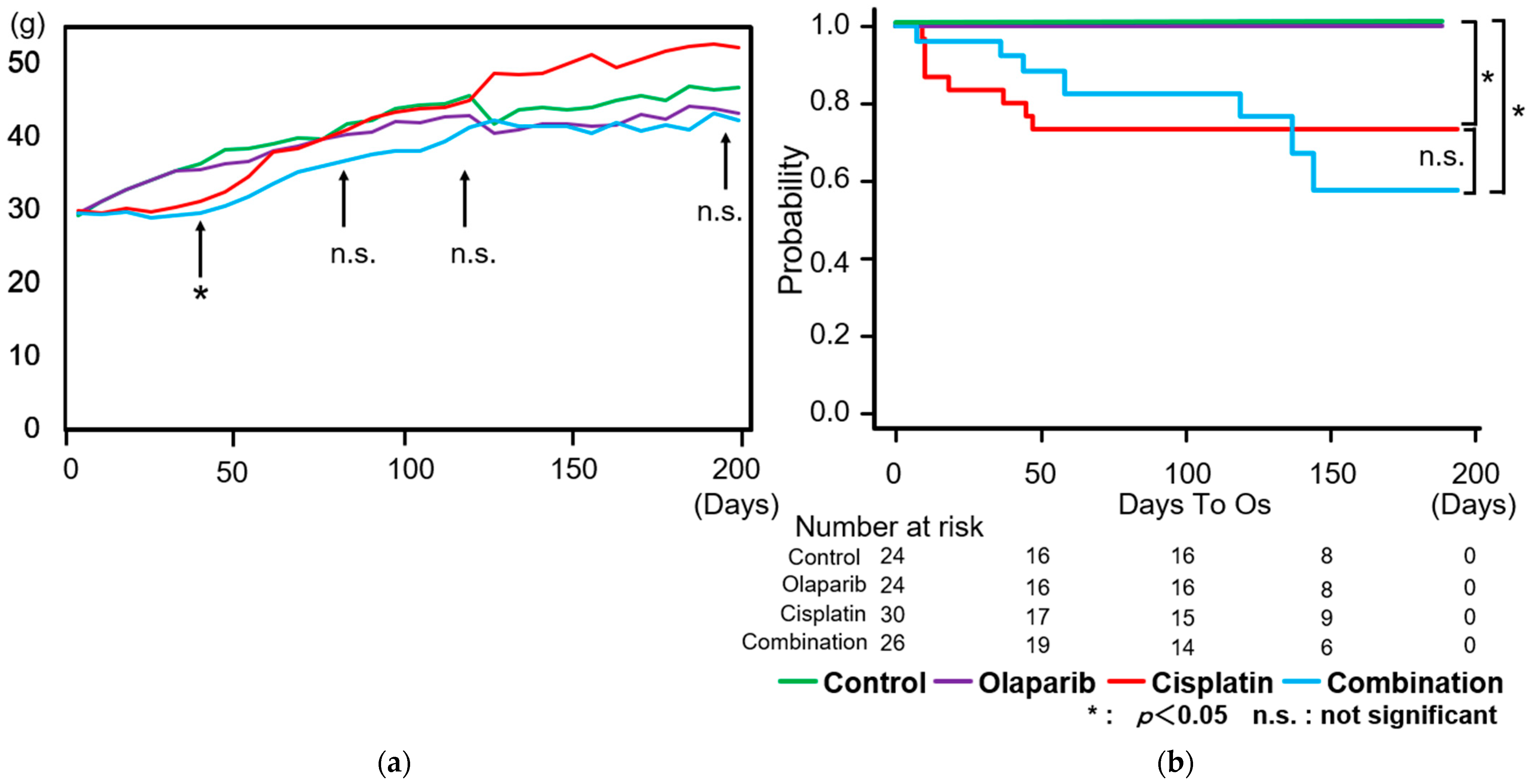
2.1. Body Weight Changes and Survival Rate
2.2. Results of Biochemical Blood Test
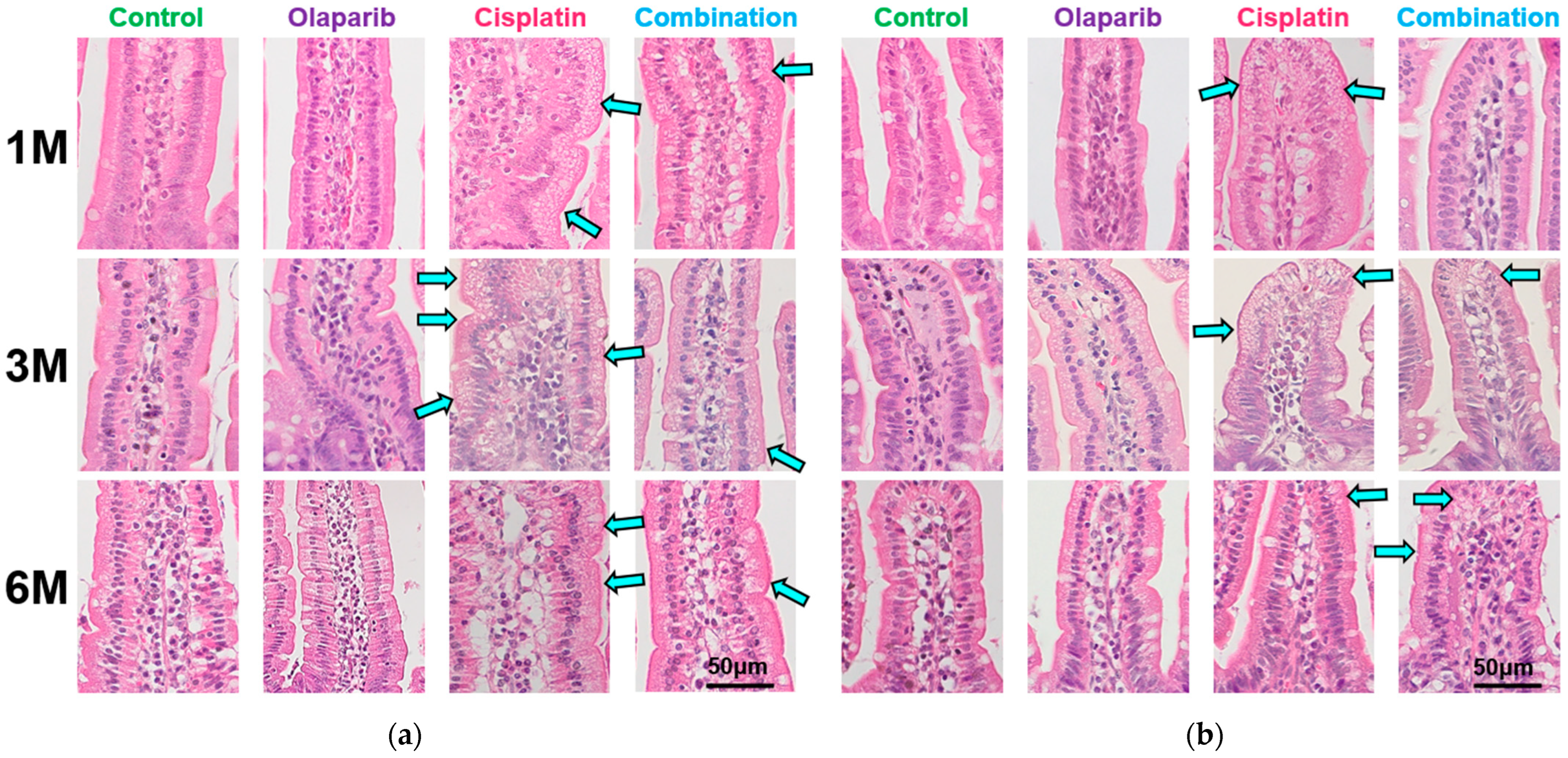
2.3. Histopathological Changes
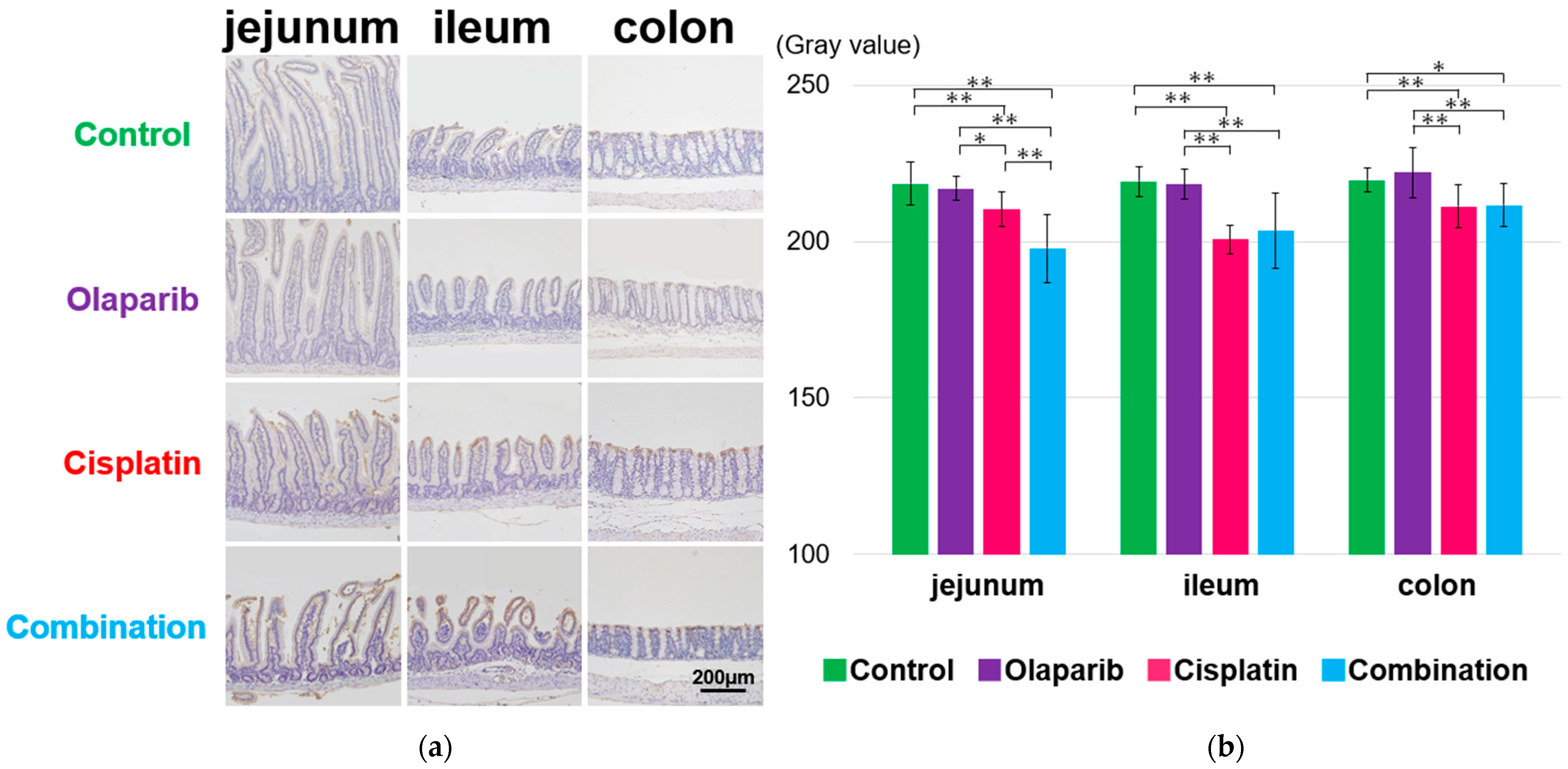
2.4. Immunohistochemical Analysis of PARP-1, EGF, and E-Cadherin
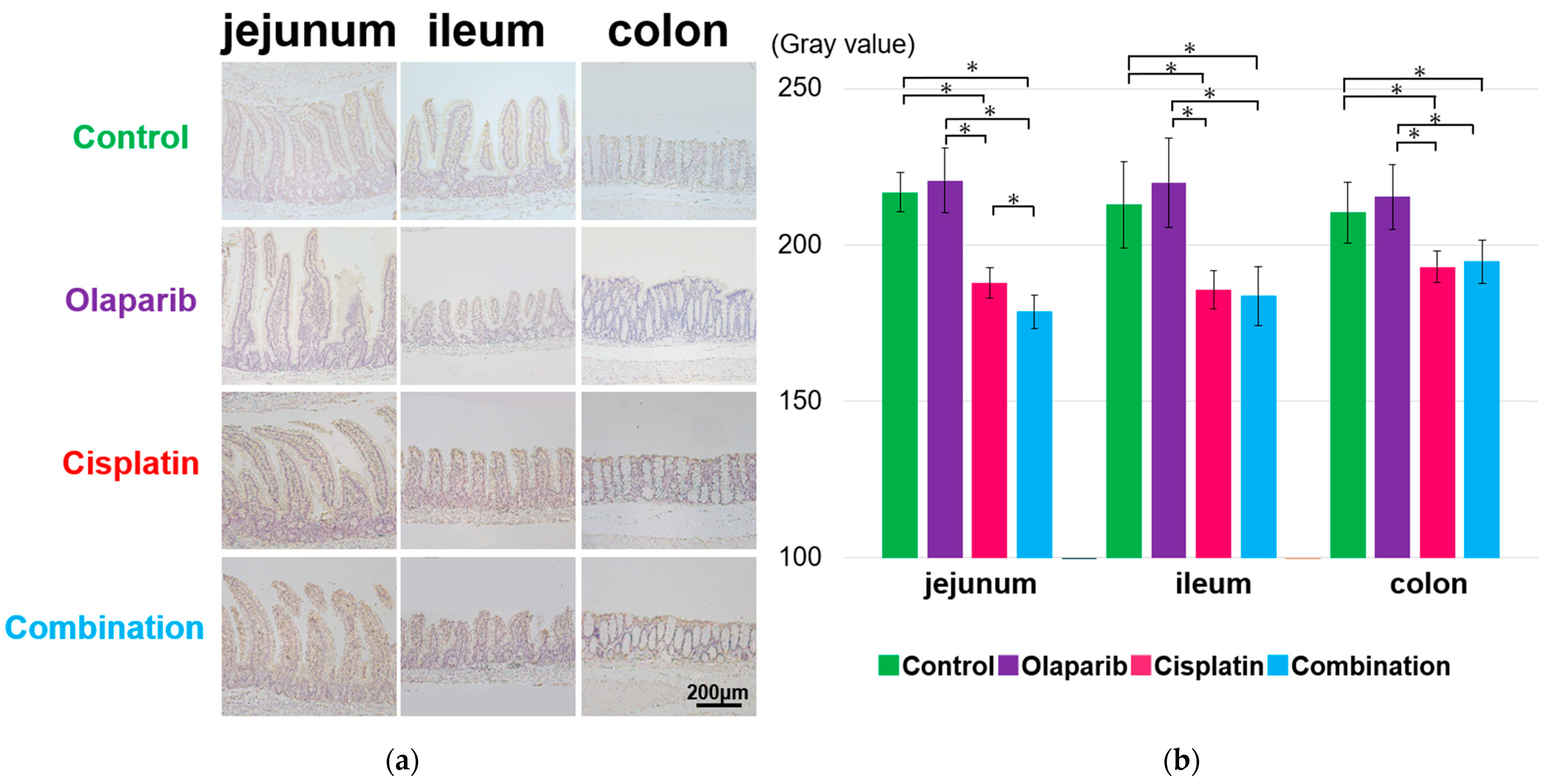
2.4.1. PARP-1 Expression
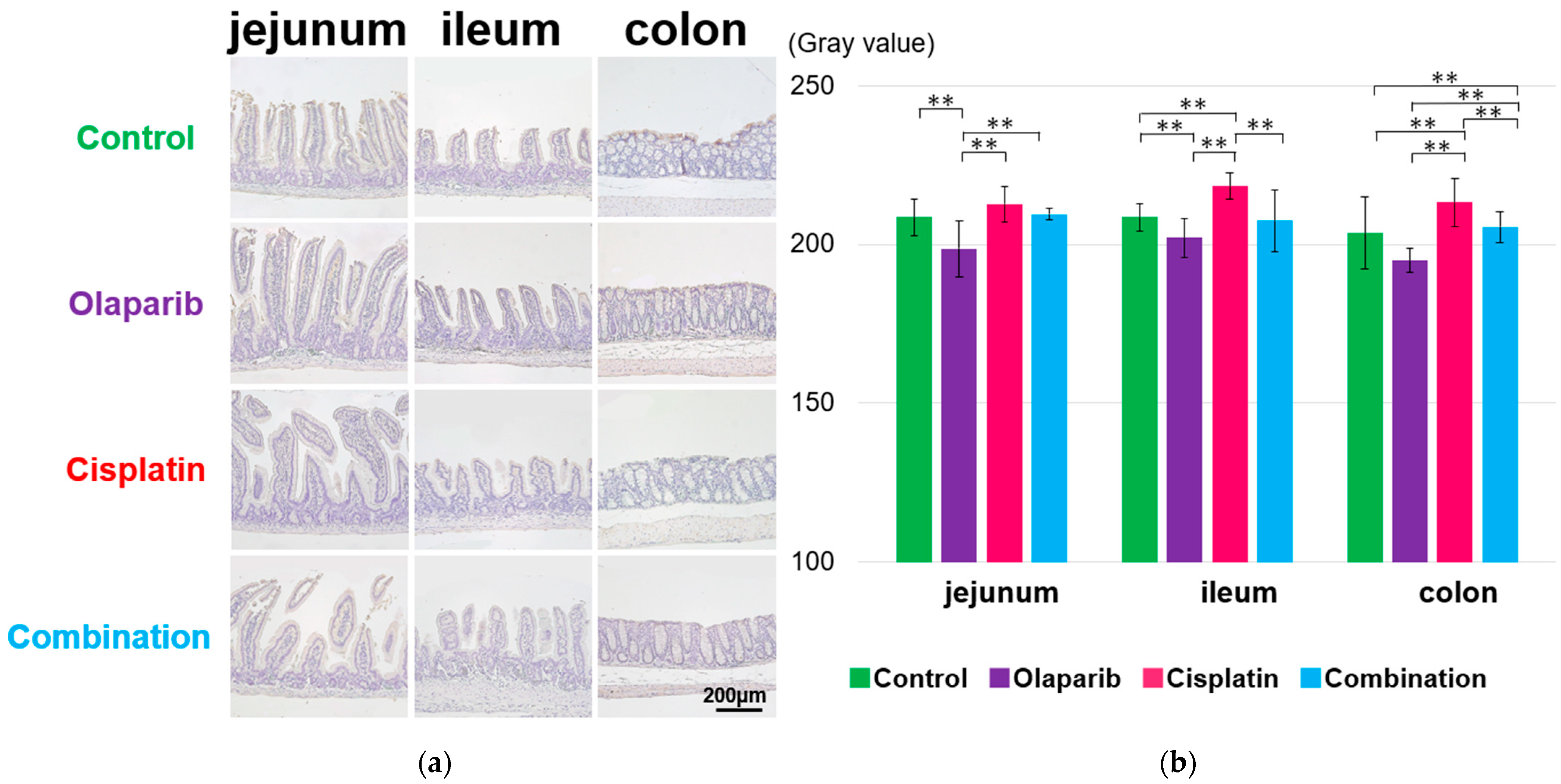
2.4.2. EGF Expression
2.4.3. E-Cadherin Expression
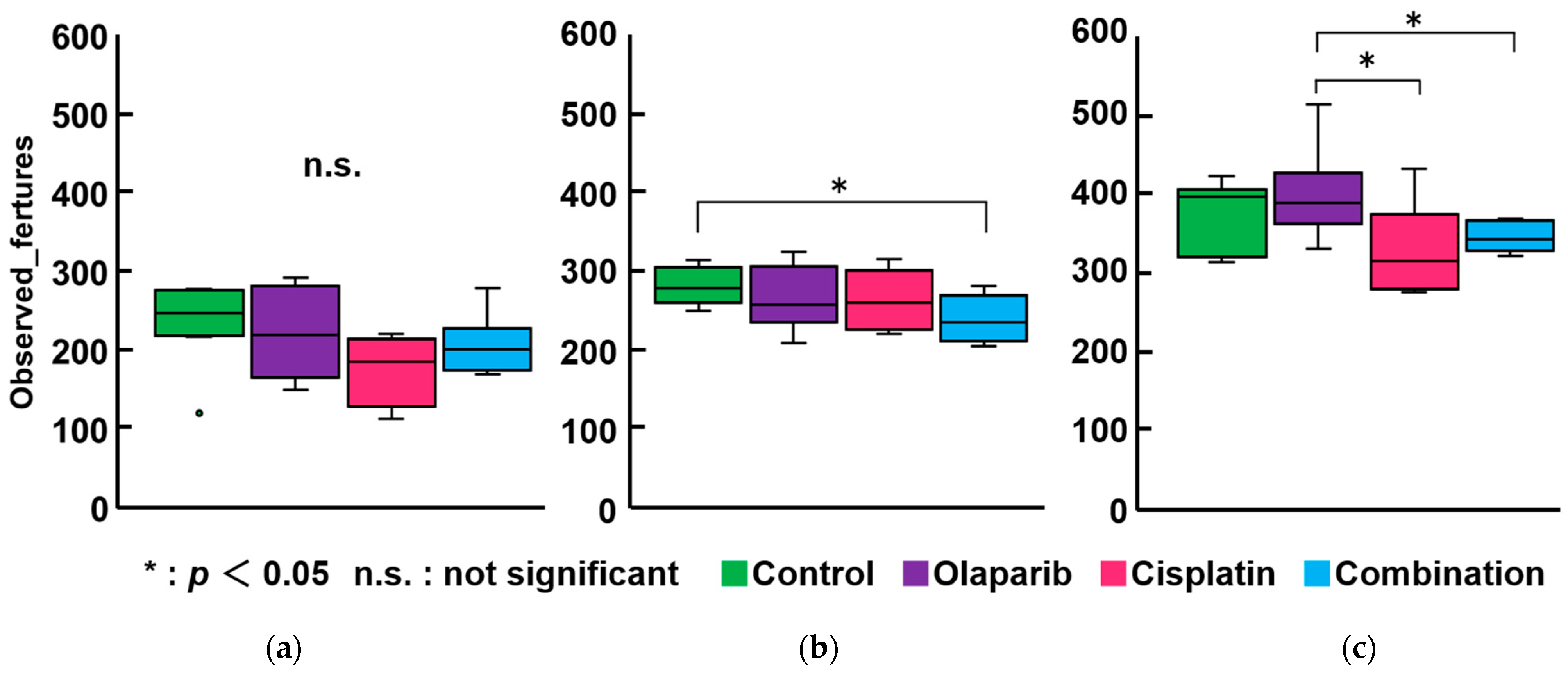
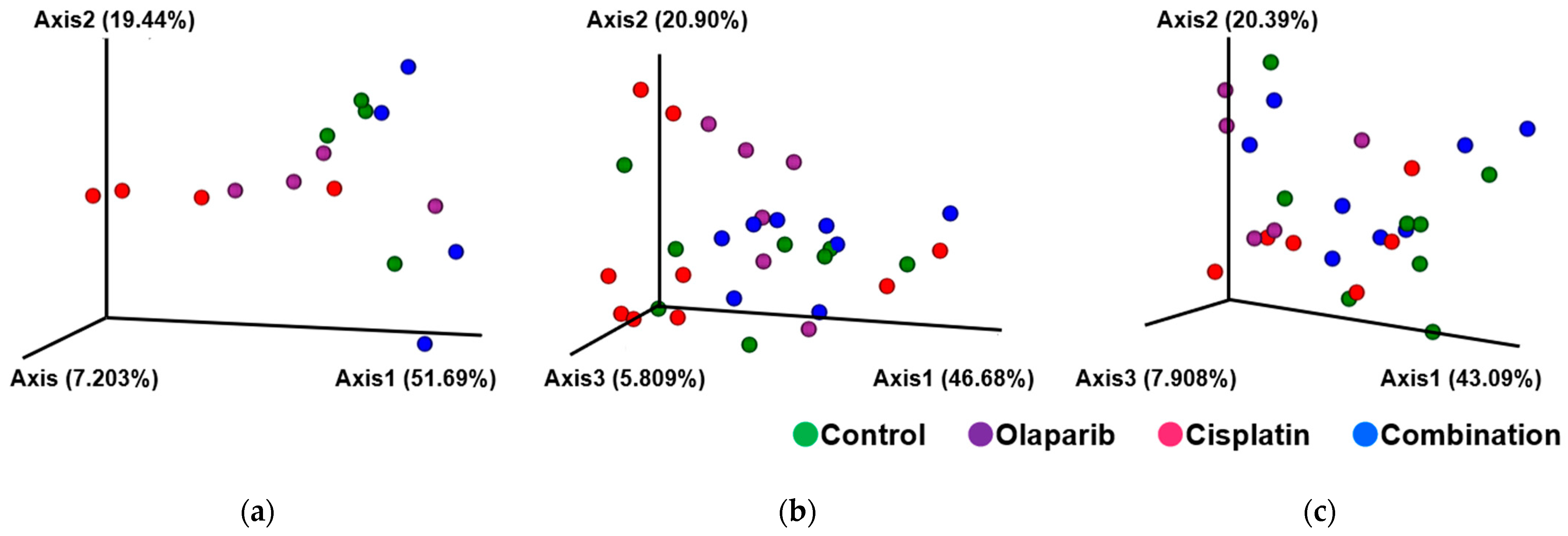
2.5. The Result of Microbiome Analysis
2.5.1. Within-Subject α-Diversity and Between-Subject β-Diversity After 1, 3, and 6 Months After Drug Administration
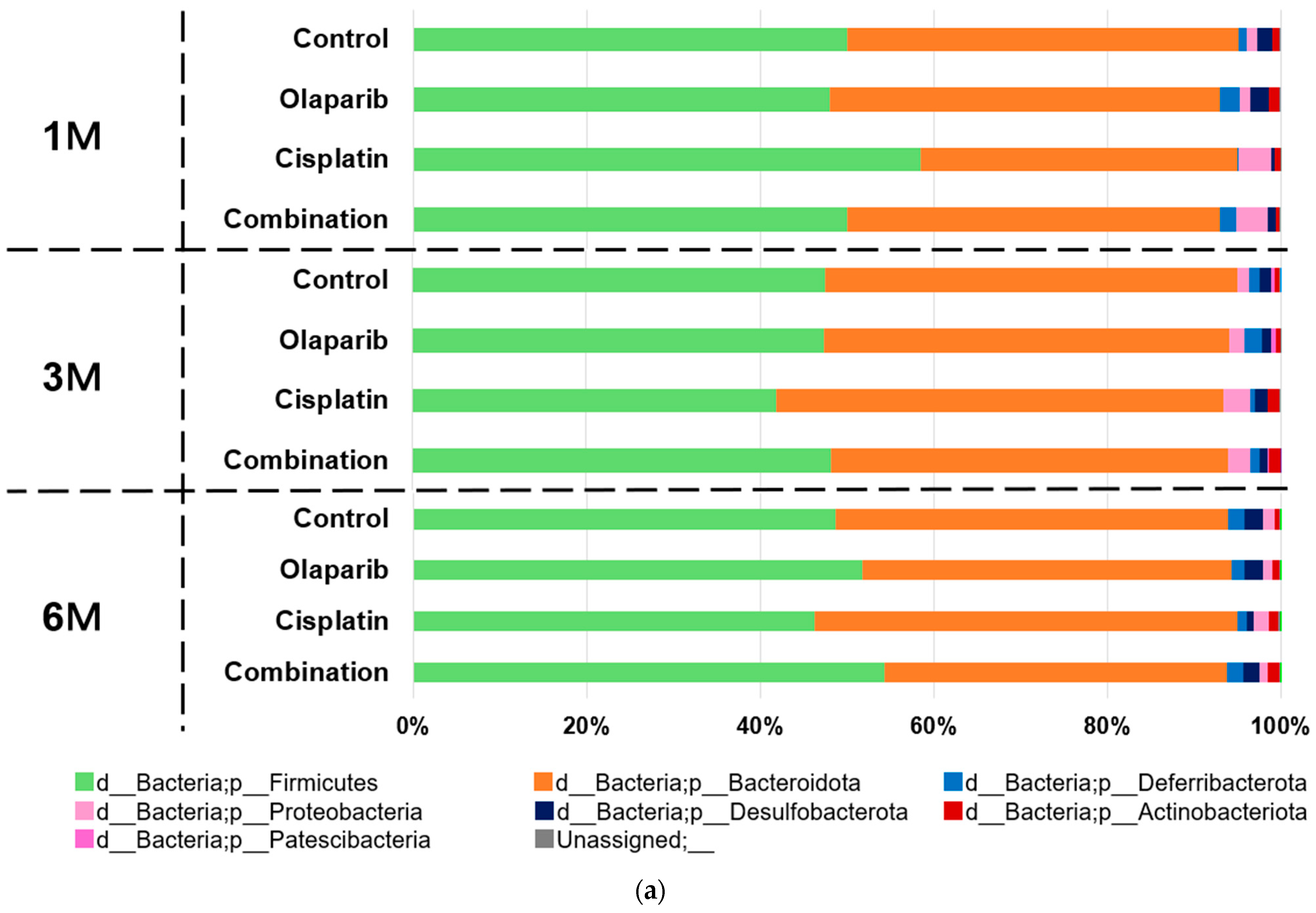
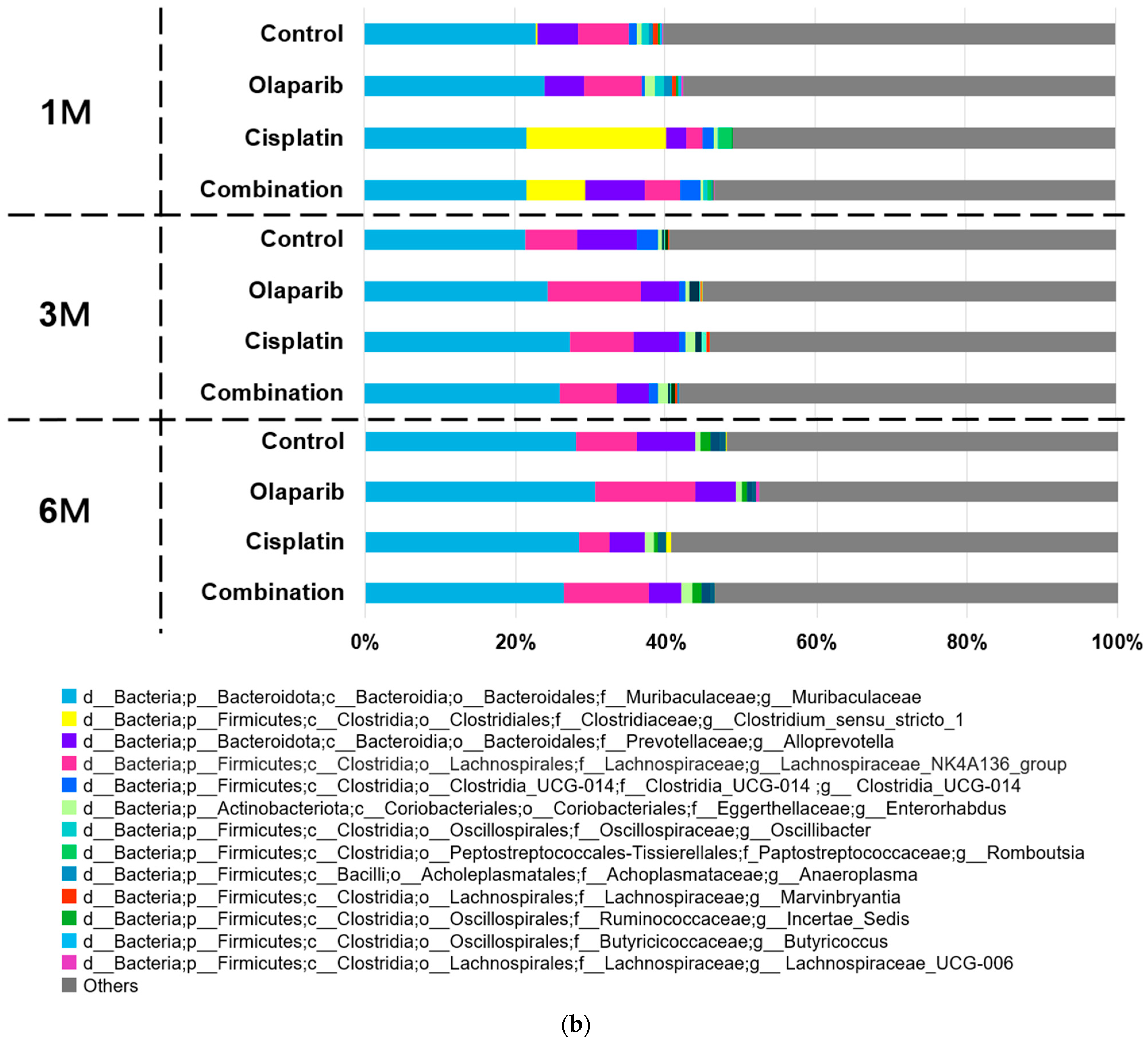
2.5.2. Microbial Taxa Analysis at Phylum and Genus Level
- f_Muribaculaceae; g_Muribaculaceae: This genus consistently represented approximately 20% of the microbiome across all experimental groups and time points. No significant differences in abundance were observed between the groups at any time point.
- f_Lachnospiraceae; g_Lachnospiraceae_NK4A136_group: Members of the Lachnospiraceae family demonstrated dynamic changes in abundance. At one month following drug administration, the relative abundances of the control, Olaparib, cisplatin, and combination groups were 5.83%, 6.18%, 1.98%, and 4.00%, respectively, with a statistically significant reduction observed in the cisplatin group (p < 0.05). By three months, these differences were no longer significant. Notably, at 6 months after drug administration, the relative abundances were 6.53%, 9.92%, 3.47%, and 8.90%, respectively, showing a significant decrease in the cisplatin group (p < 0.05) and a similar trend for the Olaparib and combination groups.
- f_Clostridiaceae; g_Clostridium_sensu_stricto_1: This genus exhibited marked group-specific abundance patterns. At 1 month after drug administration, its mean relative abundance was significantly higher in the cisplatin (16.78%) and combination (6.62%) groups compared to the control (0.23%) and Olaparib (0.00%) groups (p < 0.01) (Figure 9b).
2.5.3. Microbiome Alterations Induced by Cisplatin and PARPis Identified by LEfSe Analysis
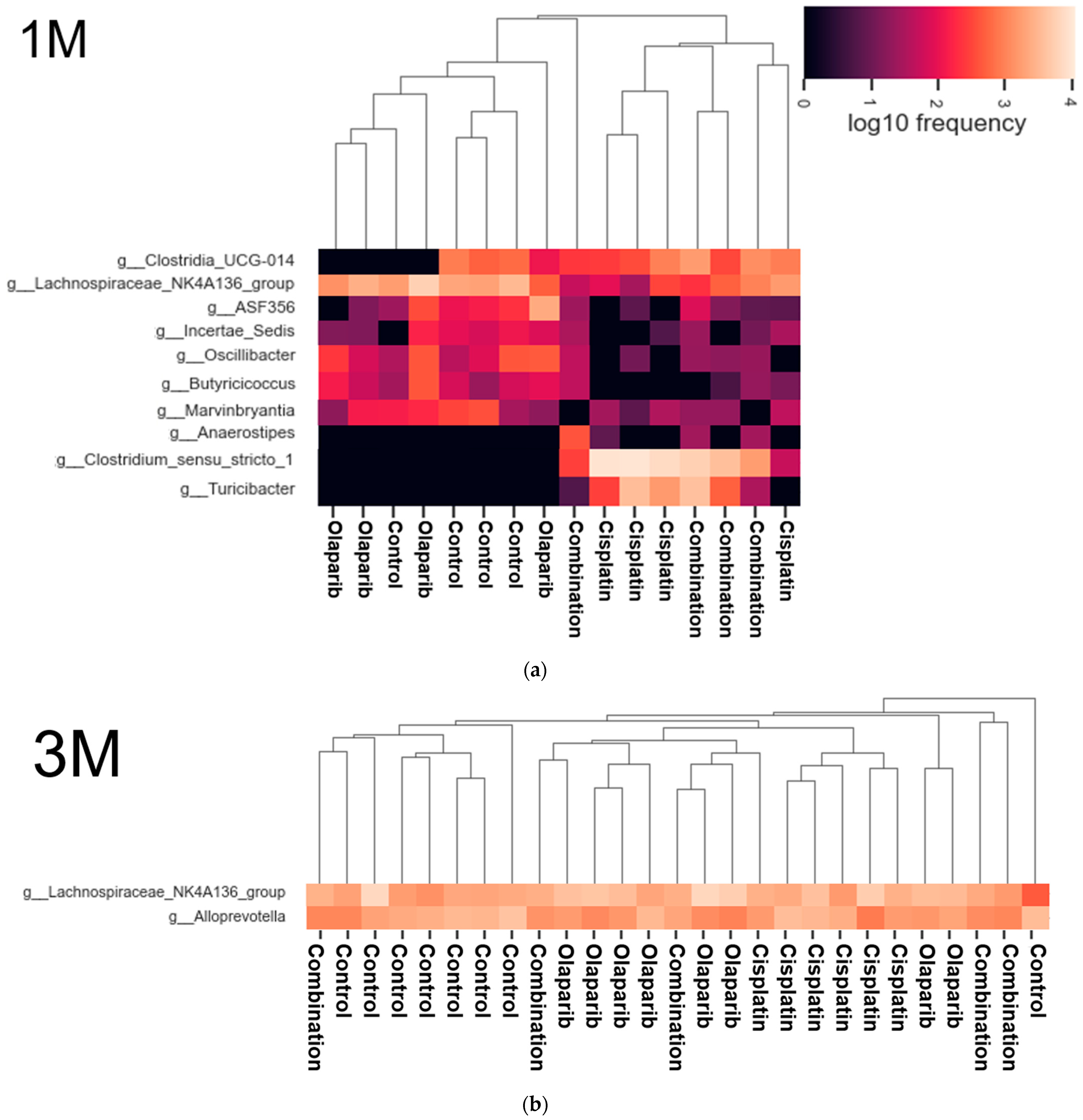
2.5.4. Heatmap Analysis
2.5.5. Analysis of Composition of Microbiomes (ANCOM)
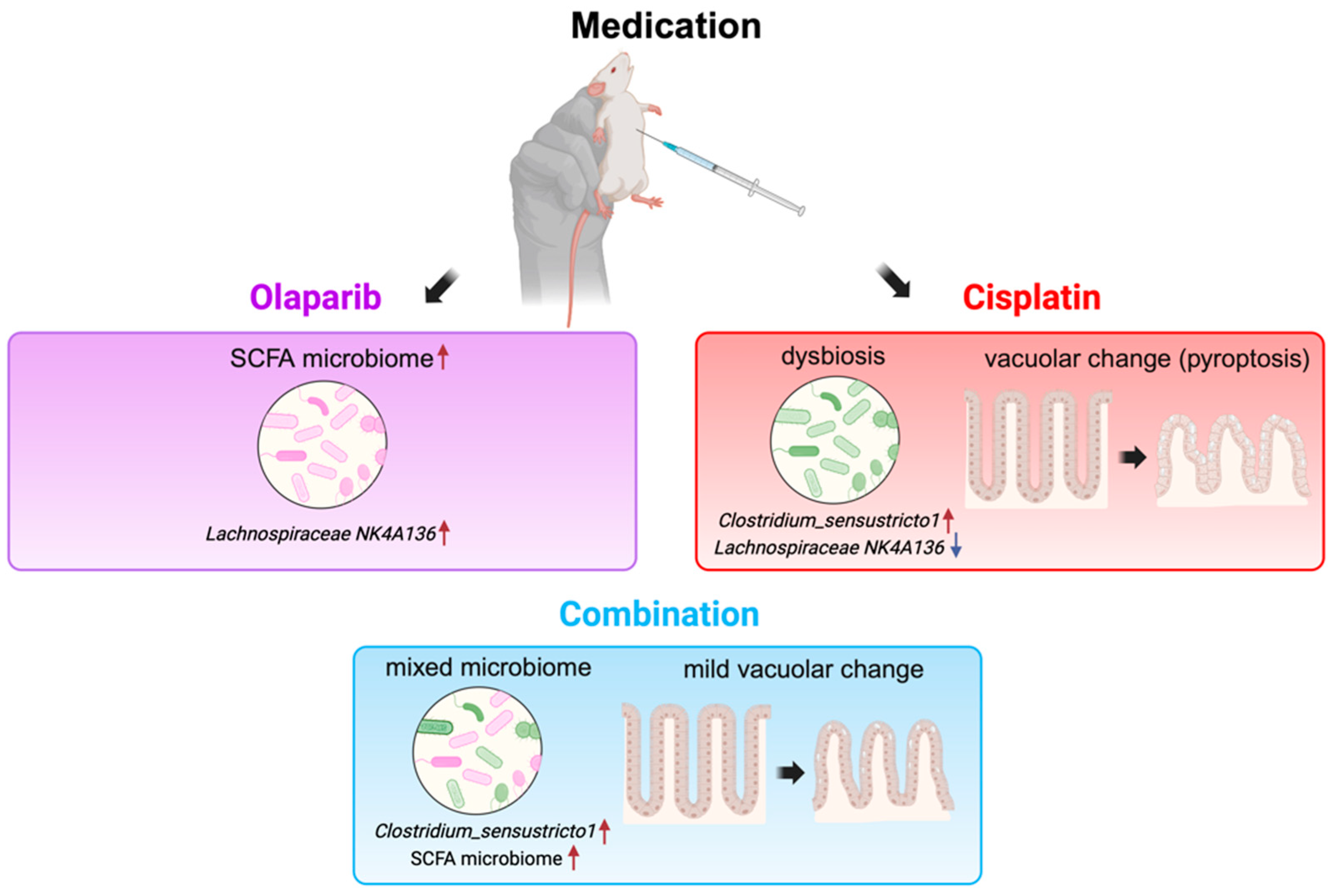
3. Discussion
4. Materials and Methods
4.1. Animal Models
4.2. Procedure for Animal Experiments
4.3. Measurement of Body Weight and Survival Ratio
4.4. Preparation of Blood Samples and Biochemical Blood Test
4.5. Preparation of Fecal and Intestinal Samples
4.6. Histopathological Analysis—Villi Length, Thickness, and Area
4.7. Immunohistochemical Analysis and Its Semi-Quantitative Analysis
4.8. DNA Extraction and 16S rRNA Gene Amplicon Sequencing
4.8.1. DNA Extraction from Fecal Samples
4.8.2. 16S rRNA Gene Amplification and Library Preparation
4.9. Analysis of Experimental Data
4.9.1. Microbiome Data
4.9.2. Procedure of Microbiome Analysis
4.9.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pignata, S.; Cecere, S.C.; Du Bois, A.; Harter, P.; Heitz, F. Treatment of Recurrent Ovarian Cancer. Ann. Oncol. 2017, 28, viii51–viii56. [Google Scholar] [CrossRef]
- Lheureux, S.; Braunstein, M.; Oza, A.M. Epithelial Ovarian Cancer: Evolution of Management in the Era of Precision Medicine. CA Cancer J. Clin. 2019, 69, 280–304. [Google Scholar] [CrossRef] [PubMed]
- Chelariu-Raicu, A.; Trillsch, F.; Burges, A.; Czogalla, B.; Hester, A.; Wuerstlein, R.; Harbeck, N.; Mahner, S. PARP Inhibitors: Risk Factors for Toxicity and Matching Patients to the Proper Poly (ADP-Ribose) Polymerase Inhibitor (PARPi) Therapy. Int. J. Gynecol. Cancer 2023, 33, 812–822. [Google Scholar] [CrossRef]
- Pujade-Lauraine, E.; Selle, F.; Scambia, G.; Asselain, B.; Marmé, F.; Lindemann, K.; Colombo, N.; Mądry, R.; Glasspool, R.; Vergote, I.; et al. Maintenance Olaparib Rechallenge in Patients with Platinum-Sensitive Relapsed Ovarian Cancer Previously Treated with a PARP Inhibitor (OReO/ENGOT-Ov38): A Phase IIIb Trial. Ann. Oncol. 2023, 34, 1152–1164. [Google Scholar] [CrossRef]
- Uekusa, R.; Yokoi, A.; Watanabe, E.; Yoshida, K.; Yoshihara, M.; Tamauchi, S.; Shimizu, Y.; Ikeda, Y.; Yoshikawa, N.; Niimi, K.; et al. Safety Assessments and Clinical Features of PARP Inhibitors from Real-World Data of Japanese Patients with Ovarian Cancer. Sci. Rep. 2024, 14, 12595. [Google Scholar] [CrossRef] [PubMed]
- Tonti, N.; Golia D’Augè, T.; Cuccu, I.; De Angelis, E.; D’Oria, O.; Perniola, G.; Laganà, A.S.; Etrusco, A.; Ferrari, F.; Saponara, S.; et al. The Role of Tumor Biomarkers in Tailoring the Approach to Advanced Ovarian Cancer. Int. J. Mol. Sci. 2024, 25, 11239. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, L.; Palluzzi, E.; Di Napoli, M.; Lauria, R.; Ferrandina, G.; Angioli, R.; Bergamini, A.; Corrado, G.; Perniola, G.; Cassani, C.; et al. Real World Data of Niraparib in Platinum Sensitive Relapsed Ovarian Cancer: A Multicenter Experience of the MITO Group. Gynecol. Oncol. 2024, 184, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.; Li, L.; Fattah, F.J.; Dong, Y.; Bey, E.A.; Patel, M.; Gao, J.; Boothman, D.A. Review of Poly (ADP-Ribose) Polymerase (PARP) Mechanisms of Action and Rationale for Targeting in Cancer and Other Diseases. Crit. Rev. Eukaryot. Gene Expr. 2014, 24, 15–28. [Google Scholar] [CrossRef]
- Lee, E.K.; Konstantinopoulos, P.A. PARP Inhibition and Immune Modulation: Scientific Rationale and Perspectives for the Treatment of Gynecologic Cancers. Ther. Adv. Vaccines 2018, 12, 1758835920944116. [Google Scholar] [CrossRef] [PubMed]
- Keung, M.Y.T.; Wu, Y.; Vadgama, J.V. PARP Inhibitors as a Therapeutic Agent for Homologous Recombination Deficiency in Breast Cancers. J. Clin. Med. 2019, 8, 435. [Google Scholar] [CrossRef]
- Schiewer, M.J.; Mandigo, A.C.; Gordon, N.; Huang, F.; Gaur, S.; de Leeuw, R.; Zhao, S.G.; Evans, J.; Han, S.; Parsons, T.; et al. PARP-1 Regulates DNA Repair Factor Availability. EMBO Mol. Med. 2018, 10, e8816. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, M.; Fujihara, H.; Fujimori, H.; Kawaguchi, K.; Yamada, H.; Nakayama, R.; Yamamoto, N.; Kishi, Y.; Hamada, Y.; Masutani, M. Synergetic Effects of PARP Inhibitor AZD2281 and Cisplatin in Oral Squamous Cell Carcinoma in Vitro and in Vivo. Int. J. Mol. Sci. 2016, 17, 272. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, Y.; Habeebu, S.S.M.; Klaassen, C.D. Metallothionein (MT)-Null Mice Are Sensitive to Cisplatin-Induced Hepatotoxicity. Toxicol. Appl. Pharmacol. 1998, 149, 24–31. [Google Scholar] [CrossRef]
- Sheikh-Hamad, D.; Timmins, K.; Jalali, Z. Cisplatin-Induced Renal Toxicity. J. Am. Soc. Nephrol. 1997, 8, 1640–1644. [Google Scholar] [CrossRef]
- Sharman, R.; Harris, Z.; Ernst, B.; Mussallem, D.; Larsen, A.; Gowin, K. Lifestyle Factors and Cancer: A Narrative Review. Lifestyle Med. 2024, 8, 166–183. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, X.; Lin, J.; Wang, R.; Zhang, B.; Zhang, X.; He, W.; Gao, F.; Song, D.; Zhao, K.; et al. Natural Flavonoid Sinensetin Inhibits Cisplatin-Induced Pyroptosis and Attenuates Intestinal Injury. Biochim. Biophys. Acta—Mol. Basis Dis. 2023, 1869, 166637. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Z.; Ruan, J.; Pan, Y.; Magupalli, V.G.; Wu, H.; Lieberman, J. Inflammasome-Activated Gasdermin D Causes Pyroptosis by Forming Membrane Pores. Nature 2016, 535, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Wallach, D.; Kang, T.B.; Dillon, C.P.; Green, D.R. Programmed Necrosis in Inflammation: Toward Identification of the Effector Molecules. Science 2016, 352, aaf2154. [Google Scholar] [CrossRef]
- Kiss, R.C.; Xia, F.; Acklin, S. Targeting DNA Damage Response and Repair to Enhance Therapeutic Index in Cisplatin-Based Cancer Treatment. Int. J. Mol. Sci. 2021, 22, 8199. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.-W.; Hsu, S.-C.; Xia, W.; Cao, X.; Shih, J.-Y.; Wei, Y.; James, L.; Abbruzzese, G.N.H.; Hung, M.-C. Epidermal Growth Factor Receptor Cooperates with Signal Transducer and Activator of Transcription 3 to Induce Epithelial-Mesenchymal Transition in Cancer Cells via Up-Regulation of TWIST Gene Expression. Cancer Res. 2007, 67, 9066–9076. [Google Scholar] [CrossRef]
- Brüser, L.; Bogdan, S. Adherens Junctions on the Move—Membrane Trafficking of E-Cadherin. Cold Spring Harb. Perspect. Biol. 2017, 9, a029140. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Q.; Liu, Z.; Li, H.; Liu, X.; Yu, H. EGF Protects Epithelial Cells from Barrier Damage in Chronic Rhinosinusitis with Nasal Polyps. J. Inflamm. Res. 2022, 15, 439–450. [Google Scholar] [CrossRef]
- Winkles, J.A. Serum- and Polypeptide Growth Factor-Inducible Gene Expression in Mouse Fibroblasts. Prog. Nucleic Acid. Res. Mol. Biol. 1997, 58, 41–78. [Google Scholar] [CrossRef]
- Ramesh, V.; Brabletz, T.; Ceppi, P. Targeting EMT in Cancer with Repurposed Metabolic Inhibitors. Trends in Cancer 2020, 6, 942–950. [Google Scholar] [CrossRef] [PubMed]
- Schacke, M.; Kumar, J.; Colwell, N.; Hermanson, K.; Folle, G.A.; Nechaev, S.; Dhasarathy, A.; Lafon-Hughes, L. PARP-1/2 Inhibitor Olaparib Prevents or Partially Reverts EMT Induced by TGF-β in NMuMG Cells. Int. J. Mol. Sci. 2019, 20, 518. [Google Scholar] [CrossRef]
- Wang, X.; Sun, J.; Shao, K.; Zhi, H.; Lin, Y.; Fu, Y. Study on the Interactions between Cisplatin and Cadherin. bioRxiv 2022. [Google Scholar] [CrossRef]
- McPhee, T.R.; McDonald, P.C.; Oloumi, A.; Dedhar, S. Integrin-Linked Kinase Regulates E-Cadherin Expression through PARP-1. Dev. Dyn. 2008, 237, 2737–2747. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Kumar, S.; Chauhan, S.S.; Bhatla, N.; Kumar, S.; Bakhshi, S.; Gupta, R.; Sharma, A.; Kumar, L. Correction: PARP-1 Inhibitor Modulate β-Catenin Signaling to Enhance Cisplatin Sensitivity in Cancer Cervix. Oncotarget 2019, 10, 4802. [Google Scholar] [CrossRef]
- Song, Y.; Ye, M.; Zhou, J.; Wang, Z.W.; Zhu, X. Restoring E-Cadherin Expression by Natural Compounds for Anticancer Therapies in Genital and Urinary Cancers. Mol. Ther. Oncolytics 2019, 14, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Süsse, S.; Scholz, C.J.; Bürkle, A.; Wiesmüller, L. Poly(ADP-Ribose) Polymerase (PARP-1) and P53 Independently Function in Regulating Double-Strand Break Repair in Primate Cells. Nucleic Acids Res. 2004, 32, 669–680. [Google Scholar] [CrossRef][Green Version]
- Oikawa, T.; Otsuka, Y.; Onodera, Y.; Horikawa, M.; Handa, H.; Hashimoto, S.; Suzuki, Y.; Sabe, H. Necessity of P53-Binding to the CDH1 Locus for Its Expression Defines Two Epithelial Cell Types Differing in Their Integrity. Sci. Rep. 2018, 8, 1595. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.J.; Li, X.; Lehmler, H.J.; Wang, D.; Gu, H.; Cui, J.Y. Gut Microbiome Critically Impacts PCB-Induced Changes in Metabolic Fingerprints and the Hepatic Transcriptome in Mice. Toxicol. Sci. 2020, 177, 168–187. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.Y.; Cui, Z.Y.; Zhao, M.Q.; Zhang, M.J.; Jiang, Q.L.; Wang, J.J.; Lu, L.G.; Lu, Y.Y. The Impact of Helicobacter Pylori Infection and Eradication Therapy Containing Minocycline and Metronidazole on Intestinal Microbiota. BMC Microbiol. 2022, 22, 321. [Google Scholar] [CrossRef]
- Wang, Y.; Xia, X.; Zhou, X.; Zhan, T.; Dai, Q.; Zhang, Y.; Zhang, W.; Shu, Y.; Li, W.; Xu, H. Association of Gut Microbiome and Metabolites with Onset and Treatment Response of Patients with Pemphigus Vulgaris. Front. Immunol. 2023, 14, 1114586. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.C.; Lin, S.F.; Chen, S.T.; Chang, P.Y.; Yeh, Y.M.; Lo, F.S.; Lu, J.J. Alterations of Gut Microbiota in Patients With Graves’ Disease. Front. Cell. Infect. Microbiol. 2021, 11, 663131. [Google Scholar] [CrossRef]
- Huang, D.; Chen, Y.; Chen, H.; Deng, X.; Huang, J.; Lu, S.; Li, P.; Du, B. Supplementation of Bacillus Sp. DU-106 Alleviates Antibiotic-Associated Diarrhea in Association with the Regulation of Intestinal Microbiota in Mice. Probiotics Antimicrob. Proteins 2022, 14, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Larmonier, C.B.; Shehab, K.W.; Laubitz, D.; Jamwal, D.R.; Ghishan, F.K.; Kiela, P.R. Transcriptional Reprogramming and Resistance to Colonic Mucosal Injury in Poly (ADP-Ribose) Polymerase 1 (PARP1) Deficient Mice. J. Biol. Chem. 2016, 291, 8918–8930. [Google Scholar] [CrossRef]
- Vida, A.; Kardos, G.; Kovács, T.; Bodrogi, B.L.; Bai, P. Deletion of Poly(ADP-ribose) Polymerase-1 Changes the Composition of the Microbiome in the Gut. Mol. Med. Rep. 2018, 18, 4335–4341. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Fang, Z.; Li, L.; Wang, H.; Zhu, J.; Zhang, P.; Lee, Y.K.; Zhao, J.; Zhang, H.; Lu, W.; et al. Lactobacillus Mucosae Exerted Different Antiviral Effects on Respiratory Syncytial Virus Infection in Mice. Front. Microbiol. 2022, 13, 1001313. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Yuan, X.; Liu, J.; Shi, Z.; Cao, L.; Yang, L.; Wu, K.; Lou, Y.; Tong, H.; Jiang, L.; et al. Albuca Bracteata Polysaccharides Attenuate AOM/DSS Induced Colon Tumorigenesis via Regulating Oxidative Stress, Inflammation and Gut Microbiota in Mice. Front. Pharmacol. 2022, 13, 833077. [Google Scholar] [CrossRef] [PubMed]
- Cheung, S.G.; Goldenthal, A.R.; Uhlemann, A.C.; Mann, J.J.; Miller, J.M.; Sublette, M.E. Systematic Review of Gut Microbiota and Major Depression. Front. Psychiatry 2019, 10, 34. [Google Scholar] [CrossRef]
- Daniel, N.; Lécuyer, E.; Chassaing, B. Host/Microbiota Interactions in Health and Diseases—Time for Mucosal Microbiology! Mucosal Immunol. 2021, 14, 1006–1016. [Google Scholar] [CrossRef]
- Fernandes, M.R.; Aggarwal, P.; Costa, R.G.F.; Cole, A.M.; Trinchieri, G. Targeting the Gut Microbiota for Cancer Therapy. Nat. Rev. Cancer 2022, 22, 703–722. [Google Scholar] [CrossRef]
- Wen, X.; Wang, H.G.; Zhang, M.N.; Zhang, M.H.; Wang, H.; Yang, X.Z. Fecal Microbiota Transplantation Ameliorates Experimental Colitis via Gut Microbiota and T-Cell Modulation. World J. Gastroenterol. 2021, 27, 2834–2849. [Google Scholar] [CrossRef]
- Wang, X.; Yi, W.; He, L.; Luo, S.; Wang, J.; Jiang, L.; Long, H.; Zhao, M.; Lu, Q. Abnormalities in Gut Microbiota and Metabolism in Patients With Chronic Spontaneous Urticaria. Front. Immunol. 2021, 12, 691304. [Google Scholar] [CrossRef] [PubMed]
- Getachew, B.; Aubee, J.I.; Schottenfeld, R.S.; Csoka, A.B.; Thompson, K.M.; Tizabi, Y. Ketamine Interactions with Gut-Microbiota in Rats: Relevance to Its Antidepressant and Anti-Inflammatory Properties. BMC Microbiol. 2018, 18, 222. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Stražar, M.; Mohamed, A.M.T.; Pacheco, J.A.; Walker, R.L.; Lebar, T.; Zhao, S.; Lockart, J.; Dame, A.; Thurimella, K.; et al. Gut Microbiome and Metabolome Profiling in Framingham Heart Study Reveals Cholesterol-Metabolizing Bacteria. Cell 2024, 187, 1834–1852. [Google Scholar] [CrossRef] [PubMed]
- Ordoñez-Rodriguez, A.; Roman, P.; Rueda-Ruzafa, L.; Campos-Rios, A.; Cardona, D. Changes in Gut Microbiota and Multiple Sclerosis: A Systematic Review. Int. J. Environ. Res. Public Health 2023, 20, 4624. [Google Scholar] [CrossRef] [PubMed]
- Aarnoutse, R.; Ziemons, J.; Hillege, L.E.; de Vos-Geelen, J.; de Boer, M.; Bisschop, S.M.P.; Vriens, B.E.P.J.; Vincent, J.; van de Wouw, A.J.; Le, G.N.; et al. Changes in Intestinal Microbiota in Postmenopausal Oestrogen Receptor-Positive Breast Cancer Patients Treated with (Neo)Adjuvant Chemotherapy. npj Breast Cancer 2022, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Kawai, S.; Takagi, Y.; Kaneko, S.; Kurosawa, T. Effect of Three Types of Mixed Anesthetic Agents Alternate to Ketamine in Mice. Exp. Anim. 2011, 60, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Hoff, J. Methods of Blood Collection in the Mouse. Lab Anim. 2000, 29, 47–53. [Google Scholar]
- Yuan, X.; Liu, J.; Hu, X.; Yang, S.; Zhong, S.; Yang, T.; Zhou, Y.; Zhao, G.; Jiang, Y.; Li, Y. Alterations in the Jejunal Microbiota and Fecal Metabolite Profiles of Rabbits Infected with Eimeria Intestinalis. Parasites Vectors 2022, 15, 231. [Google Scholar] [CrossRef] [PubMed]
- Bialkowska, A.B.; Ghaleb, A.M.; Nandan, M.O.; Yang, V.W. Improved Swiss-Rolling Technique for Intestinal Tissue Preparation for Immunohistochemical and Immunofluorescent Analyses. J. Vis. Exp. 2016, 2016, 54161. [Google Scholar] [CrossRef]
- Le Naour, J.; Montégut, L.; Joseph, A.; Garbin, K.; Vacchelli, E.; Kroemer, G.; Pol, J.G.; Maiuri, M.C. Improved Swiss-Rolling Method for Histological Analyses of Colon Tissue. MethodsX 2022, 9, 101630. [Google Scholar] [CrossRef] [PubMed]
- Landini, G.; Martinelli, G.; Piccinini, F. Colour Deconvolution: Stain Unmixing in Histological Imaging. Bioinformatics 2021, 37, 1485–1487. [Google Scholar] [CrossRef] [PubMed]
- Tomikawa, D.; Okuda, H. Analysis of the Bacterial Flora of Substrates Used for the Cultivation of Agaricus Bisporus: Relationship between Clostridia and Yield. Microbes Environ. 2023, 38, 2–5. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Zhang, W.; Zhang, T.; He, Q.; Kwok, L.Y.; Tan, Y.; Zhang, H. Heat-Killed Bifidobacterium Bifidum B1628 May Alleviate Dextran Sulfate Sodium-Induced Colitis in Mice, and the Anti-Inflammatory Effect Is Associated with Gut Microbiota Modulation. Nutrients 2022, 14, 5233. [Google Scholar] [CrossRef]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M.; et al. Fusobacterium Nucleatum in Colorectal Carcinoma Tissue and Patient Prognosis. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef]
- Teofani, A.; Marafini, I.; Laudisi, F.; Pietrucci, D.; Salvatori, S.; Unida, V.; Biocca, S.; Monteleone, G.; Desideri, A. Intestinal Taxa Abundance and Diversity in Inflammatory Bowel Disease Patients: An Analysis Including Covariates and Confounders. Nutrients 2022, 14, 260. [Google Scholar] [CrossRef]
- Pascal, V.; Pozuelo, M.; Borruel, N.; Casellas, F.; Campos, D.; Santiago, A.; Martinez, X.; Varela, E.; Sarrabayrouse, G.; Machiels, K.; et al. A Microbial Signature for Crohn’s Disease. Gut 2017, 66, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High- Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Kato-Kogoe, N.; Sakaguchi, S.; Kamiya, K.; Omori, M.; Gu, Y.H.; Ito, Y.; Nakamura, S.; Nakano, T.; Tamaki, J.; Ueno, T.; et al. Characterization of Salivary Microbiota in Patients with Atherosclerotic Cardiovascular Disease: A Case-Control Study. J. Atheroscler. Thromb. 2022, 29, 403–421. [Google Scholar] [CrossRef]
- An, Y.; Li, Y.; Wang, X.; Chen, Z.; Xu, H.; Wu, L.; Li, S.; Wang, C.; Luan, W.; Wang, X.; et al. Cordycepin Reduces Weight through Regulating Gut Microbiota in High-Fat Diet-Induced Obese Rats. Lipids Health Dis. 2018, 17, 276. [Google Scholar] [CrossRef] [PubMed]
- Shamsaddini, A.; Dadkhah, K.; Gillevet, P.M. BiomMiner: An Advanced Exploratory Microbiome Analysis and Visualization Pipeline. PLoS ONE 2020, 15, e0234860. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of Composition of Microbiomes: A Novel Method for Studying Microbial Composition. Microb. Ecol. Health Dis. 2015, 29, 27663. [Google Scholar] [CrossRef]
- Kanda, Y. Investigation of the Freely Available Easy-to-Use Software “EZR” for Medical Statistics. Bone Marrow Transpl. 2013, 48, 452–458. [Google Scholar] [CrossRef]



| 1 M | 3 M | 6 M | |
|---|---|---|---|
| Control Group | 8 | 8 | 8 |
| Olaparib Group | 8 | 8 | 8 |
| Cisplatin Group | 7 | 6 | 9 |
| Combination Group (Both Cisplatin and Olaparib) | 8 | 5 | 6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsumura, M.; Fujihara, H.; Maita, K.; Miyakawa, M.; Sakai, Y.; Nakayama, R.; Ito, Y.; Hasebe, M.; Kawaguchi, K.; Hamada, Y. Combinatorial Effects of Cisplatin and PARP Inhibitor Olaparib on Survival, Intestinal Integrity, and Microbiome Modulation in Murine Model. Int. J. Mol. Sci. 2025, 26, 1191. https://doi.org/10.3390/ijms26031191
Matsumura M, Fujihara H, Maita K, Miyakawa M, Sakai Y, Nakayama R, Ito Y, Hasebe M, Kawaguchi K, Hamada Y. Combinatorial Effects of Cisplatin and PARP Inhibitor Olaparib on Survival, Intestinal Integrity, and Microbiome Modulation in Murine Model. International Journal of Molecular Sciences. 2025; 26(3):1191. https://doi.org/10.3390/ijms26031191
Chicago/Turabian StyleMatsumura, Mitsuki, Hisako Fujihara, Kanna Maita, Moeko Miyakawa, Yushi Sakai, Ryoko Nakayama, Yumi Ito, Mitsuhiko Hasebe, Koji Kawaguchi, and Yoshiki Hamada. 2025. "Combinatorial Effects of Cisplatin and PARP Inhibitor Olaparib on Survival, Intestinal Integrity, and Microbiome Modulation in Murine Model" International Journal of Molecular Sciences 26, no. 3: 1191. https://doi.org/10.3390/ijms26031191
APA StyleMatsumura, M., Fujihara, H., Maita, K., Miyakawa, M., Sakai, Y., Nakayama, R., Ito, Y., Hasebe, M., Kawaguchi, K., & Hamada, Y. (2025). Combinatorial Effects of Cisplatin and PARP Inhibitor Olaparib on Survival, Intestinal Integrity, and Microbiome Modulation in Murine Model. International Journal of Molecular Sciences, 26(3), 1191. https://doi.org/10.3390/ijms26031191






