Multi-Omics Analysis Decodes Biosynthesis of Specialized Metabolites Constituting the Therapeutic Terrains of Magnolia obovata
Abstract
1. Introduction
2. Results
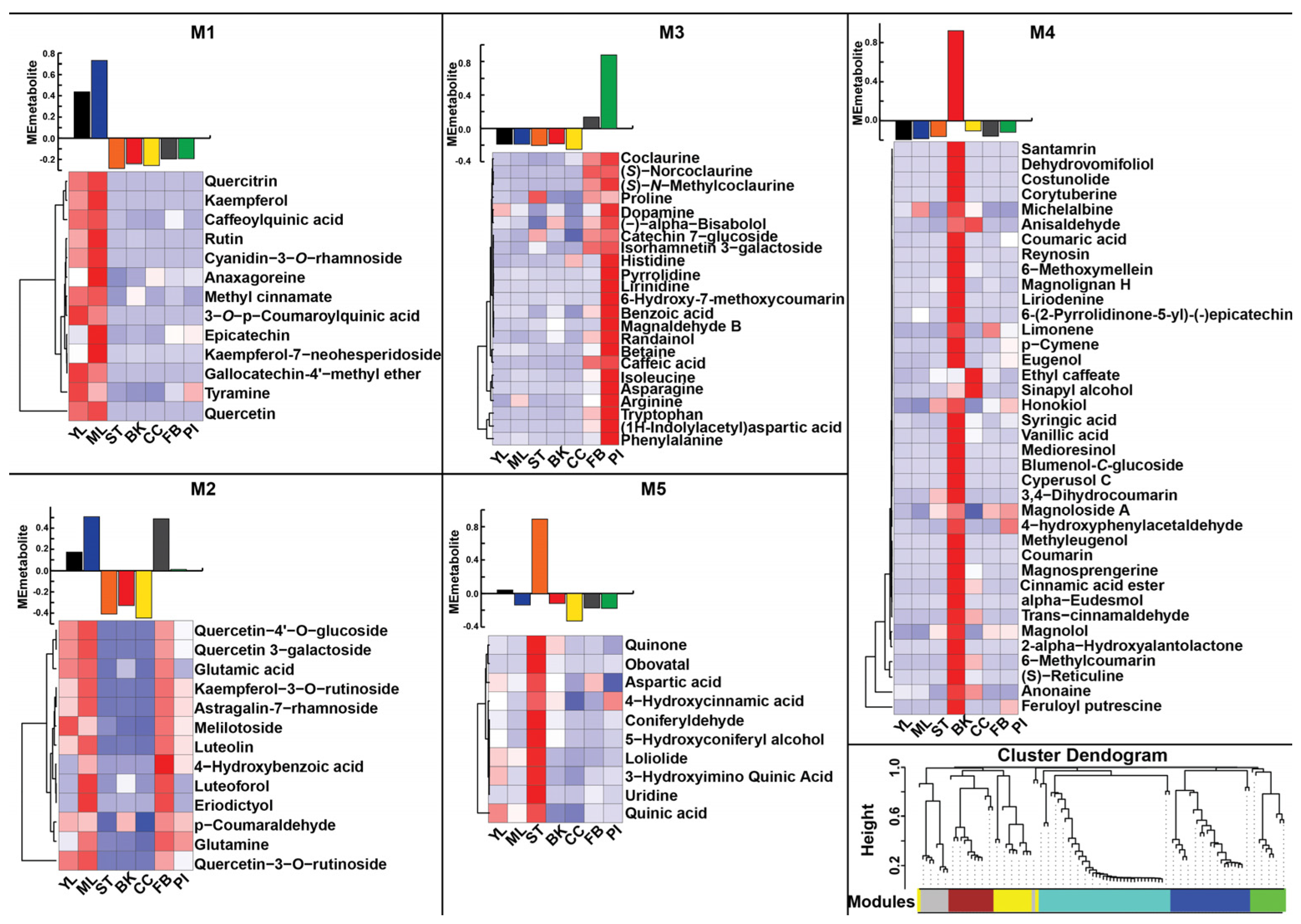
2.1. Metabo-Constituents of Magnolia obovata and Its Chemo-Diversity Revealed by Untargeted Metabolomics
2.2. Distinctive Metabolic Signatures of Magnolia obovata Tissues
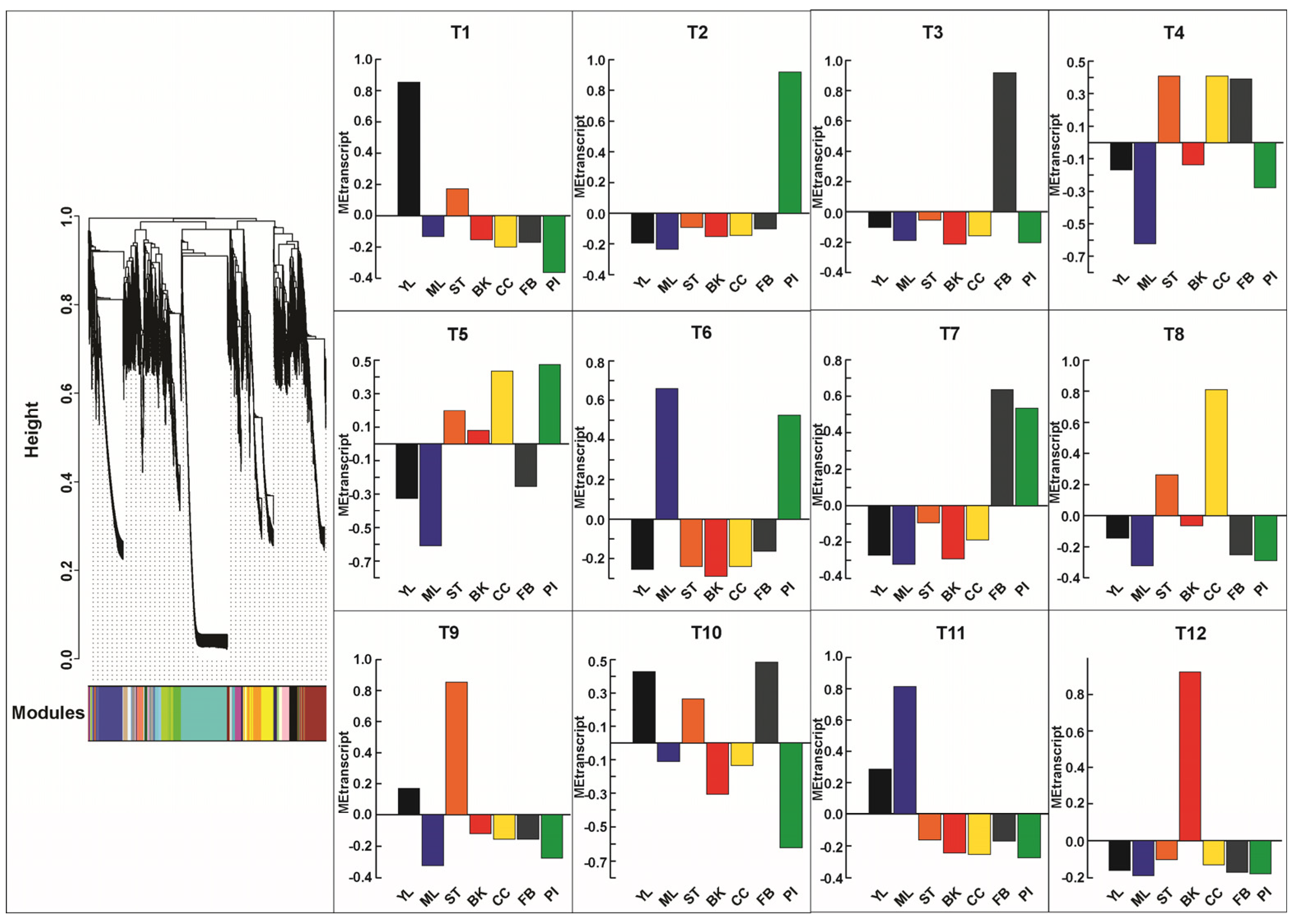
2.3. Genomic Features Characteristics Through De Novo Transcriptome Assembly of Magnolia obovata
2.4. Transcripts Expression Analysis Reveals the Tissue-Specific Molecular Signatures in Magnolia obovata
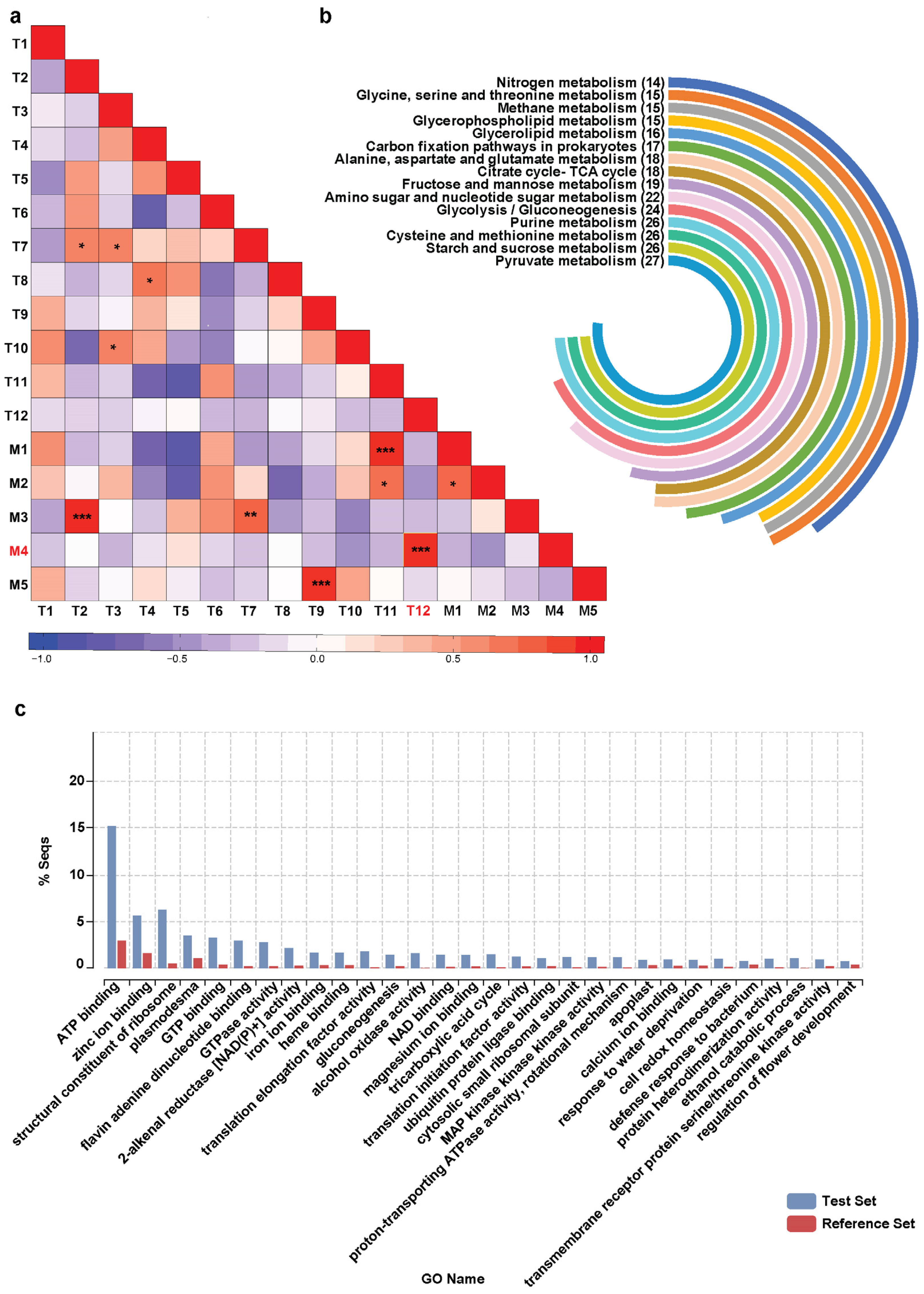
2.5. Construction of Gene-Metabolite Co-Expression Network Uncovers Tissue-Specific Transcriptome Signature and Specialized Metabolites Biosynthesis Pathways in Magnolia obovata
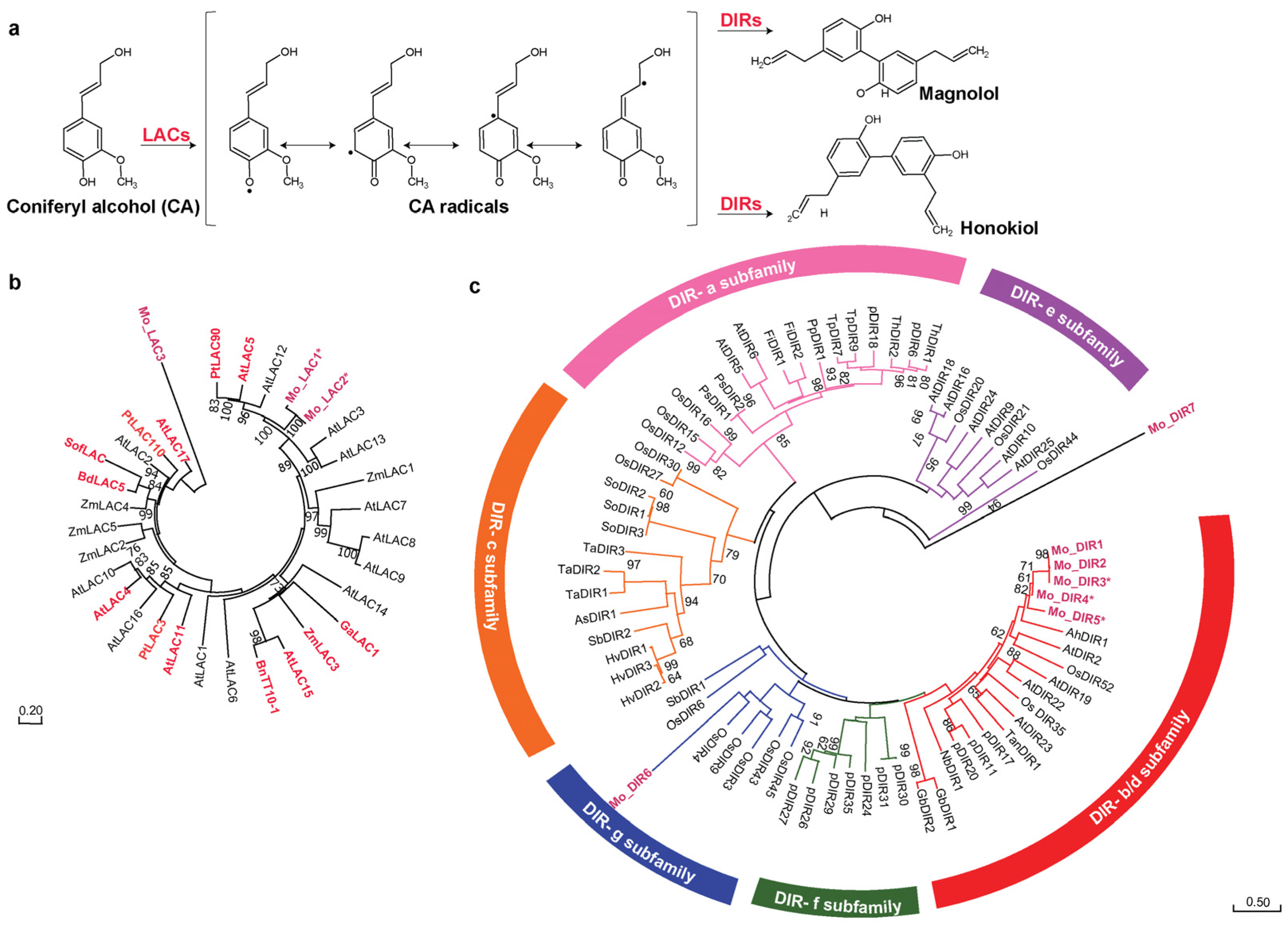
2.6. Merging Omics Landscapes to Decode Neolignans Biosynthesis in Magnolia obovata
3. Materials and Methods
3.1. Plant Materials
3.2. Untargeted Metabolite Analysis for Seven Tissues of Magnolia obovata
3.3. RNA Extraction, and cDNA Library Preparation
3.4. Illumina Sequencing
3.5. RNA-seq Raw Reads Pre-Processing, De Novo Transcriptome Assembly, and Functional Classification of Transcripts
3.6. Module Construction Using Omics Dataset of Magnolia obovata
3.7. Integrative Omics Analysis Through Correlation of Metabolite and Transcript Modules of Magnolia obovata
3.8. Phylogenetic Analysis of Candidate Laccase and Dirigent Proteins in Magnolia obovata
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kwon, O.J.; Oh, C.H. Naturalization of landscaping woody plant, Magnolia obovata potentially invasive species. J. Mt. Sci. 2015, 12, 30–38. [Google Scholar] [CrossRef]
- Lovecka, P.; Svobodova, A.; Macurkova, A.; Vrchotova, B.; Demnerova, K.; Wimmer, Z. Decorative Magnolia Plants: A Comparison of the Content of Their Biologically Active Components Showing Antimicrobial Effects. Plants 2020, 9, 879. [Google Scholar] [CrossRef]
- Lee, Y.J.; Lee, Y.M.; Lee, C.K.; Jung, J.K.; Han, S.B.; Hong, J.T. Therapeutic applications of compounds in the Magnolia family. Pharmacol. Ther. 2011, 130, 157–176. [Google Scholar] [CrossRef]
- Poivre, M.; Duez, P. Biological activity and toxicity of the Chinese herb Magnolia officinalis Rehder & E. Wilson (Houpo) and its constituents. J. Zhejiang Univ. Sci. B 2017, 18, 194–214. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, Y.; Kuribara, H. Overview of the Pharmacological Features of Honokiol. CNS Drug Rev. 2000, 6, 35–44. [Google Scholar] [CrossRef]
- Zhou, L.; Hou, F.; Wang, L.; Zhang, L.; Wang, Y.; Yin, Y.; Pei, J.; Peng, C.; Qin, X.; Gao, J. The genome of Magnolia hypoleuca provides a new insight into cold tolerance and the evolutionary position of magnoliids. Front. Plant Sci. 2023, 14, 1108701. [Google Scholar] [CrossRef]
- Oshima, N.; Kume, H.; Umeda, T.; Takito, H.; Tsukimoto, M.; Hada, N. Structures and Inhibitory Activities for Interleukin-2 Production of Seasonally Variable Constituents in Flower Parts of Magnolia kobus at Different Growth Stages. Chem. Pharm. Bull. 2020, 68, 91–95. [Google Scholar] [CrossRef]
- Sarker, S.D.; Maruyama, Y.J. Magnolia: The Genus Magnolia; Taylor & Francis: New York, NY, USA, 2002; p. xi. 187p. [Google Scholar]
- Luo, H.; Wu, H.; Yu, X.; Zhang, X.; Lu, Y.; Fan, J.; Tang, L.; Wang, Z. A review of the phytochemistry and pharmacological activities of Magnoliae officinalis cortex. J. Ethnopharmacol. 2019, 236, 412–442. [Google Scholar] [CrossRef]
- Dominiak, K.; Gostynska, A.; Szulc, M.; Stawny, M. The Anticancer Application of Delivery Systems for Honokiol and Magnolol. Cancers 2024, 16, 2257. [Google Scholar] [CrossRef] [PubMed]
- Shinbo, Y.; Nakamura, Y.; Altaf-Ul-Amin, M.; Asahi, H.; Kurokawa, K.; Arita, M.; Saito, K.; Ohta, D.; Shibata, D.; Kanaya, S. KNApSAcK: A Comprehensive Species-Metabolite Relationship Database. In Plant Metabolomics; Saito, K., Dixon, R.A., Willmitzer, L., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 165–181. [Google Scholar]
- Rai, A.; Saito, K.; Yamazaki, M. Integrated omics analysis of specialized metabolism in medicinal plants. Plant J. 2017, 90, 764–787. [Google Scholar] [CrossRef]
- Rai, A.; Yamazaki, M.; Saito, K. A new era in plant functional genomics. Curr. Opin. Syst. Biol. 2019, 15, 58–67. [Google Scholar] [CrossRef]
- Rai, A.; Kamochi, H.; Suzuki, H.; Nakamura, M.; Takahashi, H.; Hatada, T.; Saito, K.; Yamazaki, M. De novo transcriptome assembly and characterization of nine tissues of Lonicera japonica to identify potential candidate genes involved in chlorogenic acid, luteolosides, and secoiridoid biosynthesis pathways. J. Nat. Med. 2017, 71, 1–15. [Google Scholar] [CrossRef]
- Rai, A.; Nakamura, M.; Takahashi, H.; Suzuki, H.; Saito, K.; Yamazaki, M. High-throughput sequencing and de novo transcriptome assembly of Swertia japonica to identify genes involved in the biosynthesis of therapeutic metabolites. Plant Cell Rep. 2016, 35, 2091–2111. [Google Scholar] [CrossRef]
- Rai, A.; Yamazaki, M.; Takahashi, H.; Nakamura, M.; Kojoma, M.; Suzuki, H.; Saito, K. RNA-seq Transcriptome Analysis of Panax japonicus, and Its Comparison with Other Panax Species to Identify Potential Genes Involved in the Saponins Biosynthesis. Front. Plant Sci. 2016, 7, 481. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Rai, A.; Saito, K.; Nakabayashi, R. Metabolomics and complementary techniques to investigate the plant phytochemical cosmos. Nat. Prod. Rep. 2021, 38, 1729–1759. [Google Scholar] [CrossRef]
- Rai, M.; Rai, A.; Mori, T.; Nakabayashi, R.; Yamamoto, M.; Nakamura, M.; Suzuki, H.; Saito, K.; Yamazaki, M. Gene-Metabolite Network Analysis Revealed Tissue-Specific Accumulation of Therapeutic Metabolites in Mallotus japonicus. Int. J. Mol. Sci. 2021, 22, 8835. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.; Rai, M.; Kamochi, H.; Mori, T.; Nakabayashi, R.; Nakamura, M.; Suzuki, H.; Saito, K.; Yamazaki, M. Multiomics-based characterization of specialized metabolites biosynthesis in Cornus officinalis. DNA Res. 2020, 27, dsaa009. [Google Scholar] [CrossRef]
- Rai, A.; Nakaya, T.; Shimizu, Y.; Rai, M.; Nakamura, M.; Suzuki, H.; Saito, K.; Yamazaki, M. De Novo Transcriptome Assembly and Characterization of Lithospermum officinale to Discover Putative Genes Involved in Specialized Metabolites Biosynthesis. Planta Med. 2018, 84, 920–934. [Google Scholar] [CrossRef]
- Dharmananda, S. Safety Issues Affecting Chinese Herbs: The Case of Kava; ITM: Portland, OR, USA, 2002. [Google Scholar]
- Yan, R.; Wang, W.; Guo, J.; Liu, H.; Zhang, J.; Yang, B. Studies on the alkaloids of the bark of Magnolia officinalis: Isolation and on-line analysis by HPLC-ESI-MS(n). Molecules 2013, 18, 7739. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.X.; Yan, R.Y.; Liang, R.X.; Wang, W.; Yang, B. Bioactive polar compounds from stem bark of Magnolia officinalis. Fitoterapia 2012, 83, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Seo, K.H.; Lee, D.Y.; Lee, D.S.; Park, J.H.; Jeong, R.H.; Jung, Y.J.; Shrestha, S.; Chung, I.S.; Kim, G.S.; Kim, Y.C.; et al. Neolignans from the fruits of Magnolia obovata and their inhibition effect on NO production in LPS-induced RAW 264.7 cells. Planta Med. 2013, 79, 1335–1340. [Google Scholar] [CrossRef]
- Barros, L.F.; Barison, A.; Salvador, M.J.; de Mello-Silva, R.; Cabral, E.C.; Eberlin, M.N.; Stefanello, M.E. Constituents of the leaves of Magnolia ovata. J. Nat. Prod. 2009, 72, 1529–1532. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, N.H.; Nam, J.W.; Lee, Y.M.; Jang, D.S.; Kim, Y.S.; Nam, S.H.; Seo, E.K.; Yang, M.S.; Kim, J.S. Scopoletin from the flower buds of Magnolia fargesii inhibits protein glycation, aldose reductase, and cataractogenesis ex vivo. Arch. Pharm. Res. 2010, 33, 1317–1323. [Google Scholar] [CrossRef] [PubMed]
- Park, C.H.; Park, S.Y.; Lee, S.Y.; Kim, J.K.; Park, S.U. Analysis of Metabolites in White Flowers of Magnolia denudata Desr. and Violet Flowers of Magnolia Liliiflora Desr. Molecules 2018, 23, 1558. [Google Scholar] [CrossRef] [PubMed]
- Dhandapani, S.; Jin, J.; Sridhar, V.; Sarojam, R.; Chua, N.H.; Jang, I.C. Integrated metabolome and transcriptome analysis of Magnolia champaca identifies biosynthetic pathways for floral volatile organic compounds. BMC Genom. 2017, 18, 463. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, H.; Hyun, H.B.; Go, B.; Kim, S.C.; Jung, Y.H.; Ham, Y.M. Profiles of Essential Oils and Correlations with Phenolic Acids and Primary Metabolites in Flower Buds of Magnolia heptapeta and Magnolia denudata var. purpurascens. Molecules 2021, 27, 221. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Kind, T.; Nakabayashi, R.; Yukihira, D.; Tanaka, W.; Cajka, T.; Saito, K.; Fiehn, O.; Arita, M. Hydrogen Rearrangement Rules: Computational MS/MS Fragmentation and Structure Elucidation Using MS-FINDER Software. Anal. Chem. 2016, 88, 7946–7958. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Yang, L.; Wang, D.; Shu, Z.; Yang, Y.; Chen, S.; Song, C. 1 K Medicinal Plant Genome Database: An integrated database combining genomes and metabolites of medicinal plants. Hortic. Res. 2022, 9, uhac075. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.; Hirakawa, H.; Nakabayashi, R.; Kikuchi, S.; Hayashi, K.; Rai, M.; Tsugawa, H.; Nakaya, T.; Mori, T.; Nagasaki, H.; et al. Chromosome-level genome assembly of Ophiorrhiza pumila reveals the evolution of camptothecin biosynthesis. Nat. Commun. 2021, 12, 405. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Rai, A.; Rai, M.; Nakamura, M.; Kawano, N.; Yoshimatsu, K.; Suzuki, H.; Kawahara, N.; Saito, K.; Yamazaki, M. Comparative transcriptome analyses of three medicinal Forsythia species and prediction of candidate genes involved in secondary metabolisms. J. Nat. Med. 2018, 72, 867–881. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.; Rai, A.; Kawano, N.; Yoshimatsu, K.; Takahashi, H.; Suzuki, H.; Kawahara, N.; Saito, K.; Yamazaki, M. De Novo RNA Sequencing and Expression Analysis of Aconitum carmichaelii to Analyze Key Genes Involved in the Biosynthesis of Diterpene Alkaloids. Molecules 2017, 22, 2155. [Google Scholar] [CrossRef] [PubMed]
- Udomsom, N.; Rai, A.; Suzuki, H.; Okuyama, J.; Imai, R.; Mori, T.; Nakabayashi, R.; Saito, K.; Yamazaki, M. Function of AP2/ERF Transcription Factors Involved in the Regulation of Specialized Metabolism in Ophiorrhiza pumila Revealed by Transcriptomics and Metabolomics. Front. Plant Sci. 2016, 7, 1861. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kumar, S.; Blaxter, M.L. Comparing de novo assemblers for 454 transcriptome data. BMC Genom. 2010, 11, 571. [Google Scholar] [CrossRef] [PubMed]
- Nakasugi, K.; Crowhurst, R.; Bally, J.; Waterhouse, P. Combining transcriptome assemblies from multiple de novo assemblers in the allo-tetraploid plant Nicotiana benthamiana. PLoS ONE 2014, 9, e91776. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Xie, Y.; Wu, G.; Tang, J.; Luo, R.; Patterson, J.; Liu, S.; Huang, W.; He, G.; Gu, S.; Li, S.; et al. SOAPdenovo-Trans: De novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 2014, 30, 1660–1666. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol. Biol. 2019, 1962, 227–245. [Google Scholar] [CrossRef]
- Cristea, R.M.; Sava, C.; Căpățână, C.; Kanellou, A. Phytochemical Analysis and Specific Activities of Bark and Flower Extracts from Four Magnolia Plant Species. Horticulturae 2024, 10, 141. [Google Scholar] [CrossRef]
- Kelm, M.A.; Nair, M.G. A Brief Summary of Biologically Active Compounds from Magnolia spp. In Studies in Natural Products Chemistry; Attaur, R., Ed.; Elsevier: Amsterdam, The Netherlands, 2000; Volume 24, pp. 845–873. [Google Scholar]
- Li, D.; Liu, K.; Zhao, C.; Liang, S.; Yang, J.; Peng, Z.; Xia, A.; Yang, M.; Luo, L.; Huang, C.; et al. GWAS Combined with WGCNA of Transcriptome and Metabolome to Excavate Key Candidate Genes for Rice Anaerobic Germination. Rice 2023, 16, 49. [Google Scholar] [CrossRef] [PubMed]
- Abid, M.; Gu, S.; Zhang, Y.J.; Sun, S.; Li, Z.; Bai, D.F.; Sun, L.; Qi, X.J.; Zhong, Y.P.; Fang, J.B. Comparative transcriptome and metabolome analysis reveal key regulatory defense networks and genes involved in enhanced salt tolerance of Actinidia (kiwifruit). Hortic. Res. 2022, 9, uhac189. [Google Scholar] [CrossRef] [PubMed]
- Dowle, E.J.; Powell, T.H.Q.; Doellman, M.M.; Meyers, P.J.; Calvert, M.B.; Walden, K.K.O.; Robertson, H.M.; Berlocher, S.H.; Feder, J.L.; Hahn, D.A.; et al. Genome-wide variation and transcriptional changes in diverse developmental processes underlie the rapid evolution of seasonal adaptation. Proc. Natl. Acad. Sci. USA 2020, 117, 23960–23969. [Google Scholar] [CrossRef]
- Zheng, J.; He, C.; Qin, Y.; Lin, G.; Park, W.D.; Sun, M.; Li, J.; Lu, X.; Zhang, C.; Yeh, C.T.; et al. Co-expression analysis aids in the identification of genes in the cuticular wax pathway in maize. Plant J. 2019, 97, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Shahan, R.; Zawora, C.; Wight, H.; Sittmann, J.; Wang, W.; Mount, S.M.; Liu, Z. Consensus Coexpression Network Analysis Identifies Key Regulators of Flower and Fruit Development in Wild Strawberry. Plant Physiol. 2018, 178, 202–216. [Google Scholar] [CrossRef]
- Xu, Y.; Magwanga, R.O.; Yang, X.; Jin, D.; Cai, X.; Hou, Y.; Wei, Y.; Zhou, Z.; Wang, K.; Liu, F. Genetic regulatory networks for salt-alkali stress in Gossypium hirsutum with differing morphological characteristics. BMC Genom. 2020, 21, 15. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Wang, B.; Yang, J.; Wang, W.; Liu, A.; Leng, L.; Xiang, L.; Song, C.; Chen, S. Weighted Gene Co-expression Network Analysis of the Dioscin Rich Medicinal Plant Dioscorea nipponica. Front. Plant Sci. 2017, 8, 789. [Google Scholar] [CrossRef]
- Umezawa, T.; Yamamura, M.; Nakatsubo, T.; Suzuki, S.; Hattori, T. Stereoselectivity of the Biosynthesis of Norlignans and Related Compounds. In The Biological Activity of Phytochemicals; Gang, D.R., Ed.; Springer: New York, NY, USA, 2011; pp. 179–197. [Google Scholar]
- Suzuki, S.; Umezawa, T. Biosynthesis of lignans and norlignans. J. Wood Sci. 2007, 53, 273–284. [Google Scholar] [CrossRef]
- Davin, L.B.; Lewis, N.G. An historical perspective on lignan biosynthesis: Monolignol, allylphenol and hydroxycinnamic acid coupling and downstream metabolism. Phytochem. Rev. 2003, 2, 257–288. [Google Scholar] [CrossRef]
- Umezawa, T. Diversity in lignan biosynthesis. Phytochem. Rev. 2003, 2, 371–390. [Google Scholar] [CrossRef]
- Katayama, T.; Kado, Y. Formation of optically active neolignans from achiral coniferyl alcohol by cell-free extracts of Eucommia ulmoides. J. Wood Sci. 1998, 44, 244–246. [Google Scholar] [CrossRef]
- Lourith, N.; Katayama, T.; Ishikawa, K.; Suzuki, T. Biosynthesis of a syringyl 8-O-4′ neolignan in Eucommia ulmoides: Formation of syringylglycerol-8-O-4′-(sinapyl alcohol) ether from sinapyl alcohol. J. Wood Sci. 2005, 51, 379–386. [Google Scholar] [CrossRef]
- Orr, J.D.; Lynn, D.G. Biosynthesis of dehydrodiconiferyl alcohol glucosides: Implications for the control of tobacco cell growth. Plant Physiol. 1992, 98, 343–352. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sartorelli, P.c.; Benevides, P.J.C.; Ellensohn, R.M.; Rocha, M.V.A.F.; Moreno, P.R.H.; Kato, M.J. Enantioselective conversion of p-hydroxypropenylbenzene to (+)-conocarpan in Piper regnellii. Plant Sci. 2001, 161, 1083–1088. [Google Scholar] [CrossRef]
- Cardullo, N.; Muccilli, V.; Tringali, C. Laccase-mediated synthesis of bioactive natural products and their analogues. RSC Chem. Biol. 2022, 3, 614–647. [Google Scholar] [CrossRef] [PubMed]
- Janusz, G.; Pawlik, A.; Swiderska-Burek, U.; Polak, J.; Sulej, J.; Jarosz-Wilkolazka, A.; Paszczynski, A. Laccase Properties, Physiological Functions, and Evolution. Int. J. Mol. Sci. 2020, 21, 966. [Google Scholar] [CrossRef] [PubMed]
- Niemetz, R.; Gross, G.G. Oxidation of pentagalloylglucose to the ellagitannin, tellimagrandin II, by a phenol oxidase from Tellima grandiflora leaves. Phytochemistry 2003, 62, 301–306. [Google Scholar] [CrossRef]
- Niemetz, R.; Gross, G.G. Ellagitannin biosynthesis: Laccase-catalyzed dimerization of tellimagrandin II to cornusiin E in Tellima grandiflora. Phytochemistry 2003, 64, 1197–1201. [Google Scholar] [CrossRef] [PubMed]
- Davin, L.B.; Wang, H.B.; Crowell, A.L.; Bedgar, D.L.; Martin, D.M.; Sarkanen, S.; Lewis, N.G. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 1997, 275, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Yamamura, M.; Matsuda, F.; Ono, E.; Nakabayashi, R.; Sugawara, S.; Mori, T.; Tobimatsu, Y.; Umezawa, T.; Saito, K. Seed-coat protective neolignans are produced by the dirigent protein AtDP1 and the laccase AtLAC5 in Arabidopsis. Plant Cell 2021, 33, 129–152. [Google Scholar] [CrossRef] [PubMed]
- Ranocha, P.; McDougall, G.; Hawkins, S.; Sterjiades, R.; Borderies, G.; Stewart, D.; Cabanes-Macheteau, M.; Boudet, A.M.; Goffner, D. Biochemical characterization, molecular cloning and expression of laccases—A divergent gene family—in poplar. Eur. J. Biochem. 1999, 259, 485–495. [Google Scholar] [CrossRef]
- Razna, K.; Harencar, L.; Kucka, M. The Involvement of microRNAs in Plant Lignan Biosynthesis-Current View. Cells 2022, 11, 2151. [Google Scholar] [CrossRef]
- Corbin, C.; Drouet, S.; Markulin, L.; Auguin, D.; Laine, E.; Davin, L.B.; Cort, J.R.; Lewis, N.G.; Hano, C. A genome-wide analysis of the flax (Linum usitatissimum L.) dirigent protein family: From gene identification and evolution to differential regulation. Plant Mol. Biol. 2018, 97, 73–101. [Google Scholar] [CrossRef]
- Yadav, V.; Wang, Z.; Yang, X.; Wei, C.; Changqing, X.; Zhang, X. Comparative Analysis, Characterization and Evolutionary Study of Dirigent Gene Family in Cucurbitaceae and Expression of Novel Dirigent Peptide against Powdery Mildew Stress. Genes 2021, 12, 326. [Google Scholar] [CrossRef]
- Meng, Q.; Kim, S.J.; Costa, M.A.; Moinuddin, S.G.A.; Celoy, R.M.; Smith, C.A.; Cort, J.R.; Davin, L.B.; Lewis, N.G. Dirigent protein subfamily function and structure in terrestrial plant phenol metabolism. Methods Enzym. 2023, 683, 101–150. [Google Scholar] [CrossRef]
- Luo, R.; Pan, W.; Liu, W.; Tian, Y.; Zeng, Y.; Li, Y.; Li, Z.; Cui, L. The barley DIR gene family: An expanded gene family that is involved in stress responses. Front. Genet. 2022, 13, 1042772. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Li, B.; Zhu, T.; Xue, B. Genome-wide identification and expression profiling analysis of DIR gene family in Setaria italica. Front. Plant Sci. 2023, 14, 1243806. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]


| Assembler | Kmer | No. of Contigs | N50 | Average Length | Median Length | Max Length |
|---|---|---|---|---|---|---|
| CLC | 20 | 126,234 | 1126 | 719 | 408 | 15,296 |
| Trinity | 25 | 404,394 | 1230 | 734 | 403 | 15,776 |
| SOAPdenovo | 31 | 147,639 | 1239 | 720 | 384 | 15,241 |
| 41 | 148,423 | 1257 | 715 | 373 | 15,334 | |
| 51 | 146,829 | 1212 | 687 | 355 | 15,635 | |
| 63 | 135,683 | 1171 | 661 | 334 | 15,335 | |
| 71 | 109,186 | 1214 | 695 | 353 | 15,321 | |
| 91 | 28,652 | 1415 | 936 | 650 | 12,222 | |
| CLC_Trinity_SOAPdenovo (kmer31) CD-HIT-EST | N.A. | 284,165 | 1280 | 762 | 424 | 17,676 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rai, M.; Rai, A.; Yokosaka, T.; Mori, T.; Nakabayashi, R.; Nakamura, M.; Suzuki, H.; Saito, K.; Yamazaki, M. Multi-Omics Analysis Decodes Biosynthesis of Specialized Metabolites Constituting the Therapeutic Terrains of Magnolia obovata. Int. J. Mol. Sci. 2025, 26, 1068. https://doi.org/10.3390/ijms26031068
Rai M, Rai A, Yokosaka T, Mori T, Nakabayashi R, Nakamura M, Suzuki H, Saito K, Yamazaki M. Multi-Omics Analysis Decodes Biosynthesis of Specialized Metabolites Constituting the Therapeutic Terrains of Magnolia obovata. International Journal of Molecular Sciences. 2025; 26(3):1068. https://doi.org/10.3390/ijms26031068
Chicago/Turabian StyleRai, Megha, Amit Rai, Towa Yokosaka, Tetsuya Mori, Ryo Nakabayashi, Michimi Nakamura, Hideyuki Suzuki, Kazuki Saito, and Mami Yamazaki. 2025. "Multi-Omics Analysis Decodes Biosynthesis of Specialized Metabolites Constituting the Therapeutic Terrains of Magnolia obovata" International Journal of Molecular Sciences 26, no. 3: 1068. https://doi.org/10.3390/ijms26031068
APA StyleRai, M., Rai, A., Yokosaka, T., Mori, T., Nakabayashi, R., Nakamura, M., Suzuki, H., Saito, K., & Yamazaki, M. (2025). Multi-Omics Analysis Decodes Biosynthesis of Specialized Metabolites Constituting the Therapeutic Terrains of Magnolia obovata. International Journal of Molecular Sciences, 26(3), 1068. https://doi.org/10.3390/ijms26031068






