1. Introduction
Cervical cancer is a widespread disease in women, responsible for significant global morbidity and mortality. Among the various risk factors, HPV infection is the leading cause, being implicated in approximately 99.7% of all cases [
1].
The human immune system plays a pivotal role in defending against infections, including those caused by HPV. However, in the context of HPV infection, certain immune responses can inadvertently contribute to tumor progression. For instance, an increase in neutrophil populations has been observed to suppress T-cell-mediated antitumor activity, thereby facilitating the development of malignancies and impairing overall immune function [
2]. HPV-infected cells also exhibit elevated expression of genes associated with pro-inflammatory mediators, such as interleukin-1 beta (
IL-1β). The inflammasome, a multi-protein complex, is responsible for the activation of caspase-1, which in turn processes and secretes
IL-1β and
IL-18. These cytokines play crucial roles in initiating and propagating inflammatory responses [
3]. On the other hand, the
NLRP3 inflammasome acts as a molecular sensor, triggering pyroptosis, which can both promote and hinder cancer progression depending on the tumor microenvironment. Activation of
NLRP3 can enhance immune responses by recruiting immune cells to the tumor, but chronic activation leads to sustained inflammation, promoting tumor growth, invasion, and metastasis. This dual role highlights
NLRP3 as a potential therapeutic target in cancer, as well as its relevance in immune modulation during viral infections like SARS-CoV-2 [
4].
Despite significant advances in the study and treatment of cervical cancer caused by HPV infection, there are still considerable gaps in understanding the precise mechanisms involved in the interactions between the virus and the immune system. Recent studies have demonstrated the importance of metabolic and immune-related gene signatures in cancer progression and treatment response. For example, a 12-gene signature related to OXPHOS (oxidative phosphorylation) has been shown to predict ovarian cancer patients’ response to immune checkpoint inhibitors, linking metabolic pathways with immune regulation in cancer [
5]. Additionally, the role of exosomal miRNAs in modulating immune responses and inflammation further highlights the complexity of immune evasion mechanisms in cancers, including cervical cancer [
6].
A recent transcriptomic study revealed that
EGCG and
myricetin exhibited synergistic antiproliferative effects on cervical cancer cell lines (ME180 and SiHa), with strong molecular binding to the
HK2 and
MAP7 targets and associated regulatory miRNAs [
7]. Furthermore, another study reported a lectin protein (AEL) derived from Abelmoschus esculentus, which was fully sequenced and functionally characterized. This protein demonstrated selective cytotoxicity against cervical (HeLa) and colon (T84) cancer cell lines, while showing minimal effects on normal HEK293 cells [
8].
One of the main challenges is the complexity of the TME, which includes intricate interactions between tumor and immune cells. Recent studies have shown that tumor-associated fibroblasts (CAFs) play a crucial role in cancer progression and resistance to treatments, but the diversity of these cells and their interactions with other immune cells have not been fully elucidated [
9]. In addition, our incomplete understanding of immune pathway activation and its regulation in response to HPV infection, particularly at different stages of infection, requires further research. Pathways such as AIM2, which are activated in the early stages of infection, are suppressed in later stages of HPV infection, reflecting the complexity of the virus–immune system interactions [
10]. Furthermore, HPV exploits oncogenes like E6 and E7 to challenge the immune system by inhibiting various immune pathways, such as the cGAS-STING pathway, ultimately leading to reduced immune and inflammatory responses [
11]. Understanding these mechanisms and examining their impact on the progression of cervical cancer, particularly in advanced stages, remains critically important and necessitates further in-depth investigation. These scientific gaps highlight the need for additional studies on various immune mechanisms and molecular pathways in order to gain a better understanding of HPV-driven cervical cancer development and design more effective therapeutic strategies.
The primary objective of this study is to accurately identify and analyze the role of immune hub genes and molecular pathways in cervical cancer caused by HPV infection. This research aims to examine the complex and precise effects of HPV on the transcriptomic changes and immune responses within the TME using transcriptomic data and advanced analyses. The main innovation of this study lies in focusing on identifying immune hub genes that play crucial roles in processes such as leukocyte migration and immune regulation, which are affected by HPV infection. Additionally, by utilizing protein–protein interaction networks and pathway analyses, the relationship between genetic changes and immune impacts in cervical cancer is comprehensively examined. This research identifies and investigates immune pathways influenced by HPV, revealing their effects on tumor development at both the molecular and cellular levels. These innovations could pave the way for the design of new, targeted therapeutic strategies to combat HPV-related cervical cancer, contributing to more effective prevention and treatment approaches for the disease.
2. Results
Transcriptional alterations linked to cervical cancer and HPV infection were investigated through comparative analysis of three independent datasets: GSE127265 (cervical cancer), GSE166466 (HPV-positive cervical cancer), and GSE227550 (normal cervical tissues). Following normalization and differential expression analysis, principal component analysis (PCA) was performed to evaluate the global transcriptional variation among the three datasets. As shown in
Figure 1A, the first three principal components accounted for 83.9% of the total variance (PC1: 64.9%, PC2: 10.1%, PC3: 8.8%). The PCA plot revealed a clear separation between cancer samples and normal controls, indicating robust transcriptional differences associated with malignant transformation. Moreover, HPV-positive and HPV-negative cervical cancers exhibited partial divergence in their expression profiles, suggesting HPV-dependent modulation of gene expression in addition to cancer-related alterations.
To identify DEGs and their overlaps between groups, a Venn diagram was generated (
Figure 1B). Comparative analysis revealed 669 DEGs unique to HPV-negative cervical cancer compared with controls, whereas 314 genes were uniquely dysregulated in HPV-positive cervical cancers. A total of 572 genes were commonly deregulated between both cancer groups relative to normal tissues, representing a shared oncogenic transcriptional signature. Interestingly, 108 genes were shared between HPV-negative cancers and HPV-positive cancers, while 97 genes were unique to the HPV-negative versus HPV-positive comparison. Only four genes were consistently altered across all three datasets (Detailed results are provided in
Supplementary Files S1–S3). These findings indicate both shared and HPV-specific transcriptional programs in cervical cancer progression. To further delineate HPV-specific transcriptional alterations, we focused on genes that were uniquely dysregulated between HPV-positive and HPV-negative cervical cancers. These gene subsets represent molecular differences potentially attributable to viral oncogenic activity rather than the malignant process per se.
Molecular interactions underlying HPV-associated transcriptional changes were explored by constructing a PPI network using Cytoscape (
Figure 1C). Application of stringent filtering criteria (adjusted
p < 0.05 and |log2FC| ≥ 2) reduced the candidate gene set to 100 hub genes with the highest degree of connectivity. Notably, all hub genes identified in this analysis were downregulated, indicating that HPV infection is associated with a global repression of central regulatory genes within the cervical cancer transcriptome. In the PPI network, node color intensity corresponded to the degree parameter, highlighting highly connected genes that may act as key regulators (
Figure 1D).
Gephi software (v0.9.2) was utilized for advanced network topology analysis, enabling further characterization of the functional relevance of identified hub genes, with a specific focus on immune-related genes and RTKs. Network clustering based on centrality measures (degree, betweenness, closeness, and eigenvector) enabled the identification of the most influential hub genes (
Figure 1E). These genes, predominantly associated with immune surveillance and RTK signaling, emerged as potential key drivers in HPV-positive cervical cancer.
Heatmap visualization further demonstrated the expression patterns of the top-ranked hub genes (
Figure 1F). Consistent with the PPI and centrality analyses, these genes displayed a marked reduction in expression in HPV-positive samples compared with HPV-negative cancers and normal tissues. The uniform downregulation of immune-related and RTK-associated hub genes in the presence of HPV strongly suggests a virus-mediated suppression of tumor immune responsiveness and tyrosine kinase signaling, which may have implications for both disease progression and therapeutic intervention.
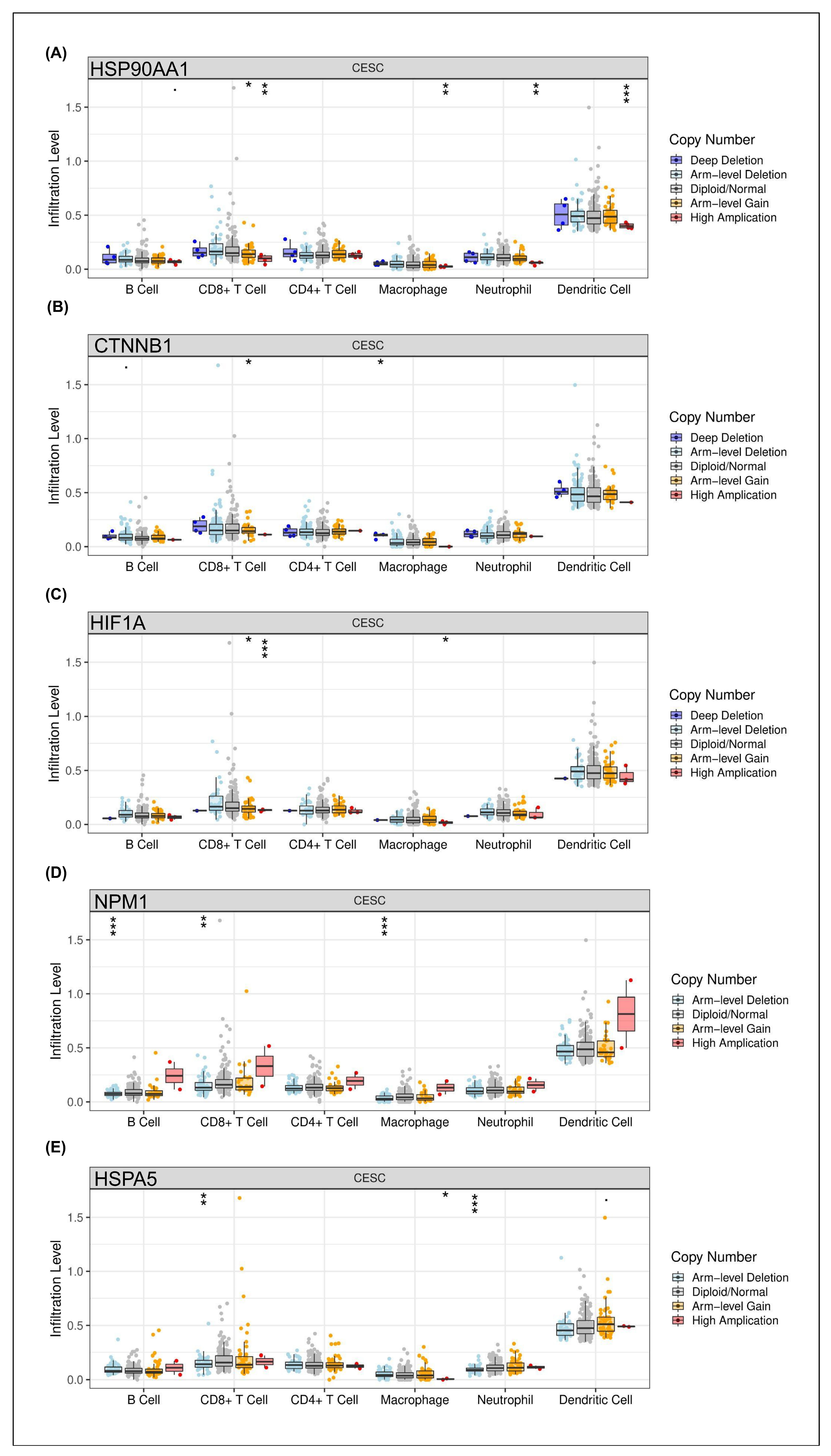
The impact of copy number alterations (CNAs) in the identified hub genes on the tumor immune microenvironment was evaluated by analyzing their association with immune cell infiltration levels in CESC using the TIMER database. The results revealed distinct patterns of immune modulation depending on the specific gene and CNA status.
Specifically, a significant association between
HSPA90AA1 amplification and increased dendritic cell infiltration was observed, whereas relatively lower immune cell levels were found in diploid/normal and deletion states (
Figure 2A). Similarly, strong correlations between
CTNNB1 CNAs and dendritic cell recruitment were detected, suggesting that antigen presentation capacity in CESC may be influenced by alterations in Wnt/β-catenin signaling (
Figure 2B). For
HIF1A, known as a hypoxia-inducible transcription factor, copy number variation was modestly linked to neutrophil and dendritic cell infiltration, indicating partial regulation of innate immune responses within the tumor microenvironment by hypoxic signaling (
Figure 2C). In contrast, significant correlations between
NPM1 alterations and changes in B cell and CD8
+ T cell infiltration were identified, implicating this nucleolar protein in adaptive immune modulation (
Figure 2D). Interestingly, consistent associations between
HSPA5 CNAs and dendritic cell as well as CD8
+ T cell infiltration were observed (
Figure 2E).
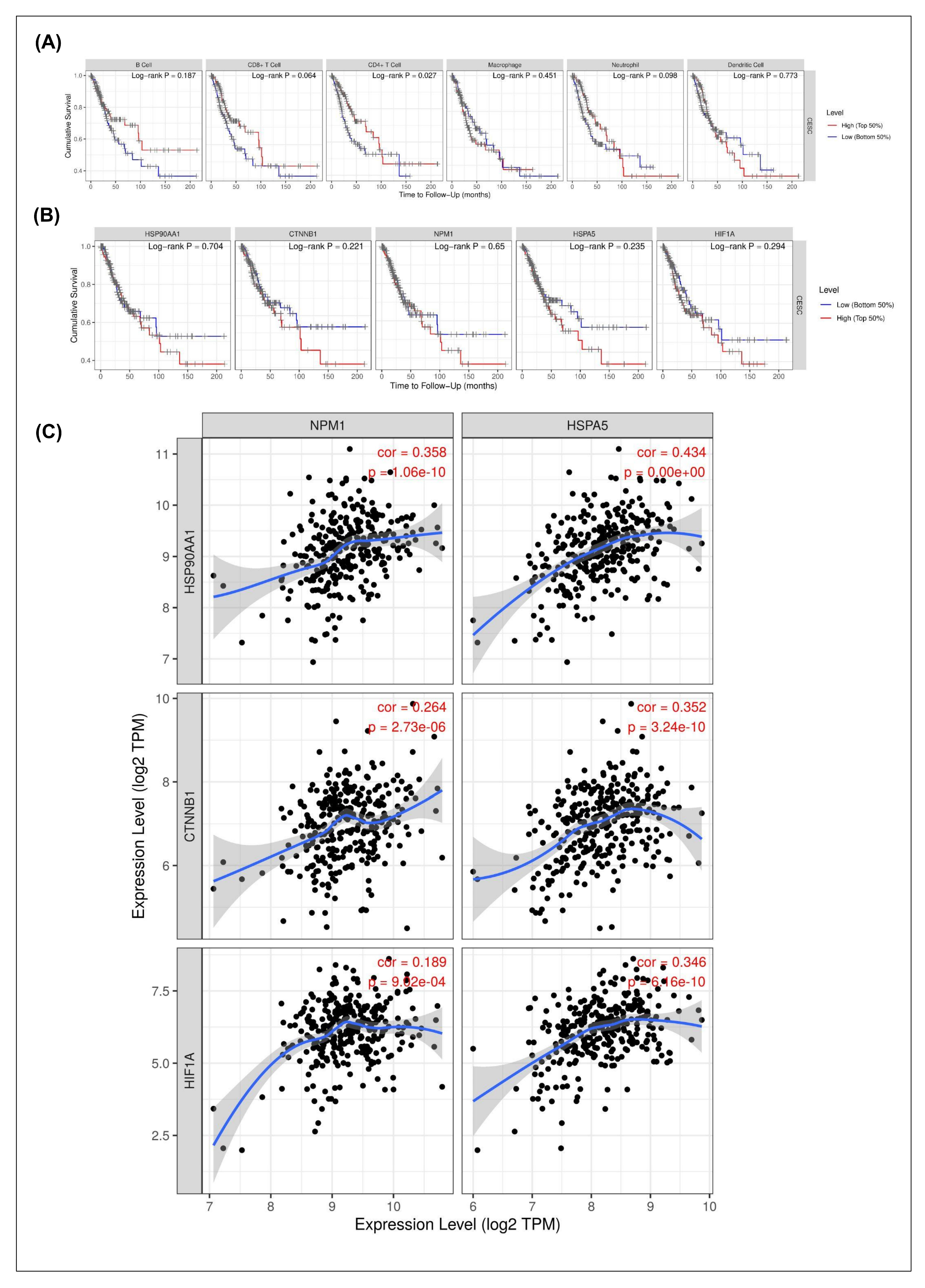
The prognostic relevance of immune infiltration and hub gene expression in CESC was assessed using Kaplan–Meier survival analyses (
Figure 3A,B). Among immune cell subsets, only CD4
+ T cell infiltration showed a significant association with patient survival (log-rank
p = 0.027), whereas B cells, CD8
+ T cells, macrophages, neutrophils, and dendritic cells did not display statistically significant effects (
Figure 3A). This finding suggests that CD4
+ T cells may play a more prominent role in shaping clinical outcomes compared to other immune populations in CESC.
With respect to hub genes, none of the five candidates (
HSPA90AA1,
CTNNB1,
NPM1,
HSPA5, and
HIF1A) demonstrated a statistically significant correlation with overall survival in the TCGA-CESC cohort (all log-rank
p > 0.05;
Figure 3B). These results indicate that although these genes are differentially expressed and immune-associated, their transcriptomic levels alone may not serve as robust independent prognostic markers in cervical cancer.
Co-expression correlation analysis was conducted to further explore the molecular interrelationships among the hub genes (
Figure 3C).
NPM1 and
HSPA5 both showed a strong positive correlation with
HSPA90AA1 expression (r = 0.358 and r = 0.434, respectively;
p < 1.0 × 10
−10), suggesting coordinated regulation of stress-related chaperone pathways. In contrast,
CTNNB1 exhibited a weak negative correlation with
NPM1 (r = −0.264;
p = 2.73 × 10
−6) but a positive correlation with
HSPA5 (r = 0.352;
p = 3.24 × 10
−10), highlighting divergent regulatory dynamics within the Wnt/β-catenin signaling context. Similarly,
HIF1A expression was moderately correlated with both
NPM1 (r = 0.189;
p = 8.92 × 10
−4) and
HSPA5 (r = 0.346;
p = 6.6 × 10
−10), consistent with hypoxia-driven stress response networks.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed on the top-ranked hub genes—identified through PPI network and centrality analyses—to gain mechanistic insights into the functional implications of HPV-associated transcriptional alterations. As illustrated in
Figure 4, chord plots revealed that these hub genes were significantly enriched in multiple oncogenic and immune-related pathways.
Specifically, KEGG pathway annotation highlighted four major biological processes: antigen processing and presentation, HIF-1 signaling pathway, leukocyte transendothelial migration, and general pathways in cancer. These results underscore the interplay between viral modulation of host immune surveillance mechanisms and classical cancer-related signaling cascades. Notably, genes such as HSPA5, CTNNB1, HIF1A, and NPM1 exhibited cross-pathway involvement, suggesting a multifaceted role in HPV-driven oncogenesis.
In the GO biological process category, top enriched terms included “peptide antigen assembly with MHC class I protein complex” and “MHC class I protein complex assembly”, reinforcing the hypothesis of HPV-mediated immune evasion through impaired antigen presentation. Additional enrichment in endothelial cell adhesion and T-cell mediated immune response pathways points to alterations in tumor–immune microenvironment remodeling.
Within the cellular component ontology, the hub genes were predominantly localized to the β-catenin–TCF complex, focal adhesion sites, and cell–substrate junctions, indicating perturbations in cell adhesion, polarity, and signal transduction hubs—key hallmarks of epithelial-to-mesenchymal transition (EMT) and metastasis.From a molecular function perspective, the most enriched terms were associated with ubiquitin-like protein ligase binding, transcription coactivator binding, and protein kinase inhibitor activity (
Table 1). These functions suggest that HPV infection exerts widespread effects on proteostasis, transcriptional regulation, and intracellular signaling balance, potentially contributing to both tumor initiation and progression.
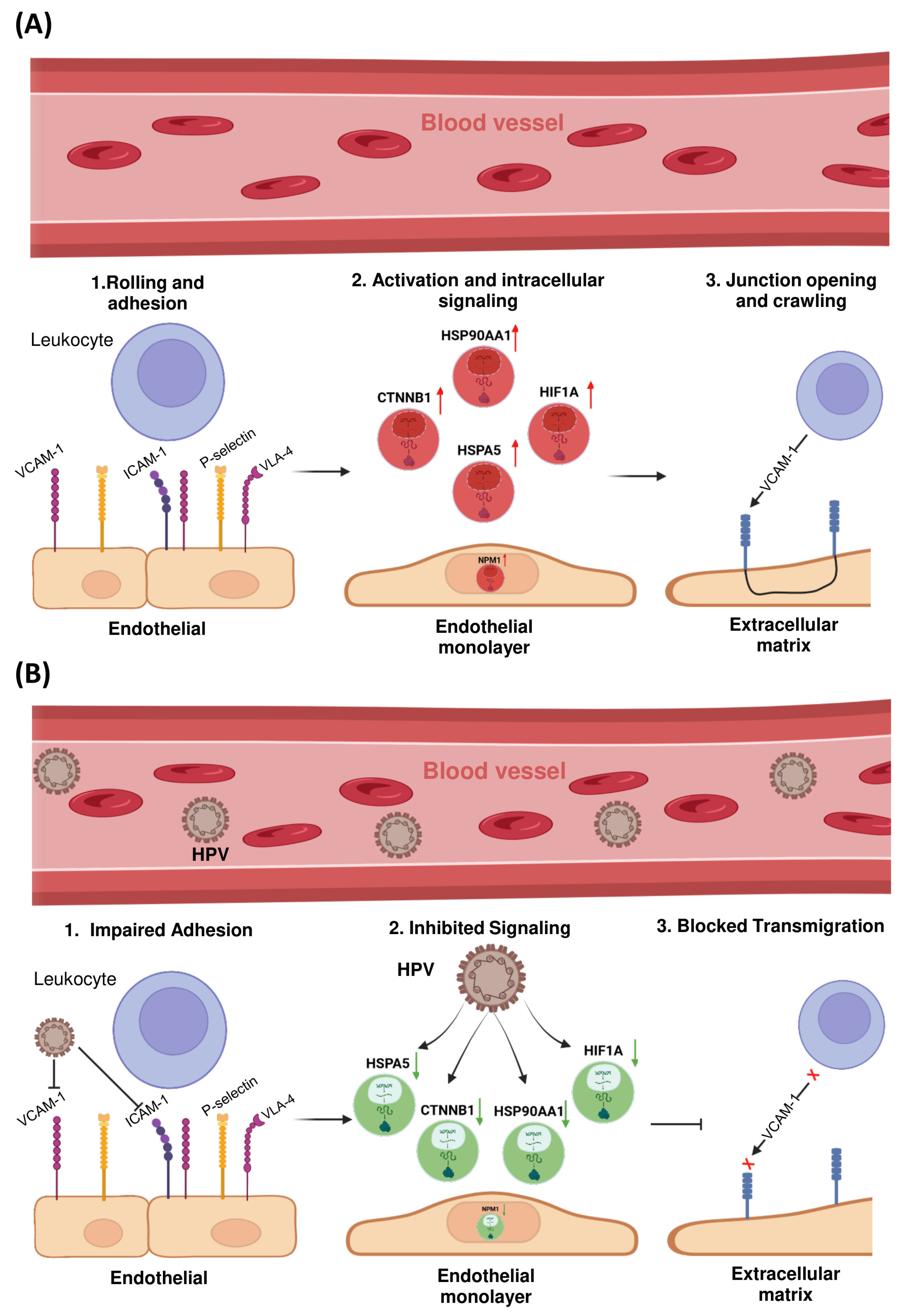
The LTM pathway was prioritized for further investigation due to the consistent enrichment of multiple HPV-associated hub genes within this axis. Given its critical role in regulating immune cell infiltration into the tumor microenvironment—and its previously reported dysregulation in HPV-positive cervical cancers—this pathway constitutes a biologically and clinically relevant focus for understanding mechanisms of viral immune evasion.
To further elucidate the mechanistic involvement of these hub genes in immune modulation, a schematic representation of the LTM pathway was constructed (
Figure 4). This visualization mapped the transcriptionally dysregulated genes onto key regulatory nodes within the pathway, revealing potential disruptions in immune cell trafficking across the endothelial barrier.
Notably, HSPA5, CTNNB1, HIF1A, and NPM1 emerged as central modulators of this axis. HSPA5, an endoplasmic reticulum chaperone involved in cellular stress responses, was found to influence the expression and trafficking of adhesion molecules such as ICAM-1 and VCAM-1, potentially impairing leukocyte adhesion and extravasation. Similarly, CTNNB1 (β-catenin), a critical component of adherens junctions and the Wnt signaling pathway, was associated with modulation of endothelial permeability and tight junction dynamics.
In addition, activation of HIF1A under hypoxic conditions may promote endothelial barrier stabilization while simultaneously altering chemokine gradients—together contributing to reduced immune cell infiltration into the tumor microenvironment. NPM1, a multifunctional nucleolar protein, was implicated in cytoskeletal remodeling and intracellular signaling crosstalk that governs transendothelial migration.
The observed downregulation of these hub genes in HPV-positive cervical cancer samples suggests that HPV infection may contribute to immune evasion not only by impairing antigen processing, but also by actively inhibiting leukocyte recruitment via suppression of the transendothelial migration pathway.
3. Discussion
This study has provided significant insights into the molecular mechanisms underlying immune evasion in HPV-driven cervical cancer. Through the transcriptomic analysis of HPV-positive and HPV-negative cervical cancer datasets, we identified a set of immune-related hub genes whose expression is downregulated in the presence of HPV. Notably, the LTM pathway emerged as a key immune evasion mechanism, highlighting how HPV infection impairs leukocyte adhesion and migration. These findings emphasize the role of specific immune pathways, such as the regulation of dendritic cell and T cell infiltration, in shaping the TME of HPV-positive cervical cancer. Furthermore, while hub gene expression did not correlate with overall survival, their regulatory interactions within the TME suggest a multifaceted role in immune modulation.
Our findings are consistent with previous studies that have highlighted immune evasion as a critical component of HPV-associated cancer progression. For instance, research by Avila et al. demonstrated that HPV infection disrupts immune surveillance through the modulation of pro-inflammatory cytokines and immune cell recruitment [
12]. Similarly, Che et al. highlighted that HPV oncoproteins E6 and E7 suppress the function of immune cells, including T-cells and dendritic cells, which are crucial for anti-tumor immunity [
13].
In support of regulatory RNA networks in cervical cancer, previous findings have shown that hsa_circ_0000021 and KPNA2 are overexpressed and act as oncogenic drivers by negatively regulating miR-3940-3p. Functional studies demonstrated that silencing either hsa_circ_0000021 or KPNA2 suppressed cervical cancer cell proliferation, invasion, and tumor growth, whereas inhibition of miR-3940-3p enhanced malignancy. Mechanistically, hsa_circ_0000021 promotes cervical cancer progression by sponging miR-3940-3p, which targets KPNA2 [
14].
Our study corroborates these findings by revealing the downregulation of immune-related hub genes such as HSPA90AA1, CTNNB1, and HIF1A in HPV-positive cervical cancer, which could impair immune cell infiltration and contribute to immune escape. Moreover, while earlier studies focused on global immune suppression, our analysis provides a deeper understanding of how specific signaling pathways, such as RTK and Wnt/β-catenin, influence immune cell infiltration.
Although the identified immune-related hub genes (
HSP90AA1,
CTNNB1,
NPM1,
HSPA5, and
HIF1A) were significantly downregulated in HPV-positive cervical cancer, none of them demonstrated a statistically significant correlation with overall survival in the TCGA-CESC cohort. Several factors may contribute to this lack of prognostic association. First, sample heterogeneity and limited cohort size can lead to underpowered survival analysis, especially when stratifying patients into high- and low-expression groups. Second, these hub genes may exhibit functional redundancy, where other genes compensate for their loss, diminishing their individual impact on clinical outcomes. Third, post-transcriptional regulation, such as changes in protein activity, localization, or degradation, might decouple gene expression levels from functional effects on survival. Finally, survival outcomes in cervical cancer are influenced by multiple variables, including treatment regimens, tumor stage, and immune contexture, which may confound single-gene prognostic signals. These findings highlight that although transcriptomic alterations of immune-related genes provide mechanistic insights into HPV-driven immune evasion, they may not serve as standalone prognostic biomarkers without integration into multi-gene or pathway-based models. Interestingly, our results also contrast with studies that have not observed significant correlations between hub gene expression and survival outcomes. For instance, previous research has identified ZNF695 as an oncogenic factor in CESC, where its overexpression was significantly associated with higher histological grade and poor prognosis, including reduced overall survival, progression-free survival, and disease-specific survival. Moreover, ZNF695 was linked to steroid hormone biosynthesis and immune infiltration, suggesting its potential as a prognostic biomarker and therapeutic target [
15]. This difference could be due to the complex, multifactorial nature of immune response regulation in cervical cancer.
The downregulation of immune-related hub genes, particularly those involved in LTM, offers novel mechanistic insights into HPV-mediated immune evasion. The LTM pathway, which regulates immune cell trafficking across the endothelial barrier, is disrupted in HPV-positive cervical cancer due to the reduced expression of key genes such as
HSPA5,
CTNNB1, and
NPM1.
HSPA5, an ER chaperone, influences the expression of adhesion molecules such as ICAM-1 and VCAM-1, impairing immune cell adhesion and extravasation [
16]. Similarly,
CTNNB1, a core component of the Wnt/β-catenin signaling pathway, affects endothelial permeability, further hindering leukocyte infiltration [
17]. Hypoxia-driven activation of
HIF1A also contributes to immune suppression by altering chemokine gradients and stabilizing the endothelial barrier, thereby reducing immune cell infiltration [
18].
These findings suggest that HPV-induced alterations in the immune microenvironment are not limited to immune cell dysfunction but also involve structural changes in the endothelial layer, impairing immune cell trafficking and subsequent immune response.
Collectively, these analyses demonstrate that although neither immune infiltration nor hub gene expression levels predict survival outcomes in CESC, their interconnected regulatory networks and coordinated transcriptional programs suggest functional crosstalk between immune-related and RTK/stress-related signaling pathways. This may provide a mechanistic link between viral oncogenesis, tumor immune evasion, and adaptive stress responses in cervical cancer.
The identification of immune-related hub genes involved in HPV-driven immune evasion provides valuable targets for therapeutic intervention. Targeting key genes in the LTM pathway, such as HSPA5 and CTNNB1, could offer novel approaches to enhance immune cell infiltration and restore immune surveillance in HPV-positive cervical cancer. Additionally, manipulating the Wnt/β-catenin and hypoxia pathways may serve as promising therapeutic strategies to counteract HPV-mediated immune suppression.
Future studies should focus on the functional validation of these hub genes in preclinical models and clinical settings. Further investigation into the role of immune checkpoint inhibitors in restoring immune function in the context of HPV-related cervical cancer is warranted. Moreover, exploring combination therapies that target both immune evasion mechanisms and cancer cell survival pathways could improve treatment outcomes for patients with HPV-positive cervical cancer.