Whole-Genome Sequencing and Functional Characterization of a Novel Kuravirus Bacteriophage with Antibiofilm Activity Against Multidrug-Resistant Avian Pathogenic Escherichia coli
Abstract
1. Introduction
2. Results
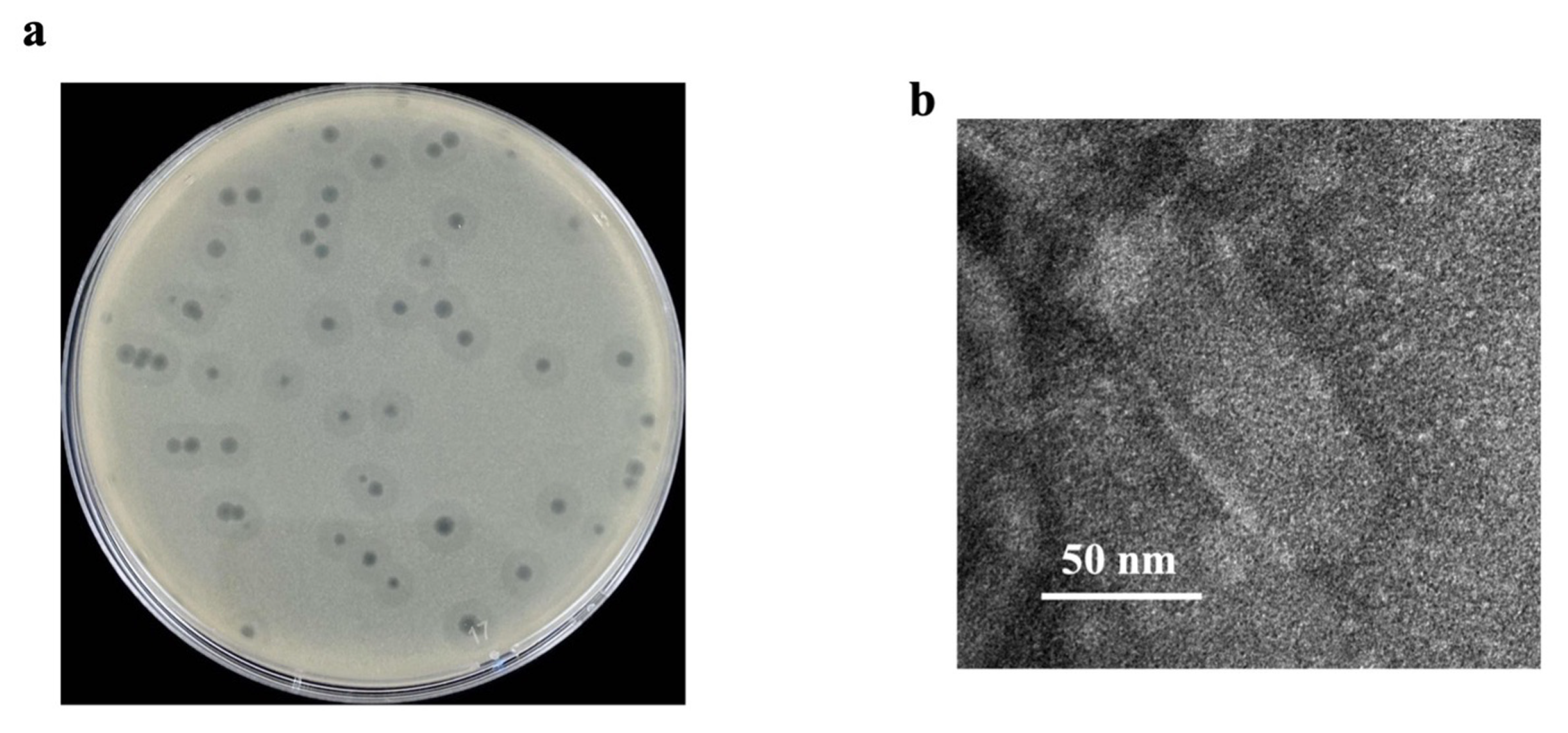
2.1. Isolation and General Features of Phage
2.2. Host Range Determination
2.3. Efficiency of Plating (EOP) of Phage
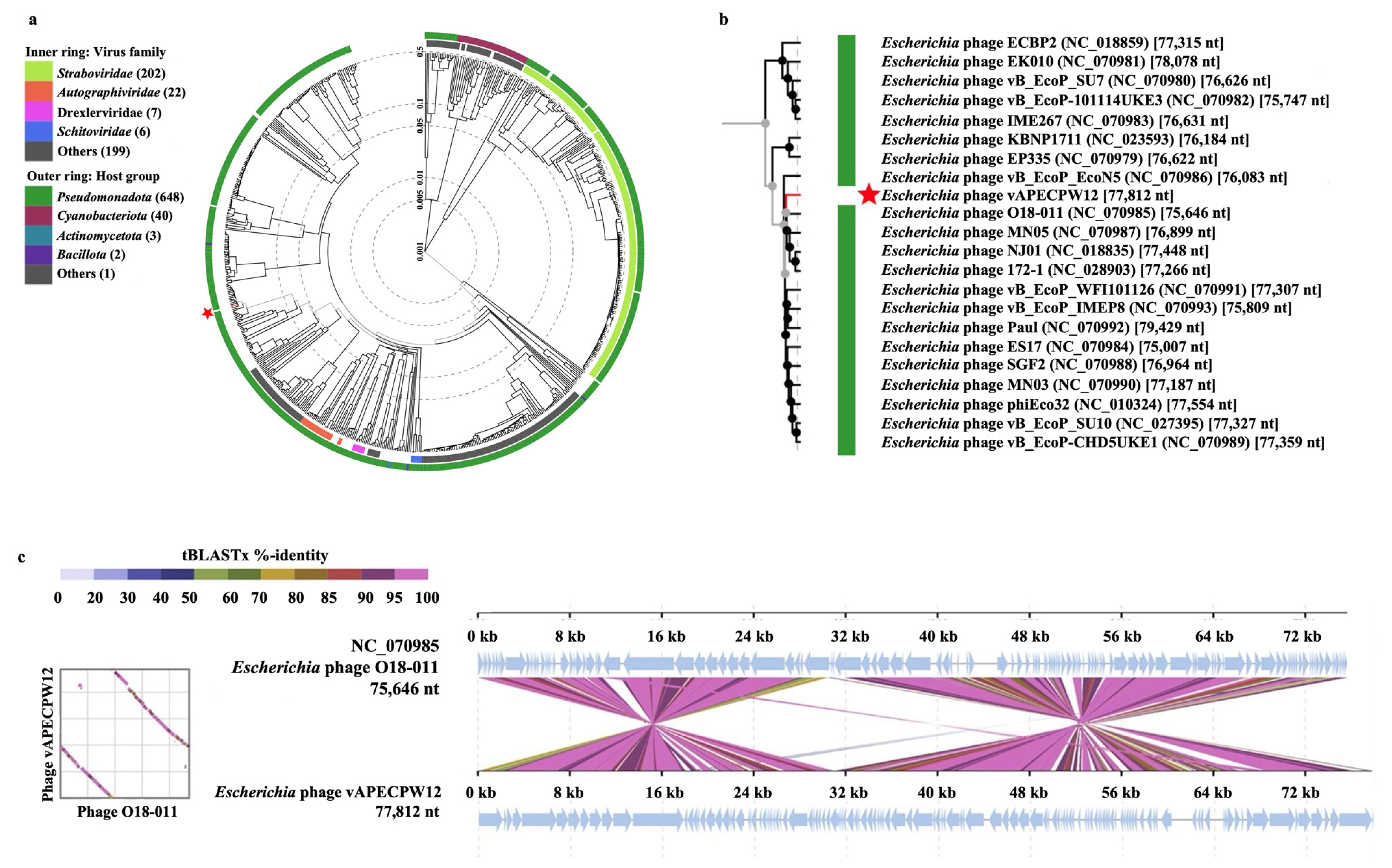
2.4. Genome Characterization of Phage
2.5. Phylogenetic and Comparative Genomic Analysis of Phage vAPECPW12
2.6. Adsorption Rate of Phage
2.7. Growth Characteristics of Phage
2.8. Lytic Activity of Phage
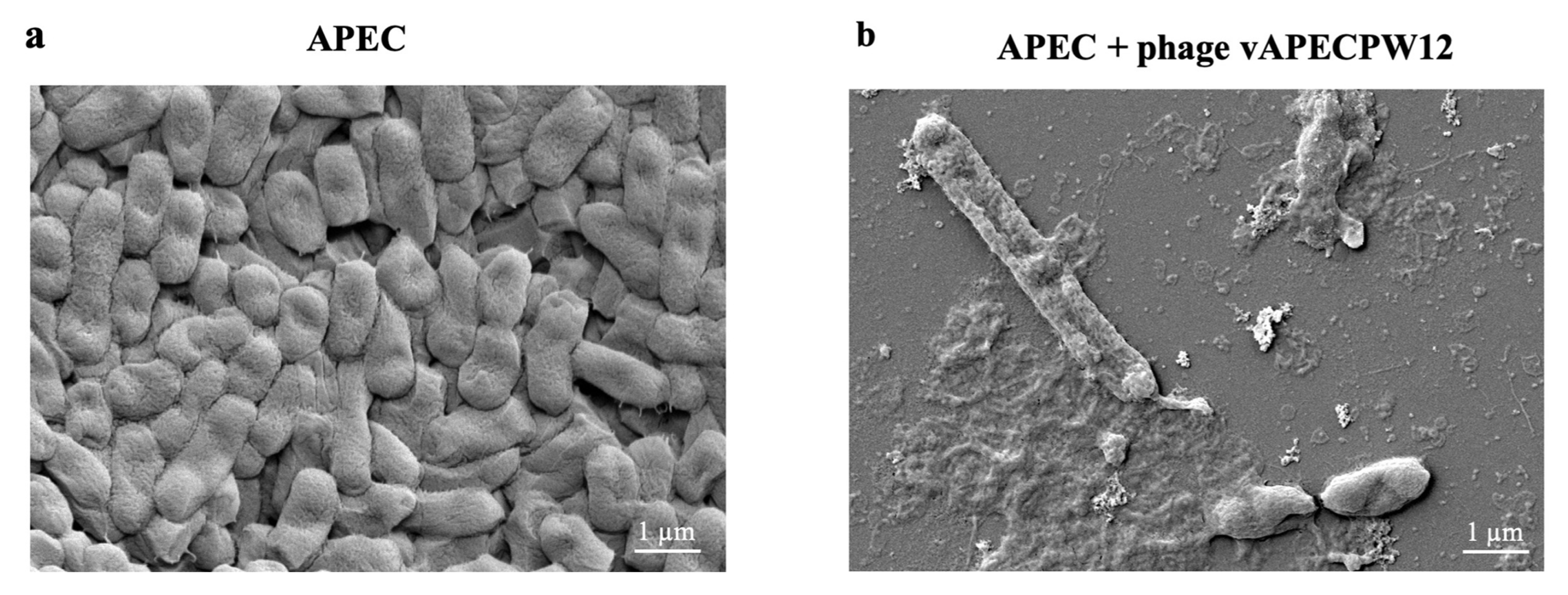
2.9. Morphology of Bacterial Cells After Phage Treatment Under Electron Microscopy
2.10. Phage Temperature, pH, and Ultraviolet (UV) Radiation Stability
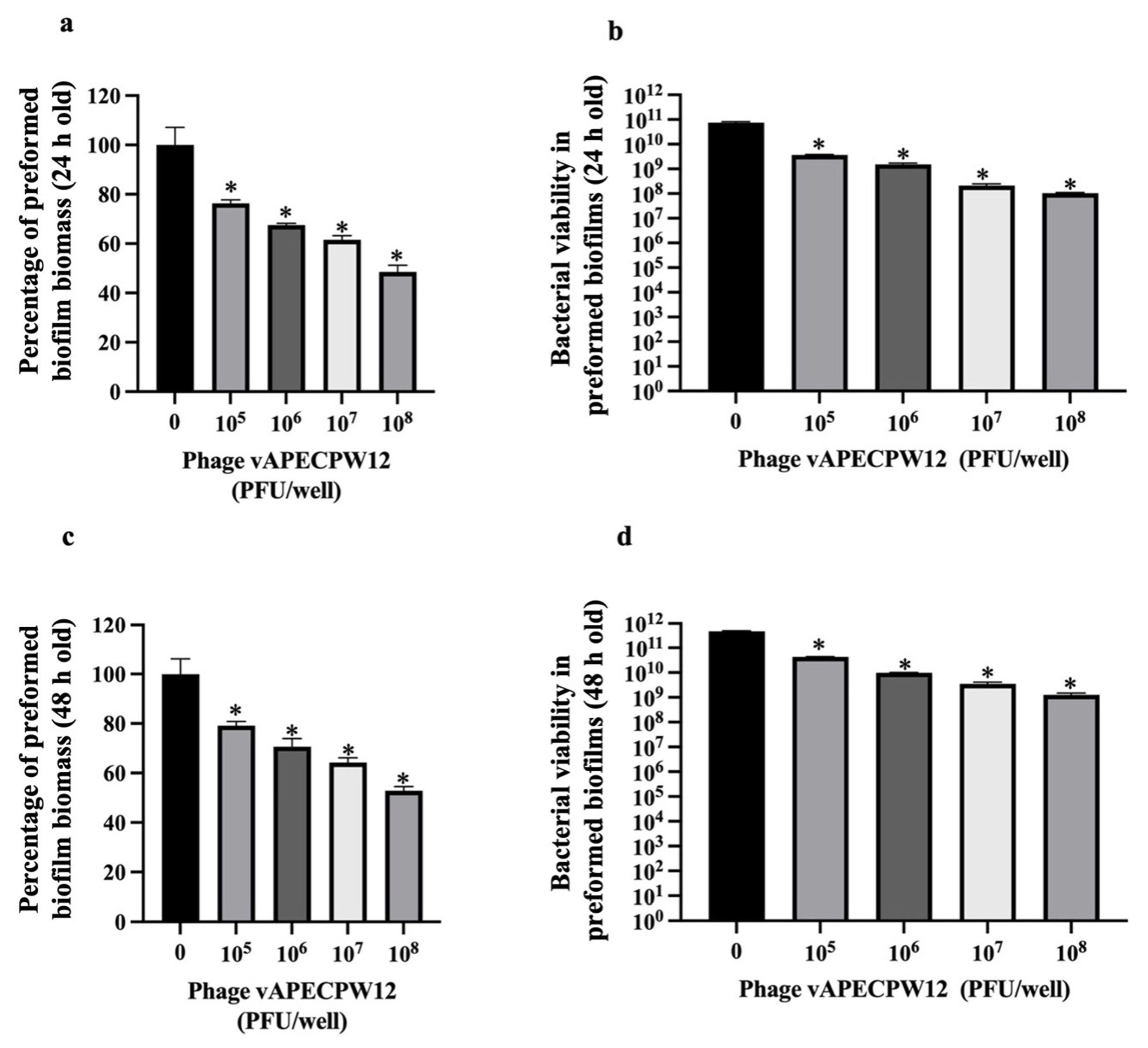
2.11. Efficacy of Phage to Remove Preformed Biofilms
2.12. Effectiveness of Phage to Reduce Biofilm Formation
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Phage Isolation and Purification
4.3. Phage Propagation
4.4. Phage Morphology Using TEM
4.5. Phage Host Range
4.6. EOP
4.7. Whole-Genome Sequencing and Bioinformatic Analysis
4.8. Phylogenetic Analysis
4.9. Phage Adsorption Assay
4.10. One-Step Growth Curve
4.11. Minimum Inhibitory Multiplicity of Infection
4.12. Bacterial Morphology After Phage Treatment Under SEM
4.13. Phage Stability Under Various Temperatures, pH Values, and UV Light
4.14. Efficacy of Phage to Remove Preformed Biofilm
4.15. Ability of Phage to Prevent Biofilm Formation
4.16. Accession Numbers of the Genome of the Phage
4.17. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antao, E.M.; Glodde, S.; Li, G.; Sharifi, R.; Homeier, T.; Laturnus, C.; Diehl, I.; Bethe, A.; Philipp, H.C.; Preisinger, R.; et al. The chicken as a natural model for extraintestinal infections caused by avian pathogenic Escherichia coli (APEC). Microb. Pathog. 2008, 45, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, S.; Wang, Z.; Zhang, Y.; Jia, Y.; Jiang, W.; Chen, Z.; Yin, H.; Huang, C.; Han, X. Avian pathogenic Escherichia coli (APEC): Current insights and future challenges. Poult. Sci. 2024, 103, 104359. [Google Scholar] [CrossRef] [PubMed]
- Dho-Moulin, M.; Fairbrother, J.M. Avian pathogenic Escherichia coli (APEC). Vet. Res. 1999, 30, 299–316. [Google Scholar] [PubMed]
- Christensen, H.; Bachmeier, J.; Bisgaard, M. New strategies to prevent and control avian pathogenic Escherichia coli (APEC). Avian Pathol. 2021, 50, 370–381. [Google Scholar] [CrossRef]
- Guerra, P.R.; Herrero-Fresno, A.; Pors, S.E.; Ahmed, S.; Wang, D.; Thofner, I.; Antenucci, F.; Olsen, J.E. The membrane transporter PotE is required for virulence in avian pathogenic Escherichia coli (APEC). Vet. Microbiol. 2018, 216, 38–44. [Google Scholar] [CrossRef]
- Johnson, J.R.; Kuskowski, M.A.; Menard, M.; Gajewski, A.; Xercavins, M.; Garau, J. Similarity between human and chicken Escherichia coli isolates in relation to ciprofloxacin resistance status. J. Infect. Dis. 2006, 194, 71–78. [Google Scholar] [CrossRef]
- Afayibo, D.J.A.; Zhu, H.; Zhang, B.; Yao, L.; Abdelgawad, H.A.; Tian, M.; Qi, J.; Liu, Y.; Wang, S. Isolation, Molecular Characterization, and Antibiotic Resistance of Avian Pathogenic Escherichia coli in Eastern China. Vet. Sci. 2022, 9, 319. [Google Scholar] [CrossRef]
- Bhattarai, R.K.; Basnet, H.B.; Dhakal, I.P.; Devkota, B. Antimicrobial resistance of avian pathogenic Escherichia coli isolated from broiler, layer, and breeder chickens. Vet. World 2024, 17, 480–499. [Google Scholar] [CrossRef]
- Yao, L.; Bao, Y.; Hu, J.; Zhang, B.; Wang, Z.; Wang, X.; Guo, W.; Wang, D.; Qi, J.; Tian, M.; et al. A lytic phage to control multidrug-resistant avian pathogenic Escherichia coli (APEC) infection. Front. Cell. Infect. Microbiol. 2023, 13, 1253815. [Google Scholar] [CrossRef]
- Sattar, S.; Bailie, M.; Yaqoob, A.; Khanum, S.; Fatima, K.; Altaf, A.; Ahmed, I.; Shah, S.T.A.; Munawar, J.; Zehra, Q.A.; et al. Characterization of two novel lytic bacteriophages having lysis potential against MDR avian pathogenic Escherichia coli strains of zoonotic potential. Sci. Rep. 2023, 13, 10043. [Google Scholar] [CrossRef] [PubMed]
- Eid, S.; Tolba, H.M.N.; Hamed, R.I.; Al-Atfeehy, N.M. Bacteriophage therapy as an alternative biocontrol against emerging multidrug resistant E. coli in broilers. Saudi J. Biol. Sci. 2022, 29, 3380–3389. [Google Scholar] [CrossRef]
- Wintachai, P.; Thaion, F.; Clokie, M.R.J.; Thomrongsuwannakij, T. Isolation and Characterization of a Novel Escherichia Bacteriophage with Potential to Control Multidrug-Resistant Avian Pathogenic Escherichia coli and Biofilms. Antibiotics 2024, 13, 1083. [Google Scholar] [CrossRef]
- Batinovic, S.; Fujii, Y.; Nittami, T. Expansion of Kuravirus-like Phage Sequences within the Past Decade, including Escherichia Phage YF01 from Japan, Prompt the Creation of Three New Genera. Viruses 2023, 15, 506. [Google Scholar] [CrossRef] [PubMed]
- Ahiwale, S.S.; Bankar, A.V.; Tagunde, S.N.; Zinjarde, S.; Ackermann, H.W.; Kapadnis, B.P. Isolation and characterization of a rare waterborne lytic phage of Salmonella enterica serovar Paratyphi B. Can. J. Microbiol. 2013, 59, 318–323. [Google Scholar] [CrossRef]
- Millard, A.; Denise, R.; Lestido, M.; Thomas, M.T.; Webster, D.; Turner, D.; Sicheritz-Pontén, T. taxmyPHAGE: Automated taxonomy of dsDNA phage genomes at the genus and species level. Phage 2025, 6, 5–11. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Prangishvili, D.; Lavigne, R. Position paper: The creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ. Microbiol. 2009, 11, 2775–2777. [Google Scholar] [CrossRef]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC-A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Pineros, G.; Millard, A.; Michniewski, S.; Scanlan, D.; Siren, K.; Reyes, A.; Petersen, B.; Clokie, M.R.J.; Sicheritz-Ponten, T. From Trees to Clouds: PhageClouds for Fast Comparison of approximately 640,000 Phage Genomic Sequences and Host-Centric Visualization Using Genomic Network Graphs. Phage 2021, 2, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Al Hakeem, W.G.; Fathima, S.; Shanmugasundaram, R.; Selvaraj, R.K. Campylobacter jejuni in Poultry: Pathogenesis and Control Strategies. Microorganisms 2022, 10, 2134. [Google Scholar] [CrossRef]
- Phu, D.H.; Narinthorn, R.; Nhung, N.T.; Chansiripornchai, N.; Blackall, P.J.; Turni, C.; Carrique-Mas, J.; Thomrongsuwannakij, T. The characterization and correlation between the phenotypic and genotypic resistance of Campylobacter spp. isolates from commercial broilers and native chickens in the south of Thailand. Avian Pathol. 2024, 53, 1–13. [Google Scholar] [CrossRef]
- Lorenzo-Rebenaque, L.; Casto-Rebollo, C.; Diretto, G.; Frusciante, S.; Rodriguez, J.C.; Ventero, M.P.; Molina-Pardines, C.; Vega, S.; Marin, C.; Marco-Jimenez, F. Modulation of Caecal Microbiota and Metabolome Profile in Salmonella-Infected Broilers by Phage Therapy. Int. J. Mol. Sci. 2023, 24, 15201. [Google Scholar] [CrossRef]
- Karami, M.; Goudarztalejerdi, A.; Mohammadzadeh, A.; Berizi, E. In vitro evaluation of two novel Escherichia bacteriophages against multiple drug resistant avian pathogenic Escherichia coli. BMC Infect. Dis. 2024, 24, 497. [Google Scholar] [CrossRef] [PubMed]
- Kazibwe, G.; Katami, P.; Alinaitwe, R.; Alafi, S.; Nanteza, A.; Nakavuma, J.L. Bacteriophage activity against and characterisation of avian pathogenic Escherichia coli isolated from colibacillosis cases in Uganda. PLoS ONE 2020, 15, e0239107. [Google Scholar] [CrossRef] [PubMed]
- Thomrongsuwannakij, T.; Narinthorn, R.; Mahawan, T.; Blackall, P.J. Molecular and phenotypic characterization of avian pathogenic Escherichia coli isolated from commercial broilers and native chickens. Poult. Sci. 2022, 101, 101527. [Google Scholar] [CrossRef]
- Chen, X.; Liu, W.; Li, H.; Yan, S.; Jiang, F.; Cai, W.; Li, G. Whole genome sequencing analysis of avian pathogenic Escherichia coli from China. Vet. Microbiol. 2021, 259, 109158. [Google Scholar] [CrossRef]
- Muhlenhoff, M.; Stummeyer, K.; Grove, M.; Sauerborn, M.; Gerardy-Schahn, R. Proteolytic processing and oligomerization of bacteriophage-derived endosialidases. J. Biol. Chem. 2003, 278, 12634–12644. [Google Scholar] [CrossRef]
- Fong, K.; Wong, C.W.Y.; Wang, S.; Delaquis, P. How Broad Is Enough: The Host Range of Bacteriophages and Its Impact on the Agri-Food Sector. Phage 2021, 2, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.J.; Gill, J.J. The habits of highly effective phages: Population dynamics as a framework for identifying therapeutic phages. Front. Microbiol. 2014, 5, 618. [Google Scholar] [CrossRef]
- Smith, K.R.; Bumunang, E.W.; Schlechte, J.; Waldner, M.; Anany, H.; Walker, M.; MacLean, K.; Stanford, K.; Fairbrother, J.M.; Alexander, T.W.; et al. The Isolation and Characterization of Bacteriophages Infecting Avian Pathogenic Escherichia coli O1, O2 and O78 Strains. Viruses 2023, 15, 2095. [Google Scholar] [CrossRef]
- Chang, C.Y.; Nam, K.; Young, R. S gene expression and the timing of lysis by bacteriophage lambda. J. Bacteriol. 1995, 177, 3283–3294. [Google Scholar] [CrossRef]
- Grabowski, L.; Lepek, K.; Stasilojc, M.; Kosznik-Kwasnicka, K.; Zdrojewska, K.; Maciag-Dorszynska, M.; Wegrzyn, G.; Wegrzyn, A. Bacteriophage-encoded enzymes destroying bacterial cell membranes and walls, and their potential use as antimicrobial agents. Microbiol. Res. 2021, 248, 126746. [Google Scholar] [CrossRef]
- Nazir, A.; Xu, X.; Liu, Y.; Chen, Y. Phage Endolysins: Advances in the World of Food Safety. Cells 2023, 12, 2169. [Google Scholar] [CrossRef]
- Sritha, K.S.; Bhat, S.G. Genomics of Salmonella phage PhiStp1: Candidate bacteriophage for biocontrol. Virus Genes 2018, 54, 311–318. [Google Scholar] [CrossRef]
- Artawinata, P.C.; Lorraine, S.; Waturangi, D.E. Isolation and characterization of bacteriophages from soil against food spoilage and foodborne pathogenic bacteria. Sci. Rep. 2023, 13, 9282. [Google Scholar] [CrossRef]
- Abedon, S.T. Bacteriophage Adsorption: Likelihood of Virion Encounter with Bacteria and Other Factors Affecting Rates. Antibiotics 2023, 12, 723. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Hong, Q.; Chang, R.Y.K.; Kwok, P.C.L.; Chan, H.K. Phage-Antibiotic Therapy as a Promising Strategy to Combat Multidrug-Resistant Infections and to Enhance Antimicrobial Efficiency. Antibiotics 2022, 11, 570. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Wang, Y.; Li, H.; Zhang, H.; Zhou, Y.; Wang, R.; Bao, H. Characterization and genomic analysis of a novel E. coli lytic phage with extended lytic activity against S. enteridis and S. typhimurium. Food Prod. Process. Nutr. 2024, 6, 14. [Google Scholar] [CrossRef]
- Ali, Z.; Dishisha, T.; El-Gendy, A.O.; Azmy, A.F. Isolation and phenotypic characterization of bacteriophage SA14 with lytic- and anti-biofilm activity against multidrug-resistant Enterococcus faecalis. Beni-Suef Univ. J. Basic. Appl. Sci. 2023, 12, 21. [Google Scholar] [CrossRef]
- Cheng, Y.; Gao, D.; Xia, Y.; Wang, Z.; Bai, M.; Luo, K.; Cui, X.; Wang, Y.; Zhang, S.; Xiao, W. Characterization of Novel Bacteriophage AhyVDH1 and Its Lytic Activity Against Aeromonas hydrophila. Curr. Microbiol. 2021, 78, 329–337. [Google Scholar] [CrossRef]
- Yoo, S.; Lee, K.M.; Kim, N.; Vu, T.N.; Abadie, R.; Yong, D. Designing phage cocktails to combat the emergence of bacteriophage-resistant mutants in multidrug-resistant Klebsiella pneumoniae. Microbiol. Spectr. 2024, 12, e0125823. [Google Scholar] [CrossRef]
- Oechslin, F. Resistance Development to Bacteriophages Occurring during Bacteriophage Therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef]
- Ngiam, L.; Weynberg, K.; Guo, J. Evolutionary and co-evolutionary phage training approaches enhance bacterial suppression and delay the emergence of phage resistance. ISME Commun. 2024, 4, ycae082. [Google Scholar] [CrossRef]
- Yan, J.; Mao, J.; Xie, J. Bacteriophage polysaccharide depolymerases and biomedical applications. BioDrugs 2014, 28, 265–274, Erratum in BioDrugs 2014, 28, 323. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.J.; Chang, K.C.; Huang, S.W.; Luo, C.H.; Chiou, P.Y.; Wu, C.C.; Lin, N.T. The Tail Associated Protein of Acinetobacter baumannii Phage PhiAB6 Is the Host Specificity Determinant Possessing Exopolysaccharide Depolymerase Activity. PLoS ONE 2016, 11, e0153361. [Google Scholar] [CrossRef] [PubMed]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [PubMed]
- Zhao, A.; Sun, J.; Liu, Y. Understanding bacterial biofilms: From definition to treatment strategies. Front. Cell. Infect. Microbiol. 2023, 13, 1137947. [Google Scholar] [CrossRef]
- Khan, M.M.; Ali, A.; Kolenda, R.; Olowe, O.A.; Weinreich, J.; Li, G.; Schierack, P. The role of AJB35136 and fdtA genes in biofilm formation by avian pathogenic Escherichia coli. BMC Vet. Res. 2023, 19, 126. [Google Scholar] [CrossRef]
- Young, M.M.; de Oliveira, A.L.; Nolan, L.K.; Barbieri, N.L.; Logue, C.M. Identification of novel genes involved in the biofilm formation process of Avian Pathogenic Escherichia coli (APEC). PLoS ONE 2022, 17, e0279206. [Google Scholar] [CrossRef]
- Guo, Z.; Liu, M.; Zhang, D. Potential of phage depolymerase for the treatment of bacterial biofilms. Virulence 2023, 14, 2273567. [Google Scholar] [CrossRef]
- Akturk, E.; Oliveira, H.; Santos, S.B.; Costa, S.; Kuyumcu, S.; Melo, L.D.R.; Azeredo, J. Synergistic Action of Phage and Antibiotics: Parameters to Enhance the Killing Efficacy Against Mono and Dual-Species Biofilms. Antibiotics 2019, 8, 103. [Google Scholar] [CrossRef]
- Lu, H.; Li, Z.; Elbaz, A.; Ni, S.Q. Synergistic action of phages and lytic proteins with antibiotics: A combination strategy to target bacteria and biofilms. BMC Microbiol. 2023, 23, 149. [Google Scholar] [CrossRef] [PubMed]
- Wintachai, P.; Naknaen, A.; Thammaphet, J.; Pomwised, R.; Phaonakrop, N.; Roytrakul, S.; Smith, D.R. Characterization of extended-spectrum-beta-lactamase producing Klebsiella pneumoniae phage KP1801 and evaluation of therapeutic efficacy in vitro and in vivo. Sci. Rep. 2020, 10, 11803. [Google Scholar] [CrossRef] [PubMed]
- Wintachai, P.; Phaonakrop, N.; Roytrakul, S.; Naknaen, A.; Pomwised, R.; Voravuthikunchai, S.P.; Surachat, K.; Smith, D.R. Enhanced antibacterial effect of a novel Friunavirus phage vWU2001 in combination with colistin against carbapenem-resistant Acinetobacter baumannii. Sci. Rep. 2022, 12, 2633. [Google Scholar] [CrossRef]
- Myers, J.; Davis, J., II; Lollo, M.; Hudec, G.; Hyman, P. More’s the Same-Multiple Hosts Do Not Select for Broader Host Range Phages. Viruses 2023, 15, 518. [Google Scholar] [CrossRef] [PubMed]
- Kutter, E. Phage host range and efficiency of plating. Methods Mol. Biol. 2009, 501, 141–149. [Google Scholar] [CrossRef]
- Wishart, D.S.; Han, S.; Saha, S.; Oler, E.; Peters, H.; Grant, J.R.; Stothard, P.; Gautam, V. PHASTEST: Faster than PHASTER, better than PHAST. Nucleic Acids Res. 2023, 51, W443–W450. [Google Scholar] [CrossRef]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Kropinski, A.M. Measurement of the rate of attachment of bacteriophage to cells. Methods Mol. Biol. 2009, 501, 151–155. [Google Scholar] [CrossRef]
- Wintachai, P.; Surachat, K.; Singkhamanan, K. Isolation and Characterization of a Novel Autographiviridae Phage and Its Combined Effect with Tigecycline in Controlling Multidrug-Resistant Acinetobacter baumannii-Associated Skin and Soft Tissue Infections. Viruses 2022, 14, 194. [Google Scholar] [CrossRef]
- Montso, P.K.; Mlambo, V.; Ateba, C.N. Characterization of Lytic Bacteriophages Infecting Multidrug-Resistant Shiga Toxigenic Atypical Escherichia coli O177 Strains Isolated From Cattle Feces. Front. Public Health 2019, 7, 355. [Google Scholar] [CrossRef]
- Wintachai, P.; Santini, J.M.; Thonguppatham, R.; Stroyakovski, M.; Surachat, K.; Atipairin, A. Isolation, Characterization, and Anti-Biofilm Activity of a Novel Kaypoctavirus Against K24 Capsular Type, Multidrug-Resistant Klebsiella pneumoniae Clinical Isolates. Antibiotics 2025, 14, 157. [Google Scholar] [CrossRef] [PubMed]
- Wintachai, P.; Surachat, K.; Chaimaha, G.; Septama, A.W.; Smith, D.R. Isolation and Characterization of a Phapecoctavirus Infecting Multidrug-Resistant Acinetobacter baumannii in A549 Alveolar Epithelial Cells. Viruses 2022, 14, 2561. [Google Scholar] [CrossRef] [PubMed]
- Wintachai, P.; Voravuthikunchai, S.P. Characterization of Novel Lytic Myoviridae Phage Infecting Multidrug-Resistant Acinetobacter baumannii and Synergistic Antimicrobial Efficacy between Phage and Sacha Inchi Oil. Pharmaceuticals 2022, 15, 291. [Google Scholar] [CrossRef] [PubMed]





| Strain | Phage vAPECPW12 | |
|---|---|---|
| Lytic Activity | EOP | |
| MDR APEC PW001 | + | + |
| MDR APEC PW002 | − | − |
| MDR APEC PW003 | − | − |
| MDR APEC PW004 | − | − |
| MDR APEC PW005 | − | − |
| MDR APEC PW006 | + | Moderate (0.25) |
| MDR APEC PW007 * | + | High (Host = 1) |
| MDR APEC PW008 | − | − |
| MDR APEC PW009 | + | High (0.86) |
| MDR APEC PW010 | − | − |
| MDR APEC PW011 | + | Moderate (0.25) |
| MDR APEC PW012 | − | − |
| MDR APEC PW013 | + | Low (0.05) |
| MDR APEC PW014 | − | − |
| MDR APEC PW015 | − | − |
| MDR APEC PW016 | − | − |
| MDR APEC PW017 | − | − |
| MDR APEC PW018 | − | − |
| MDR APEC PW019 | + | Moderate (0.46) |
| MDR APEC PW020 | − | − |
| MDR APEC PW021 | − | − |
| MDR APEC PW022 | + | High (0.65) |
| MDR APEC PW023 | − | − |
| MDR APEC PW024 | − | − |
| MDR APEC PW025 | + | Moderate (0.42) |
| C. jejuni PW001 | − | − |
| C. jejuni PW002 | − | − |
| C. jejuni PW003 | − | − |
| C. jejuni PW004 | − | − |
| C. jejuni PW005 | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wintachai, P.; Thonguppatham, R.; Clokie, M.R.J.; Thomrongsuwannakij, T. Whole-Genome Sequencing and Functional Characterization of a Novel Kuravirus Bacteriophage with Antibiofilm Activity Against Multidrug-Resistant Avian Pathogenic Escherichia coli. Int. J. Mol. Sci. 2025, 26, 11911. https://doi.org/10.3390/ijms262411911
Wintachai P, Thonguppatham R, Clokie MRJ, Thomrongsuwannakij T. Whole-Genome Sequencing and Functional Characterization of a Novel Kuravirus Bacteriophage with Antibiofilm Activity Against Multidrug-Resistant Avian Pathogenic Escherichia coli. International Journal of Molecular Sciences. 2025; 26(24):11911. https://doi.org/10.3390/ijms262411911
Chicago/Turabian StyleWintachai, Phitchayapak, Renuka Thonguppatham, Martha R. J. Clokie, and Thotsapol Thomrongsuwannakij. 2025. "Whole-Genome Sequencing and Functional Characterization of a Novel Kuravirus Bacteriophage with Antibiofilm Activity Against Multidrug-Resistant Avian Pathogenic Escherichia coli" International Journal of Molecular Sciences 26, no. 24: 11911. https://doi.org/10.3390/ijms262411911
APA StyleWintachai, P., Thonguppatham, R., Clokie, M. R. J., & Thomrongsuwannakij, T. (2025). Whole-Genome Sequencing and Functional Characterization of a Novel Kuravirus Bacteriophage with Antibiofilm Activity Against Multidrug-Resistant Avian Pathogenic Escherichia coli. International Journal of Molecular Sciences, 26(24), 11911. https://doi.org/10.3390/ijms262411911







