Cell-Free DNA and Mitochondria in Parkinson’s Disease
Abstract
1. Parkinson’s Disease
| PARK | Gene | Protein | Mitochondrial Function |
|---|---|---|---|
| PARK1 and PARK4 | SNCA | Alpha synuclein (Syn) | |
| PARK2 | PRKN | Parkin (E3 ubiquitin ligase) |
|
| PARK6 | PINK1 | PTEN-induced putative kinase 1 (mitochondrial serine/threonine protein kinase) | |
| PARK7 | DJ-1 | Protein/nucleic acid deglycase |
|
| PARK8 | LRRK2 | Leucine-rich repeat kinase 2 | |
| PARK15 | FBXO7 | F-box protein 7 |
|
| PARK22 | CHCHD2 | Coiled-coil-helix-coiled coil-helix domain 2 | |
| PARK23 | VPS13C | Vacuolar protein sorting-associated protein 13C |
|
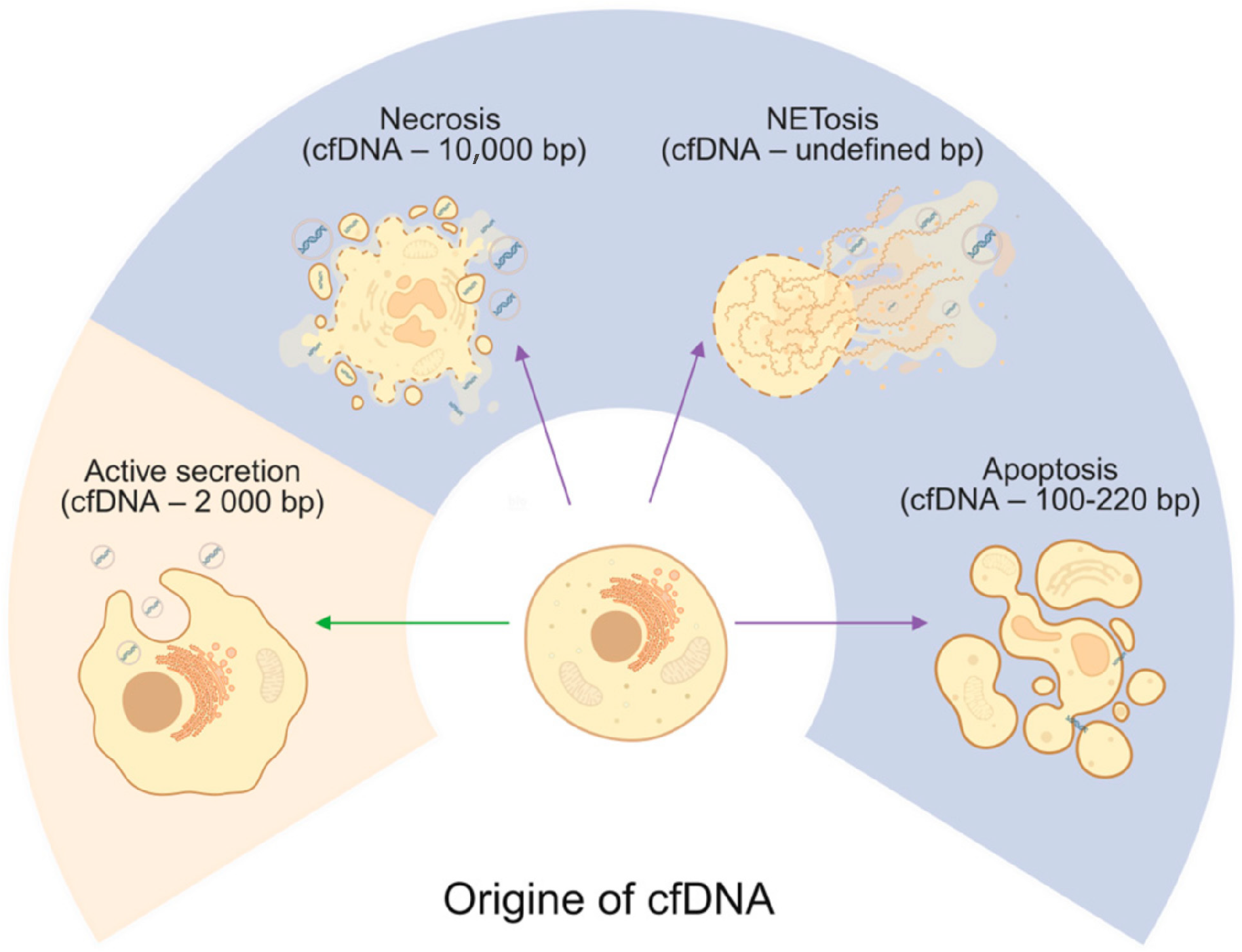
2. Cell-Free DNA Origins
2.1. Apoptosis
2.2. Necrosis
2.3. NETosis
2.4. Active Secretion
3. Mitochondria Biology
4. Mitochondria Dysfunction in Parkinson’s Disease
4.1. Mitochondrial Quality Control
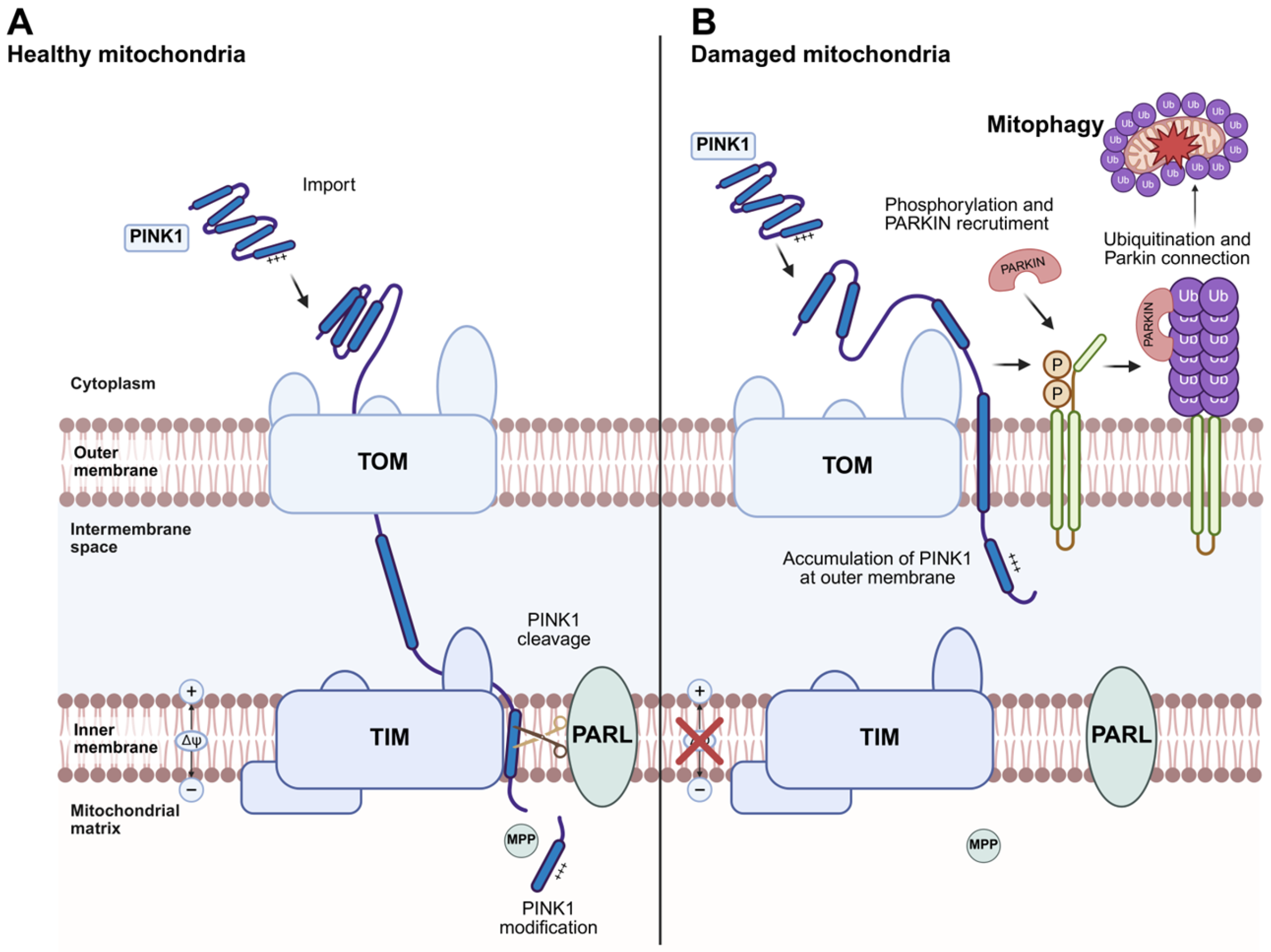
4.2. Mitophagy
4.3. PINK1/Parkin-Dependent Mitophagy
4.4. Mitochondrial-Derived Vesicles (MDVs)
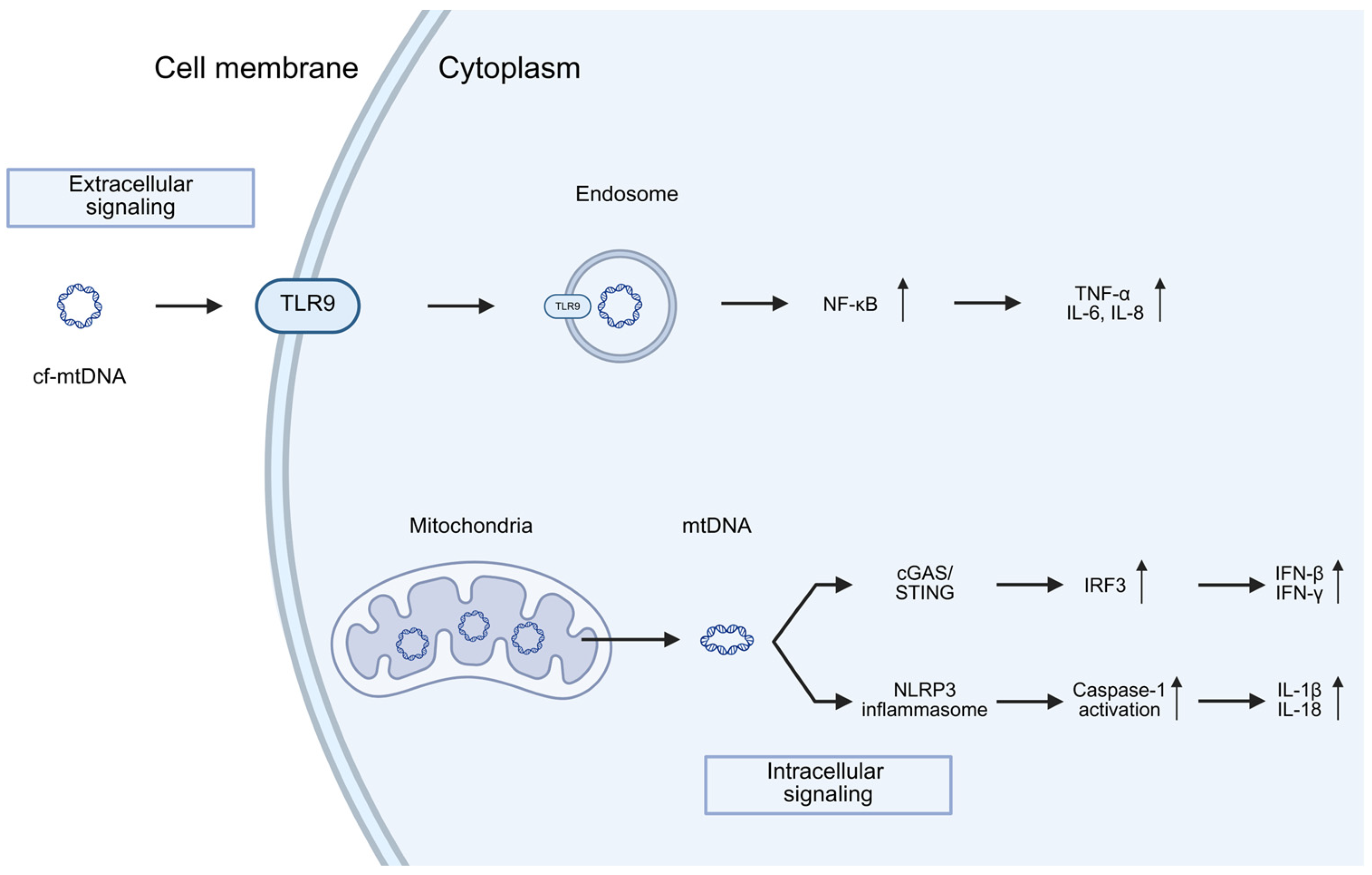
5. Mitochondria and Neuroinflammation in PD
Neuroinflammation
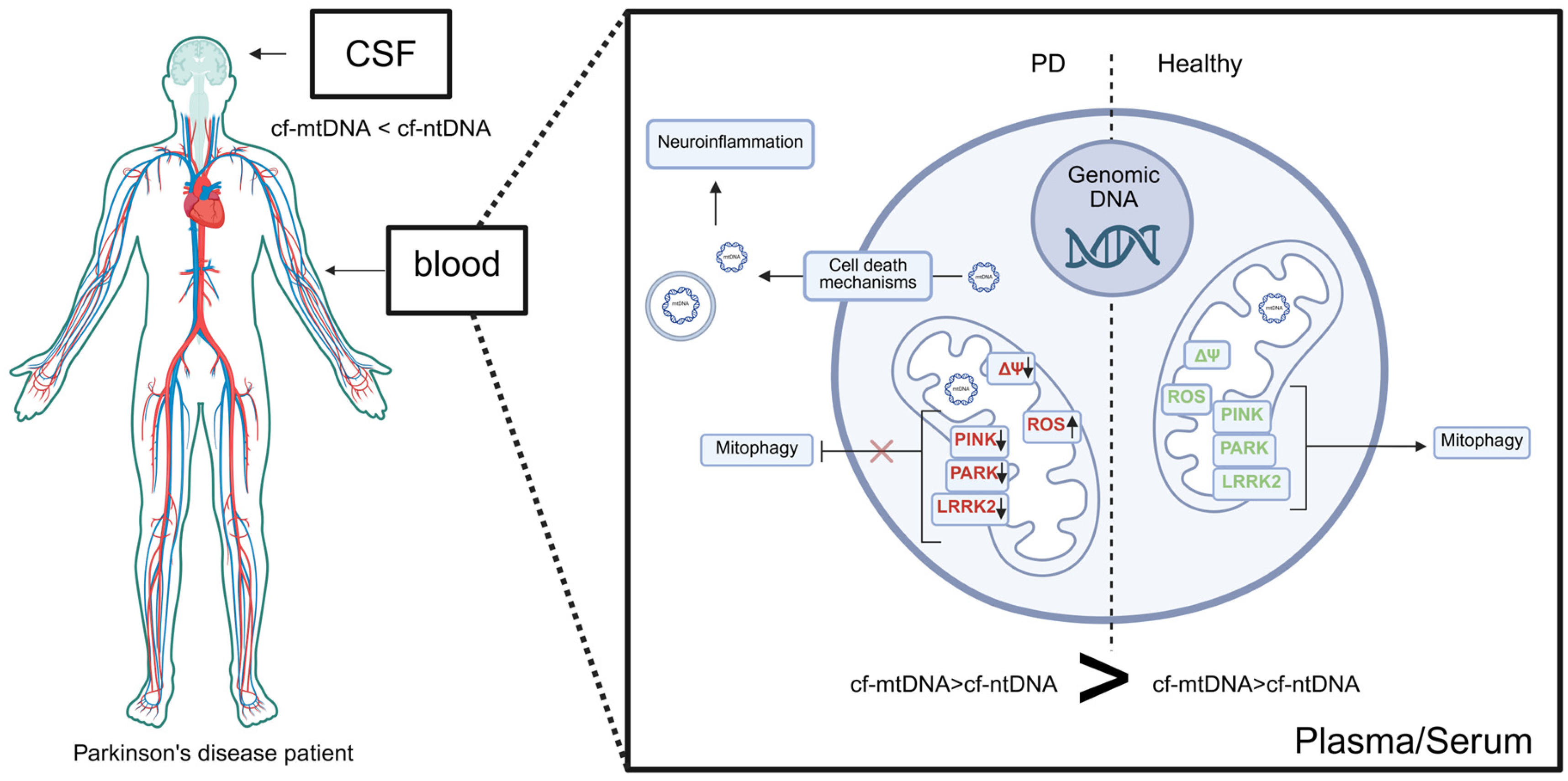
6. Cell-Free DNA in Parkinson’s Disease
6.1. cfDNA in the Serum/Plasma of PD
6.2. cfDNA Serum/Plasma Sequence Analysis in PD
6.3. cfDNA in the CSF of PD Parkisnon’s Disease
| Studies Sample/PD Subtypes | PD Subtypes | Types of cfDNA | Gene | Quantification Method | Methods of Analysis | Main Findings | References |
|---|---|---|---|---|---|---|---|
| 10 HC 53 PD | iPD | mtDNA | MTND1 MTND4 B2M | CSF | qPCR | Reduced copy number in PD patients compared to HC No correlation with cognitive impairment | [116] |
| 10 HC 26 PD of EOPD | EOPD | ntDNA | N/A | CSF | methylation | 2220 differentially methylated genes were identified; Aberrant methylation signatures were correlated with external factors | [117] |
| 372 169 after treatment 250 114 | PD | mtDNA | MTND1 (minor deletion arc mitochondrial gene) | CSF | qPCR | ccf-mtDNA levels appear significantly reduced in PD cases when compared to matched controls and are associated with cognitive impairment; comorbidities and treatment can both influence ccf-mtDNA homeostasis, | [115] |
| 262 HC 363 PD | PD | mtDNA | MTND1 MTND4 B2M | PBC | qPCR | Decreased copy number in PD patients No correlation with cognitive impairment | [109] |
| 17 HC 21 iPD 20 LRRK2 NMC * 26 | iPD LRRK2-PD | mtDNA ntDNA | mt64-D1 mt96-D5 TEFM-88 TBP1–73 | CSF | ddPCR | Reduced copy number in PD patients compared to HC Higher proportion of mtDNA molecules with deletions in PD patients | [118] |
| 57 HC 17 PD 55 HC 17 PD | mut+/+PD PRKN/PINK1 mut+/−PD PRKN/PINK1 | mtDNA | MT-ND1 B2M | serum serum | ddPCR ddPCR | cf-mtDNA is elevated in monogenic PD; results implicates inflammation due to impaired mitophagy and subsequent mtDNA release in the pathogenesis of monogenic PD | [110] |
| 3HC 6PD | iPD | ntDNA | serum | NGS; qPCR | Increase in specific cfDNA molecules in both drug-naive and drug-exposed PD serum, albeit variations between the two groups, as compared to healthy controls | [42] | |
| 5 HC 13 PD | iPD | mtDNA | COX | CSF | ddPCR | Increased cf-mtDNA vs. cf-ntDNA | [108] |
| 5 HC 13 PD | iPD | ntDNA | KRAS | CSF | ddPCR | Increased cf-ntDNA vs. control | [108] |
| 15 HC 30 PD | iPD | mtDNA | COX | serum | ddPCR | Increased level of cf-mtDNA vs. cf-ntDNA in PD; Increased cf-mtDNA vs. control; increased cfDNA vs. control | [108] |
| 15 HC 30 PD | iPD | ntDNA | KRAS | serum | ddPCR | Increased cf-ntDNA vs. control | [108] |
| 72 HC 62 PD | iPD | ntDNA | COX | plasma | NGS | cell-free DNA integrity was significantly elevated whereas cell-free DNA relative telomere length was markedly shorter | [119] |
6.4. cfDNA CSF Sequence Analysis in PD
6.5. Important Discrepancies of cfDNA Studies in PD
7. cf-mtDNA as Potential Biomarker of Parkinson’s Disease
8. Perspective and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PD | Parkinson’s disease |
| cfDNA | Cell-free DNA |
| cf-ntDNA | Nuclear derived cell-free DNA |
| cf-mtDNA | Mitochondrially derived cell-free DNA |
| CSF | Cerebrospinal fluid |
| LBs | Lewy bodies |
| SN | Substantia nigra |
| LRRK2 | Leucine rich repeat kinase 2 |
| CAD | Caspase-activated DNase |
| dsDNA | Double-stranded DNA |
| NETs | Neutrophil extracellular traps |
| ROS | Reactive oxygen species |
| AD | Alzheimer’s disease |
| RNA | Ribonucleic acids |
| ASO | Antisense oligonucleotide |
| EVs | Extracellular vesicle |
| DAMPs | Damage-Associated Molecular Patterns |
| tRNA | Transfer RNA |
| rRNA | Ribosomal RNA |
| ER | Endoplasmic reticulum |
| FIS1 | Mitochondrial fission protein 1 |
| DRP1 | Dynamic-related protein 1 |
| MFN1/2 | Mitofusion proteins 1 and 2 |
| LIR | LC3 interacting region |
| PINK1 | PTEN-induced putative kinase 1 |
| PTEN | Phosphatase and tensin homologue deleted on chromosome 10 |
| TOM | Translocase of the outer membrane |
| TIM | Translocase of the inner membrane |
| PARL | Presenilin-associated rhomboid-like protein |
| OMM | Outer mitochondrial membrane |
| OPTN | Optineurin |
| NDP52 | Nuclear dot protein 52 |
| CALCOCO2 | Calcium-binding and coiled-coil domain-containing protein 2 |
| MDVs | Mitochondrially derived vesicles |
| cGAS | Cyclic GMP/AMP synthase |
| cGAMP | Cyclic guanosine monophosphate–adenosine monophosphate |
| STING | Stimulator of interferon genes |
| TCA | Tricarboxylic acid cycle |
| PRRs | Pattern recognition receptors |
| TLRs | Toll-like receptors |
| NOD | Nucleotide-binding oligomerization domain |
| NLRs | Nucleotide-binding oligomerization domain like receptors |
| HD | Huntington’s disease |
| ALS | Amyotrophic lateral sclerosis |
| MS | Multiple sclerosis |
| IRF3 | Interferon regulatory factor 3 |
| IKK | IkappaB kinase |
| TBK1 | TANK-binding kinase 1 |
| qPCR | Quantitatively polymerase chain reaction |
| PCR | Polymerase chain reaction |
| NGS | Next Generation Sequencing |
| TNF | Tumor necrosis factor |
| IFNɣ | Interferon gamma |
| IL-6 | Interleukin 6 |
References
- Bloem, B.R.; Okun, M.S.; Klein, C. Parkinson’s disease. Lancet 2021, 397, 2284–2303. [Google Scholar] [CrossRef]
- Polymeropoulos, M.H.; Lavedan, C.; Leroy, E.; Ide, S.E.; Dehejia, A.; Dutra, A.; Pike, B.; Root, H.; Rubenstein, J.; Boyer, R.; et al. Mutation in the α-Synuclein Gene Identified in Families with Parkinson’s Disease. Science 1997, 276, 2045–2047. [Google Scholar] [CrossRef] [PubMed]
- Blauwendraat, C.; Nalls, M.A.; Singleton, A.B. The genetic architecture of Parkinson’s disease. Lancet Neurol. 2020, 19, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Wang, G. Mitochondrial dysfunction in Parkinson’s disease. Transl. Neurodegener. 2016, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Arya, R.; Haque, A.K.M.A.; Shakya, H.; Billah, M.M.; Parvin, A.; Rahman, M.M.; Sakib, K.M.; Faruquee, H.M.; Kumar, V.; Kim, J.J. Parkinson’s Disease: Biomarkers for Diagnosis and Disease Progression. Int. J. Mol. Sci. 2024, 25, 12379. [Google Scholar] [CrossRef]
- Vijiaratnam, N.; Simuni, T.; Bandmann, O.; Morris, H.R.; Foltynie, T. Progress towards therapies for disease modification in Parkinson’s disease. Lancet Neurol. 2021, 20, 559–572. [Google Scholar] [CrossRef]
- Burré, J.; Sharma, M.; Tsetsenis, T.; Buchman, V.; Etherton, M.R.; Südhof, T.C. Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science 2010, 329, 1663–1667. [Google Scholar] [CrossRef]
- Chandra, S.; Chen, X.; Rizo, J.; Jahn, R.; Südhof, T.C. A broken alpha -helix in folded alpha -Synuclein. J. Biol. Chem. 2003, 278, 15313–15318. [Google Scholar] [CrossRef]
- Varkey, J.; Isas, J.M.; Mizuno, N.; Jensen, M.B.; Bhatia, V.K.; Jao, C.C.; Petrlova, J.; Voss, J.C.; Stamou, D.G.; Steven, A.C.; et al. Membrane curvature induction and tubulation are common features of synucleins and apolipoproteins. J. Biol. Chem. 2010, 285, 32486–32493. [Google Scholar] [CrossRef]
- Stefanis, L.; Larsen, K.E.; Rideout, H.J.; Sulzer, D.; Greene, L.A. Expression of A53T mutant but not wild-type alpha-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. J. Neurosci. 2001, 21, 9549–9560. [Google Scholar] [CrossRef]
- Webb, J.L.; Ravikumar, B.; Atkins, J.; Skepper, J.N.; Rubinsztein, D.C. Alpha-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 2003, 278, 25009–25013. [Google Scholar] [CrossRef]
- Choubey, V.; Safiulina, D.; Vaarmann, A.; Cagalinec, M.; Wareski, P.; Kuum, M.; Zharkovsky, A.; Kaasik, A. Mutant A53T alpha-synuclein induces neuronal death by increasing mitochondrial autophagy. J. Biol. Chem. 2011, 286, 10814–10824. [Google Scholar] [CrossRef]
- Martínez, J.H.; Fuentes, F.; Vanasco, V.; Alvarez, S.; Alaimo, A.; Cassina, A.; Coluccio Leskow, F.; Velazquez, F. Alpha-synuclein mitochondrial interaction leads to irreversible translocation and complex I impairment. Arch. Biochem. Biophys. 2018, 651, 1–12. [Google Scholar] [CrossRef]
- Grünewald, A.; Kumar, K.R.; Sue, C.M. New insights into the complex role of mitochondria in Parkinson’s disease. Prog. Neurobiol. 2019, 177, 73–93. [Google Scholar] [CrossRef] [PubMed]
- Lesage, S.; Drouet, V.; Majounie, E.; Deramecourt, V.; Jacoupy, M.; Nicolas, A.; Cormier-Dequaire, F.; Hassoun, S.M.; Pujol, C.; Ciura, S.; et al. Loss of VPS13C Function in Autosomal-Recessive Parkinsonism Causes Mitochondrial Dysfunction and Increases PINK1/Parkin-Dependent Mitophagy. Am. J. Hum. Genet. 2016, 98, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Dagda, R.K.; Cherra, S.J.; Kulich, S.M.; Tandon, A.; Park, D.; Chu, C.T. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 2009, 284, 13843–13855. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, J.; Wang, J.; Yang, B.; He, Q.; Weng, Q. Role of DJ-1 in Immune and Inflammatory Diseases. Front. Immunol. 2020, 11, 994. [Google Scholar] [CrossRef]
- Chen, M.L.; Wu, R.M. LRRK 2 gene mutations in the pathophysiology of the ROCO domain and therapeutic targets for Parkinson’s disease: A review. J. Biomed. Sci. 2018, 25, 52. [Google Scholar] [CrossRef]
- Steger, M.; Tonelli, F.; Ito, G.; Davies, P.; Trost, M.; Vetter, M.; Wachter, S.; Lorentzen, E.; Duddy, G.; Wilson, S.; et al. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. eLife 2016, 5, e12813. [Google Scholar] [CrossRef]
- Connor-Robson, N.; Booth, H.; Martin, J.G.; Gao, B.; Li, K.; Doig, N.; Vowles, J.; Browne, C.; Klinger, L.; Juhasz, P.; et al. An integrated transcriptomics and proteomics analysis reveals functional endocytic dysregulation caused by mutations in LRRK2. Neurobiol. Dis. 2019, 127, 512–526. [Google Scholar] [CrossRef]
- Dodson, M.W.; Zhang, T.; Jiang, C.; Chen, S.; Guo, M. Roles of the Drosophila LRRK2 homolog in Rab7-dependent lysosomal positioning. Hum. Mol. Genet. 2012, 21, 1350–1363. [Google Scholar] [CrossRef]
- Godena, V.K.; Brookes-Hocking, N.; Moller, A.; Shaw, G.; Oswald, M.; Sancho, R.M.; Miller, C.C.J.; Whitworth, A.J.; De Vos, K.J. Increasing microtubule acetylation rescues axonal transport and locomotor deficits caused by LRRK2 Roc-COR domain mutations. Nat. Commun. 2014, 5, 5245. [Google Scholar] [CrossRef]
- MacLeod, D.; Dowman, J.; Hammond, R.; Leete, T.; Inoue, K.; Abeliovich, A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron 2006, 52, 587–593. [Google Scholar] [CrossRef]
- Parisiadou, L.; Yu, J.; Sgobio, C.; Xie, C.; Liu, G.; Sun, L.; Gu, X.L.; Lin, X.; Crowley, N.A.; Lovinger, D.M.; et al. LRRK2 regulates synaptogenesis and dopamine receptor activation through modulation of PKA activity. Nat. Neurosci. 2014, 17, 367–376. [Google Scholar] [CrossRef]
- Plowey, E.D.; Cherra, S.J.; Liu, Y.J.; Chu, C.T. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J. Neurochem. 2008, 105, 1048–1056. [Google Scholar] [CrossRef]
- Yue, M.; Hinkle, K.M.; Davies, P.; Trushina, E.; Fiesel, F.C.; Christenson, T.A.; Schroeder, A.S.; Zhang, L.; Bowles, E.; Behrouz, B.; et al. Progressive dopaminergic alterations and mitochondrial abnormalities in LRRK2 G2019S knock-in mice. Neurobiol. Dis. 2015, 78, 172–195. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.; Callio, J.; Otero, P.A.; Sekler, I.; Wills, Z.P.; Chu, C.T. Mitochondrial Calcium Dysregulation Contributes to Dendrite Degeneration Mediated by PD/LBD-Associated LRRK2 Mutants. J. Neurosci. 2017, 37, 11151–11165. [Google Scholar] [CrossRef] [PubMed]
- Bannwarth, S.; Ait-El-Mkadem, S.; Chaussenot, A.; Genin, E.C.; Lacas-Gervais, S.; Fragaki, K.; Berg-Alonso, L.; Kageyama, Y.; Serre, V.; Moore, D.G.; et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain 2014, 137 Pt 8, 2329–2345. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Yamashita, C.; Shiba-Fukushima, K.; Inoshita, T.; Funayama, M.; Sato, S.; Hatta, T.; Natsume, T.; Umitsu, M.; Takagi, J.; et al. Loss of Parkinson’s disease-associated protein CHCHD2 affects mitochondrial crista structure and destabilizes cytochrome c. Nat. Commun. 2017, 8, 15500. [Google Scholar] [CrossRef]
- Chen, S.; Mari, M.; Parashar, S.; Liu, D.; Cui, Y.; Reggiori, F.; Novick, P.J.; Ferro-Novick, S. Vps13 is required for the packaging of the ER into autophagosomes during ER-phagy. Proc. Natl. Acad. Sci. USA 2020, 117, 18530–18539. [Google Scholar] [CrossRef]
- Li, Z.; Okamoto, K.I.; Hayashi, Y.; Sheng, M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell 2004, 119, 873–887. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer. Nature 2017, 545, 446–451, Erratum in Nature 2018, 554, 264. [Google Scholar]
- Moufarrej, M.N.; Bianchi, D.W.; Shaw, G.M.; Stevenson, D.K.; Quake, S.R. Noninvasive Prenatal Testing Using Circulating DNA and RNA: Advances, Challenges, and Possibilities. Annu. Rev. Biomed. Data Sci. 2023, 6, 397–418. [Google Scholar] [CrossRef]
- Jiang, P.; Lo, Y.M.D. The Long and Short of Circulating Cell-Free DNA and the Ins and Outs of Molecular Diagnostics. Trends Genet. 2016, 32, 360–371. [Google Scholar] [CrossRef]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.Ø.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef]
- Alborelli, I.; Generali, D.; Jermann, P.; Cappelletti, M.R.; Ferrero, G.; Scaggiante, B.; Bortul, M.; Zanconati, F.; Nicolet, S.; Haegele, J.; et al. Cell-free DNA analysis in healthy individuals by next-generation sequencing: A proof of concept and technical validation study. Cell Death Dis. 2019, 10, 534. [Google Scholar] [CrossRef]
- Moss, J.; Magenheim, J.; Neiman, D.; Zemmour, H.; Loyfer, N.; Korach, A.; Samet, Y.; Maoz, M.; Druid, H.; Arner, P.; et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 2018, 9, 5068. [Google Scholar] [CrossRef] [PubMed]
- Ranucci, R. Cell-Free DNA: Applications in Different Diseases. Methods Mol. Biol. 2019, 1909, 3–12. [Google Scholar] [PubMed]
- Nagata, S.; Hanayama, R.; Kawane, K. Autoimmunity and the clearance of dead cells. Cell 2010, 140, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Hochreiter-Hufford, A.; Ravichandran, K.S. Clearing the dead: Apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harb. Perspect. Biol. 2013, 5, a008748. [Google Scholar] [CrossRef]
- Giacona, M.B.; Ruben, G.C.; Iczkowski, K.A.; Roos, T.B.; Porter, D.M.; Sorenson, G.D. Cell-free DNA in human blood plasma: Length measurements in patients with pancreatic cancer and healthy controls. Pancreas 1998, 17, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Patil, K.S.; Lange, J.; Constantine, A.; Maple-Grødem, J.; Tysnes, O.B.; Alves, G.W.; DiFrancisco-Donoghue, J.; Møller, S.G. Circulating cell-free DNA as predictors of Parkinson’s disease. Park. Relat. Disord. 2025, 137, 107919. [Google Scholar] [CrossRef] [PubMed]
- Manjili, M.H.; Park, J.; Facciponte, J.G.; Subjeck, J.R. HSP110 induces “danger signals” upon interaction with antigen presenting cells and mouse mammary carcinoma. Immunobiology 2005, 210, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Vorobjeva, N.V.; Chernyak, B.V. NETosis: Molecular Mechanisms, Role in Physiology and Pathology. Biochemistry 2020, 85, 1178–1190. [Google Scholar] [CrossRef]
- Pietronigro, E.C.; Della Bianca, V.; Zenaro, E.; Constantin, G. NETosis in Alzheimer’s Disease. Front. Immunol. 2017, 8, 211. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, H.; Long, Y.; Li, P.; Gu, Y. The main sources of circulating cell-free DNA: Apoptosis, necrosis and active secretion. Crit. Rev. Oncol. Hematol. 2021, 157, 103166. [Google Scholar] [CrossRef]
- Zenaro, E.; Pietronigro, E.; Bianca, V.D.; Piacentino, G.; Marongiu, L.; Budui, S.; Turano, E.; Rossi, B.; Angiari, S.; Dusi, S.; et al. Neutrophils promote Alzheimer’s disease–like pathology and cognitive decline via LFA-1 integrin. Nat. Med. 2015, 21, 880–886. [Google Scholar] [CrossRef]
- Smyth, L.C.D.; Murray, H.C.; Hill, M.; van Leeuwen, E.; Highet, B.; Magon, N.J.; Osanlouy, M.; Mathiesen, S.N.; Mockett, B.; Singh-Bains, M.K.; et al. Neutrophil-vascular interactions drive myeloperoxidase accumulation in the brain in Alzheimer’s disease. Acta Neuropathol. Commun. 2022, 10, 38. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Wentzel, J.F.; Aucamp, J.; van Dyk, E.; du Plessis, L.; Pretorius, P.J. Characterization of the cell-free DNA released by cultured cancer cells. Biochim. Biophys. Acta 2016, 1863, 157–165. [Google Scholar] [CrossRef]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Nair, R.R.; Mazza, D.; Brambilla, F.; Gorzanelli, A.; Agresti, A.; Bianchi, M.E. LPS-Challenged Macrophages Release Microvesicles Coated With Histones. Front. Immunol. 2018, 9, 1463. [Google Scholar] [CrossRef]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS ONE 2017, 12, e0183915. [Google Scholar] [CrossRef]
- Uehara, T.; Choong, C.J.; Nakamori, M.; Hayakawa, H.; Nishiyama, K.; Kasahara, Y.; Baba, K.; Nagata, T.; Yokota, T.; Tsuda, H.; et al. Amido-bridged nucleic acid (AmNA)-modified antisense oligonucleotides targeting α-synuclein as a novel therapy for Parkinson’s disease. Sci. Rep. 2019, 9, 7567. [Google Scholar] [CrossRef] [PubMed]
- Neupert, W.; Herrmann, J.M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007, 76, 723–749. [Google Scholar] [CrossRef] [PubMed]
- Topf, U.; Uszczynska-Ratajczak, B.; Chacinska, A. Mitochondrial stress-dependent regulation of cellular protein synthesis. J. Cell Sci. 2019, 132, jcs226258. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Serwa, R.A.; Samluk, L.; Suppanz, I.; Kodroń, A.; Stępkowski, T.M.; Elancheliyan, P.; Tsegaye, B.; Oeljeklaus, S.; Wasilewski, M.; et al. Immunoproteasome-specific subunit PSMB9 induction is required to regulate cellular proteostasis upon mitochondrial dysfunction. Nat. Commun. 2023, 14, 4092. [Google Scholar] [CrossRef]
- Lill, R. Function and biogenesis of iron-sulphur proteins. Nature 2009, 460, 831–838. [Google Scholar] [CrossRef]
- Bonawitz, N.D.; Clayton, D.A.; Shadel, G.S. Initiation and Beyond: Multiple Functions of the Human Mitochondrial Transcription Machinery. Mol. Cell 2006, 24, 813–825. [Google Scholar] [CrossRef]
- Suomalainen, A.; Nunnari, J. Mitochondria at the crossroads of health and disease. Cell 2024, 187, 2601–2627. [Google Scholar] [CrossRef]
- Wojtkowska, M.; Buczek, D.; Suzuki, Y.; Shabardina, V.; Makałowski, W.; Kmita, H. The emerging picture of the mitochondrial protein import complexes of Amoebozoa supergroup. BMC Genom. 2017, 18, 997. [Google Scholar] [CrossRef]
- Liu, B.H.; Xu, C.Z.; Liu, Y.; Lu, Z.L.; Fu, T.L.; Li, G.R.; Deng, Y.; Luo, G.Q.; Ding, S.; Li, N.; et al. Mitochondrial quality control in human health and disease. Mil. Med. Res. 2024, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Pfanner, N.; Warscheid, B.; Wiedemann, N. Mitochondrial proteins: From biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 2019, 20, 267–284, Erratum in Nat. Rev. Mol. Cell Biol. 2021, 22, 367. [Google Scholar] [CrossRef] [PubMed]
- Adebayo, M.; Singh, S.; Singh, A.P.; Dasgupta, S. Mitochondrial fusion and fission: The fine-tune balance for cellular homeostasis. FASEB J. 2021, 35, e21620. [Google Scholar] [CrossRef] [PubMed]
- Grel, H.; Woznica, D.; Ratajczak, K.; Kalwarczyk, E.; Anchimowicz, J.; Switlik, W.; Olejnik, P.; Zielonka, P.; Stobiecka, M.; Jakiela, S. Mitochondrial Dynamics in Neurodegenerative Diseases: Unraveling the Role of Fusion and Fission Processes. Int. J. Mol. Sci. 2023, 24, 13033. [Google Scholar] [CrossRef]
- Jenkins, B.C.; Neikirk, K.; Katti, P.; Claypool, S.M.; Kirabo, A.; McReynolds, M.R.; Hinton, A. Mitochondria in disease: Changes in shapes and dynamics. Trends Biochem. Sci. 2024, 49, 346–360. [Google Scholar] [CrossRef]
- Ng, M.Y.W.; Wai, T.; Simonsen, A. Quality control of the mitochondrion. Dev. Cell 2021, 56, 881–905. [Google Scholar] [CrossRef]
- Agarwal, S.; Muqit, M.M.K. PTEN-induced kinase 1 (PINK1) and Parkin: Unlocking a mitochondrial quality control pathway linked to Parkinson’s disease. Curr. Opin. Neurobiol. 2022, 72, 111–119. [Google Scholar] [CrossRef]
- Subramaniam, S.R.; Chesselet, M.F. Mitochondrial dysfunction and oxidative stress in Parkinson’s disease. Prog. Neurobiol. 2013, 106–107, 17–32. [Google Scholar] [CrossRef]
- Trinh, D.; Israwi, A.R.; Arathoon, L.R.; Gleave, J.A.; Nash, J.E. The multi-faceted role of mitochondria in the pathology of Parkinson’s disease. J. Neurochem. 2021, 156, 715–752. [Google Scholar] [CrossRef]
- Schapira, A.H.; Cooper, J.M.; Dexter, D.; Clark, J.B.; Jenner, P.; Marsden, C.D. Mitochondrial complex I deficiency in Parkinson’s disease. J. Neurochem. 1990, 54, 823–827. [Google Scholar] [CrossRef]
- Keeney, P.M.; Xie, J.; Capaldi, R.A.; Bennett, J.P. Parkinson’s disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J. Neurosci. 2006, 26, 5256–5264. [Google Scholar] [CrossRef]
- Michalak, S.; Florczak-Wyspiańska, J.; Rybacka-Mossakowska, J.; Ambrosius, W.; Osztynowicz, K.; Baszczuk, A.; Kozubski, W.; Wysocka, E. Mitochondrial Respiration in Intact Peripheral Blood Mononuclear Cells and Sirtuin 3 Activity in Patients with Movement Disorders. Oxid. Med. Cell Longev. 2017, 2017, 9703574. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Yang, Y.; Zhao, J.; Li, L.; Luo, D.; Hu, J.; Gao, Y.; Xie, X.; Shen, L.; Chen, S.; et al. Identification of key mitochondria-related genes and their relevance to the immune system linking Parkinson’s disease and primary Sjögren’s syndrome through integrated bioinformatics analyses. Comput. Biol Med. 2024, 175, 108511. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Sun, Y.; Bai, Z.; Li, M.; Kong, D.; Wu, G. Mitochondria-Related Genome-Wide Mendelian Randomization Identifies Putatively Causal Genes for Neurodegenerative Diseases. Mov. Disord. 2025, 40, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Dölle, C.; Flønes, I.; Nido, G.S.; Miletic, H.; Osuagwu, N.; Kristoffersen, S.; Lilleng, P.K.; Larsen, J.P.; Tysnes, O.B.; Haugarvoll, K.; et al. Defective mitochondrial DNA homeostasis in the substantia nigra in Parkinson disease. Nat. Commun. 2016, 7, 13548. [Google Scholar] [CrossRef]
- Tzoulis, C.; Tran, G.T.; Coxhead, J.; Bertelsen, B.; Lilleng, P.K.; Balafkan, N.; Payne, B.; Miletic, H.; Chinnery, P.F.; Bindoff, L.A. Molecular pathogenesis of polymerase γ-related neurodegeneration. Ann. Neurol. 2014, 76, 66–81. [Google Scholar] [CrossRef]
- Brauer, R.; Bhaskaran, K.; Chaturvedi, N.; Dexter, D.T.; Smeeth, L.; Douglas, I. Glitazone Treatment and Incidence of Parkinson’s Disease among People with Diabetes: A Retrospective Cohort Study. PLoS Med. 2015, 12, e1001854. [Google Scholar] [CrossRef]
- Quinsay, M.N.; Thomas, R.L.; Lee, Y.; Gustafsson, A.B. Bnip3-mediated mitochondrial autophagy is independent of the mitochondrial permeability transition pore. Autophagy 2010, 6, 855–862. [Google Scholar] [CrossRef]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef]
- Johansen, T.; Lamark, T. Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J. Mol. Biol. 2020, 432, 80–103. [Google Scholar] [CrossRef]
- Malpartida, A.B.; Williamson, M.; Narendra, D.P.; Wade-Martins, R.; Ryan, B.J. Mitochondrial Dysfunction and Mitophagy in Parkinson’s Disease: From Mechanism to Therapy. Trends Biochem. Sci. 2021, 46, 329–343. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.P.; Jin, S.M.; Tanaka, A.; Suen, D.F.; Gautier, C.A.; Shen, J.; Cookson, M.R.; Youle, R.J. PINK1 Is Selectively Stabilized on Impaired Mitochondria to Activate Parkin. PLoS Biol. 2010, 8, e1000298. [Google Scholar] [CrossRef] [PubMed]
- Lazarou, M.; Jin, S.M.; Kane, L.A.; Youle, R.J. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell 2012, 22, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef]
- König, T.; McBride, H.M. Mitochondrial-derived vesicles in metabolism, disease, and aging. Cell Metab. 2024, 36, 21–35. [Google Scholar] [CrossRef]
- Leggio, L.; Paternò, G.; Vivarelli, S.; Falzone, G.G.; Giachino, C.; Marchetti, B.; Iraci, N. Extracellular Vesicles as Novel Diagnostic and Prognostic Biomarkers for Parkinson’s Disease. Aging Dis. 2021, 12, 1494–1515. [Google Scholar] [CrossRef]
- McLelland, G.L.; Soubannier, V.; Chen, C.X.; McBride, H.M.; Fon, E.A. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014, 33, 282–295. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, W.; Wu, Y.; Guo, Z.; Chen, J.; Tian, C.; Wang, P.; Zeng, S.; Xu, B.; Duan, J.; et al. CNS Mitochondria-Derived Vesicle in Blood: Potential Biomarkers for Brain Mitochondria Dysfunction. Ann. Clin. Transl. Neurol. 2025, 12, 1312–1323. [Google Scholar] [CrossRef]
- Xian, H.; Watari, K.; Sanchez-Lopez, E.; Offenberger, J.; Onyuru, J.; Sampath, H.; Ying, W.; Hoffman, H.M.; Shadel, G.S.; Karin, M. Oxidized DNA fragments exit mitochondria via mPTP- and VDAC-dependent channels to activate NLRP3 inflammasome and interferon signaling. Immunity 2022, 55, 1370–1385.e8. [Google Scholar] [CrossRef]
- Matzinger, P. The danger model: A renewed sense of self. Science 2002, 296, 301–305. [Google Scholar] [CrossRef]
- Kunze, R.; Fischer, S.; Marti, H.H.; Preissner, K.T. Brain alarm by self-extracellular nucleic acids: From neuroinflammation to neurodegeneration. J. Biomed. Sci. 2023, 30, 64. [Google Scholar] [CrossRef]
- Rodríguez-Nuevo, A.; Zorzano, A. The sensing of mitochondrial DAMPs by non-immune cells. Cell Stress 2019, 3, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Moreno-García, L.; López-Royo, T.; Calvo, A.C.; Toivonen, J.M.; de la Torre, M.; Moreno-Martínez, L.; Molina, N.; Aparicio, P.; Zaragoza, P.; Manzano, R.; et al. Competing Endogenous RNA Networks as Biomarkers in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 9582. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J.C.H.; Lo, Y.M.D. Circulating nucleic acids in plasma/serum. Pathology 2007, 39, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Calvani, R.; Coelho-Junior, H.J.; Landi, F.; Bernabei, R.; Marzetti, E. Mitochondrial Dysfunction, Oxidative Stress, and Neuroinflammation: Intertwined Roads to Neurodegeneration. Antioxidants 2020, 9, 647. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lee, A.S.; Shameli, A.; Geng, X.; Finegood, D.; Santamaria, P.; Dutz, J.P. TLR9 blockade inhibits activation of diabetogenic CD8+ T cells and delays autoimmune diabetes. J. Immunol. 2010, 184, 5645–5653. [Google Scholar] [CrossRef]
- Gambardella, S.; Limanaqi, F.; Ferese, R.; Biagioni, F.; Campopiano, R.; Centonze, D.; Fornai, F. ccf-mtDNA as a Potential Link Between the Brain and Immune System in Neuro-Immunological Disorders. Front. Immunol. 2019, 10, 1064. [Google Scholar] [CrossRef]
- Zhang, K.; Fu, R.; Liu, R.; Su, Z. Circulating cell-free DNA-based multi-cancer early detection. Trends Cancer 2024, 10, 161–174. [Google Scholar] [CrossRef]
- Kumar, V. Toll-like receptors in the pathogenesis of neuroinflammation. J. Neuroimmunol. 2019, 332, 16–30. [Google Scholar] [CrossRef]
- Stephenson, J.; Nutma, E.; van der Valk, P.; Amor, S. Inflammation in CNS neurodegenerative diseases. Immunology 2018, 154, 204–219. [Google Scholar] [CrossRef]
- West, A.P.; Shadel, G.S. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol. 2017, 17, 363–375. [Google Scholar] [CrossRef]
- Zhong, Z.; Liang, S.; Sanchez-Lopez, E.; He, F.; Shalapour, S.; Lin, X.J.; Wong, J.; Ding, S.; Seki, E.; Schnabl, B.; et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature 2018, 560, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.Y.; Tian, T.; Yao, H.; Xia, X.M.; Wang, C.; Cao, L.; Hu, G.; Du, R.H.; Lu, M. The cGAS-STING-YY1 axis accelerates progression of neurodegeneration in a mouse model of Parkinson’s disease via LCN2-dependent astrocyte senescence. Cell Death Differ. 2023, 30, 2280–2292. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Li, T.; Zhang, W.; Wu, J.; Hong, H.; Quan, W.; Qiao, X.; Cui, C.; Qiao, C.; Zhao, W.; et al. The cGAS-STING-interferon regulatory factor 7 pathway regulates neuroinflammation in Parkinson’s disease. Neural Regen Res. 2025, 20, 2361–2372. [Google Scholar] [CrossRef] [PubMed]
- Newman, L.E.; Shadel, G.S. Pink1/Parkin link inflammation, mitochondrial stress, and neurodegeneration. J. Cell Biol. 2018, 217, 3327–3329. [Google Scholar] [CrossRef]
- Tresse, E.; Marturia-Navarro, J.; Sew, W.Q.G.; Cisquella-Serra, M.; Jaberi, E.; Riera-Ponsati, L.; Fauerby, N.; Hu, E.; Kretz, O.; Aznar, S.; et al. Mitochondrial DNA damage triggers spread of Parkinson’s disease-like pathology. Mol. Psychiatry 2023, 28, 4902–4914. [Google Scholar] [CrossRef]
- Hegde, A.N. Ubiquitin-proteasome-mediated local protein degradation and synaptic plasticity. Prog. Neurobiol. 2004, 73, 311–357. [Google Scholar] [CrossRef]
- Wojtkowska, M.; Karczewska, N.; Pacewicz, K.; Pacak, A.; Kopeć, P.; Florczak-Wyspiańska, J.; Popławska-Domaszewicz, K.; Małkiewicz, T.; Sokół, B. Quantification of Circulating Cell-Free DNA in Idiopathic Parkinson’s Disease Patients. Int. J. Mol. Sci. 2024, 25, 2818. [Google Scholar] [CrossRef]
- Pyle, A.; Anugrha, H.; Kurzawa-Akanbi, M.; Yarnall, A.; Burn, D.; Hudson, G. Reduced mitochondrial DNA copy number is a biomarker of Parkinson’s disease. Neurobiol. Aging 2016, 38, 216.e7–216.e10. [Google Scholar] [CrossRef]
- Borsche, M.; König, I.R.; Delcambre, S.; Petrucci, S.; Balck, A.; Brüggemann, N.; Zimprich, A.; Wasner, K.; Pereira, S.L.; Avenali, M.; et al. Mitochondrial damage-associated inflammation highlights biomarkers in PRKN/PINK1 parkinsonism. Brain 2020, 143, 3041–3051. [Google Scholar] [CrossRef]
- Vivekanantham, S.; Shah, S.; Dewji, R.; Dewji, A.; Khatri, C.; Ologunde, R. Neuroinflammation in Parkinson’s disease: Role in neurodegeneration and tissue repair. Int. J. Neurosci. 2015, 125, 717–725. [Google Scholar] [CrossRef]
- Dobbs, R.J.; Charlett, A.; Purkiss, A.G.; Dobbs, S.M.; Weller, C.; Peterson, D.W. Association of circulating TNF-alpha and IL-6 with ageing and parkinsonism. Acta Neurol. Scand. 1999, 100, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Ying, C.; Han, C.; Li, Y.; Zhang, M.; Xiao, S.; Zhao, L.; Zhang, H.; Yu, Q.; An, J.; Mao, W.; et al. Plasma circulating cell-free DNA integrity and relative telomere length as diagnostic biomarkers for Parkinson’s disease and multiple system atrophy: A cross-sectional study. Neural Regen Res. 2025, 20, 3553–3563. [Google Scholar] [CrossRef] [PubMed]
- Aucamp, J.; Bronkhorst, A.J.; Badenhorst, C.P.S.; Pretorius, P.J. The diverse origins of circulating cell-free DNA in the human body: A critical re-evaluation of the literature. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1649–1683. [Google Scholar] [CrossRef] [PubMed]
- Lowes, H.; Pyle, A.; Santibanez-Koref, M.; Hudson, G. Circulating cell-free mitochondrial DNA levels in Parkinson’s disease are influenced by treatment. Mol. Neurodegener. 2020, 15, 10. [Google Scholar] [CrossRef]
- Pyle, A.; Brennan, R.; Kurzawa-Akanbi, M.; Yarnall, A.; Thouin, A.; Mollenhauer, B.; Burn, D.; Chinnery, P.F.; Hudson, G. Reduced cerebrospinal fluid mitochondrial DNA is a biomarker for early-stage Parkinson’s disease. Ann. Neurol. 2015, 78, 1000–1004. [Google Scholar] [CrossRef]
- Meng, J.; Wang, F.; Ji, L.; Liang, Y.; Nian, W.; Song, L.; Zhu, A. Comprehensive methylation profile of CSF cfDNA revealed pathogenesis and diagnostic markers for early-onset Parkinson’s disease. Epigenomics 2021, 13, 1637–1651. [Google Scholar] [CrossRef]
- Puigròs, M.; Calderon, A.; Pérez-Soriano, A.; de Dios, C.; Fernández, M.; Colell, A.; Martí, M.-J.; Tolosa, E.; Trullas, R. Cell-free mitochondrial DNA deletions in idiopathic, but not LRRK2, Parkinson’s disease. Neurobiol. Dis. 2022, 174, 105885. [Google Scholar] [CrossRef]
- Ying, C.; Li, Y.; Zhang, H.; Pang, S.; Hao, S.; Hu, S.; Zhao, L. Probing the diagnostic values of plasma cf-nDNA and cf-mtDNA for Parkinson’s disease and multiple system atrophy. Front. Neurosci. 2024, 18, 1488820. [Google Scholar] [CrossRef]
- Zimmermann, M.; Brockmann, K. Blood and Cerebrospinal Fluid Biomarkers of Inflammation in Parkinson’s Disease. J. Parkinsons Dis. 2022, 12 (Suppl. 1), S183–S200. [Google Scholar] [CrossRef]
- Ma, Z.L.; Wang, Z.L.; Zhang, F.Y.; Liu, H.X.; Mao, L.H.; Yuan, L. Biomarkers of Parkinson’s Disease: From Basic Research to Clinical Practice. Aging Dis. 2024, 15, 1813–1830. [Google Scholar]
- Baran, A.; Bulut, M.; Kaya, M.C.; Demirpençe, Ö.; Sevim, B.; Akıl, E.; Varol, S. High-sensitivity C-reactive protein and high mobility group box-1 levels in Parkinson’s disease. Neurol. Sci. 2019, 40, 167–173. [Google Scholar] [CrossRef]
- Jin, H.; Gu, H.Y.; Mao, C.J.; Chen, J.; Liu, C.F. Association of inflammatory factors and aging in Parkinson’s disease. Neurosci. Lett. 2020, 736, 135259. [Google Scholar] [CrossRef]
- Dommershuijsen, L.J.; Ruiter, R.; Erler, N.S.; Rizopoulos, D.; Ikram, M.A.; Ikram, M.K. Peripheral Immune Cell Numbers and C-Reactive Protein in Parkinson’s Disease: Results from a Population-Based Study. J. Parkinsons Dis. 2022, 12, 667–678. [Google Scholar] [CrossRef]
- Thaler, A.; Omer, N.; Giladi, N.; Gurevich, T.; Bar-Shira, A.; Gana-Weisz, M.; Goldstein, O.; Kestenbaum, M.; Shirvan, J.C.; Cedarbaum, J.M.; et al. Mutations in GBA and LRRK2 Are Not Associated with Increased Inflammatory Markers. J. Parkinsons Dis. 2021, 11, 1285–1296. [Google Scholar] [CrossRef]
- Majbour, N.K.; Aasly, J.O.; Hustad, E.; Thomas, M.A.; Vaikath, N.N.; Elkum, N.; van de Berg, W.D.J.; Tokuda, T.; Mollenhauer, B.; Berendse, H.W.; et al. CSF total and oligomeric α-Synuclein along with TNF-α as risk biomarkers for Parkinson’s disease: A study in LRRK2 mutation carriers. Transl. Neurodegener. 2020, 9, 15. [Google Scholar] [CrossRef]
- Galper, J.; Balwani, M.; Fahn, S.; Waters, C.; Krohn, L.; Gan-Or, Z.; Dzamko, N.; Alcalay, R.N. Cytokines and Gaucher Biomarkers in Glucocerebrosidase Carriers with and Without Parkinson Disease. Mov. Disord. 2021, 36, 1451–1455. [Google Scholar] [CrossRef]
- Narendra, D.; Walker, J.E.; Youle, R. Mitochondrial quality control mediated by PINK1 and Parkin: Links to parkinsonism. Cold Spring Harb. Perspect. Biol. 2012, 4, a011338. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef] [PubMed]
- Mulas, G.; Espa, E.; Fenu, S.; Spiga, S.; Cossu, G.; Pillai, E.; Carboni, E.; Simbula, G.; Jadžić, D.; Angius, F.; et al. Differential induction of dyskinesia and neuroinflammation by pulsatile versus continuous l-DOPA delivery in the 6-OHDA model of Parkinson’s disease. Exp. Neurol. 2016, 286, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Arshad, O.; Gadawska, I.; Sattha, B.; Côté, H.C.F.; Hsieh, A.Y.Y.; Canadian Institutes of Health Research Team on Cellular Aging and HIV Comorbidities in Women and Children (CARMA). Elevated Cell-Free Mitochondrial DNA in Filtered Plasma Is Associated With HIV Infection and Inflammation. J. Acquir. Immune Defic. Syndr. 2018, 78, 111–118. [Google Scholar] [CrossRef]
- Liu, J.; Cai, X.; Xie, L.; Tang, Y.; Cheng, J.; Wang, J.; Wang, L.; Gong, J. Circulating Cell Free Mitochondrial DNA is a Biomarker in the Development of Coronary Heart Disease in the Patients with Type 2 Diabetes. Clin. Lab. 2015, 61, 661–667. [Google Scholar] [CrossRef]
- Lindqvist, D.; Wolkowitz, O.M.; Picard, M.; Ohlsson, L.; Bersani, F.S.; Fernström, J.; Westrin, Å.; Hough, C.M.; Lin, J.; Reus, V.I.; et al. Circulating cell-free mitochondrial DNA, but not leukocyte mitochondrial DNA copy number, is elevated in major depressive disorder. Neuropsychopharmacology 2018, 43, 1557–1564. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wojtkowska, M.; Ambrosius, F. Cell-Free DNA and Mitochondria in Parkinson’s Disease. Int. J. Mol. Sci. 2025, 26, 11615. https://doi.org/10.3390/ijms262311615
Wojtkowska M, Ambrosius F. Cell-Free DNA and Mitochondria in Parkinson’s Disease. International Journal of Molecular Sciences. 2025; 26(23):11615. https://doi.org/10.3390/ijms262311615
Chicago/Turabian StyleWojtkowska, Małgorzata, and Franciszek Ambrosius. 2025. "Cell-Free DNA and Mitochondria in Parkinson’s Disease" International Journal of Molecular Sciences 26, no. 23: 11615. https://doi.org/10.3390/ijms262311615
APA StyleWojtkowska, M., & Ambrosius, F. (2025). Cell-Free DNA and Mitochondria in Parkinson’s Disease. International Journal of Molecular Sciences, 26(23), 11615. https://doi.org/10.3390/ijms262311615






