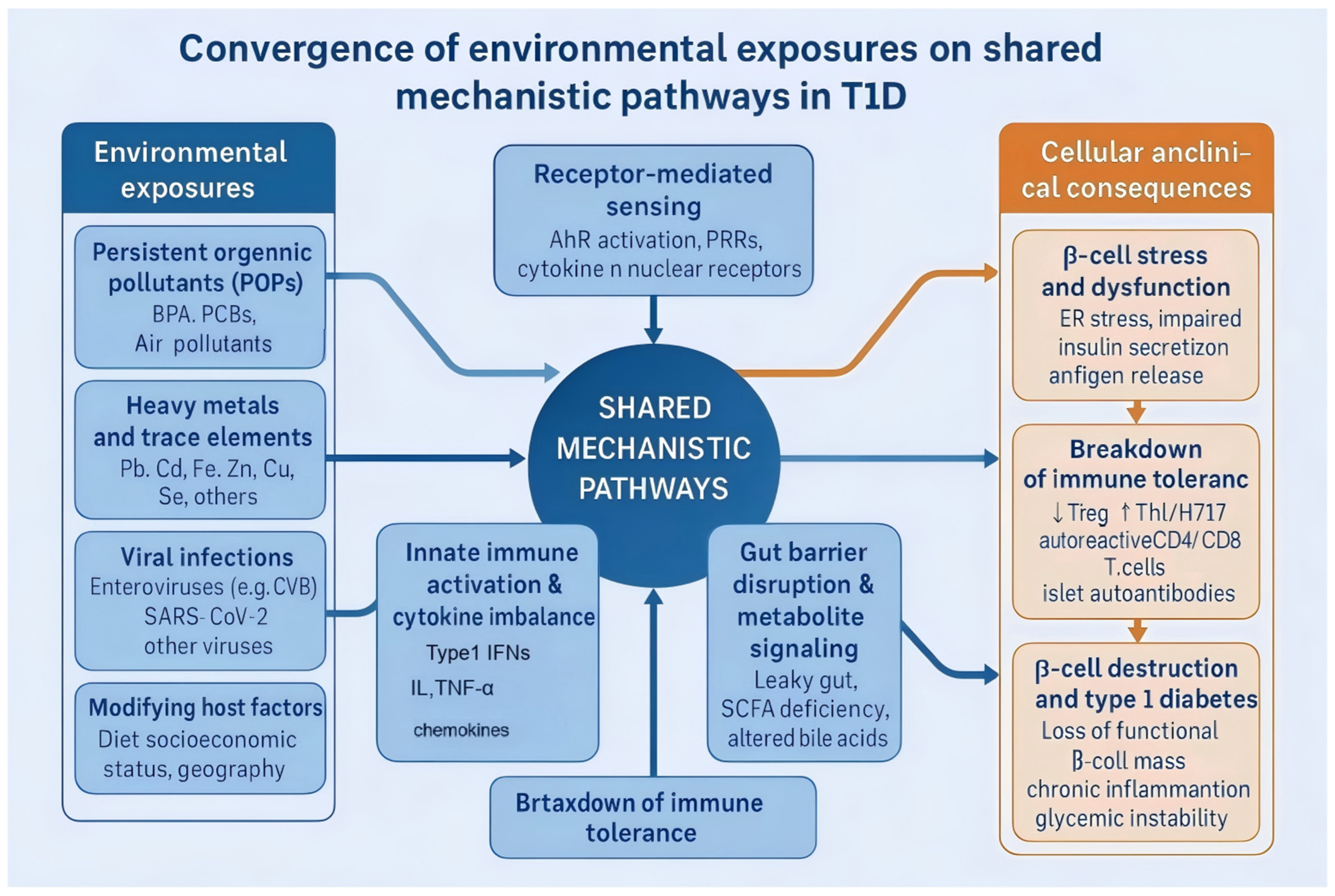
Environmental Mechanisms Influencing the Pathogenesis and Progression of Type 1 Diabetes
Abstract
1. Introduction
2. Environmental Pollutants and T1D Susceptibility
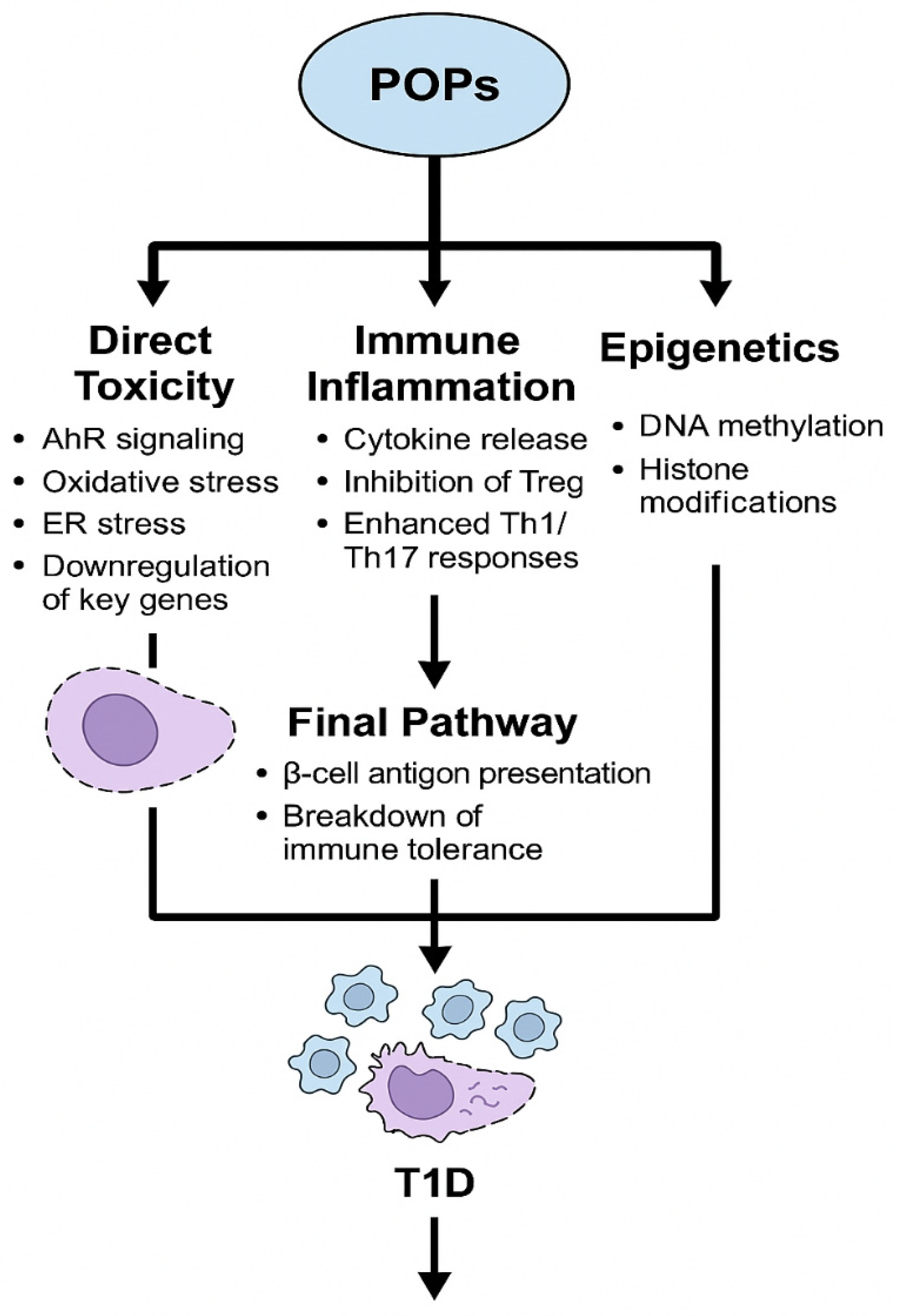
2.1. Persistent Organic Pollutants (POPs)
2.1.1. Experimental Studies
2.1.2. Human Studies
2.1.3. Mechanistic Insights
2.2. Metal and Trace Elements
2.2.1. Iron
2.2.2. Zinc
2.2.3. Copper
2.2.4. Toxic Heavy Metals
2.2.5. Other Trace Elements
2.3. Air Pollutants and Particulate Matter
2.3.1. Particulate Matter (PM2.5/PM10)
2.3.2. Ozone (O3)
2.3.3. Asian Sand Dust (ASD)
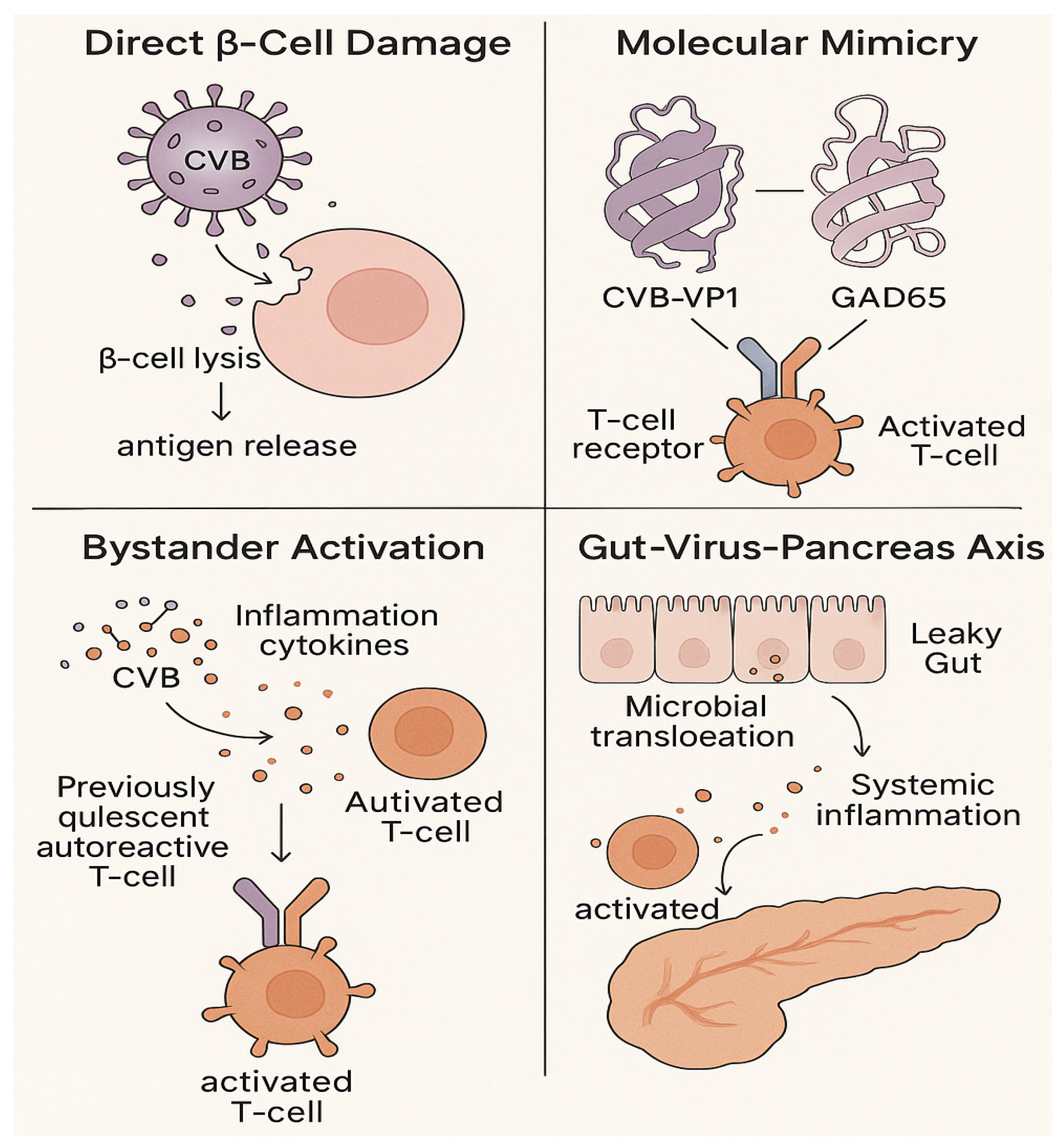
3. Viral Infections: Key Triggers of β-Cell Autoimmunity
3.1. Direct β-Cell Damage and Dysfunction
3.2. Immune-Mediated Mechanisms
3.3. Gut-Virus-Pancreas Crosstalk
3.4. Emerging Viral Triggers
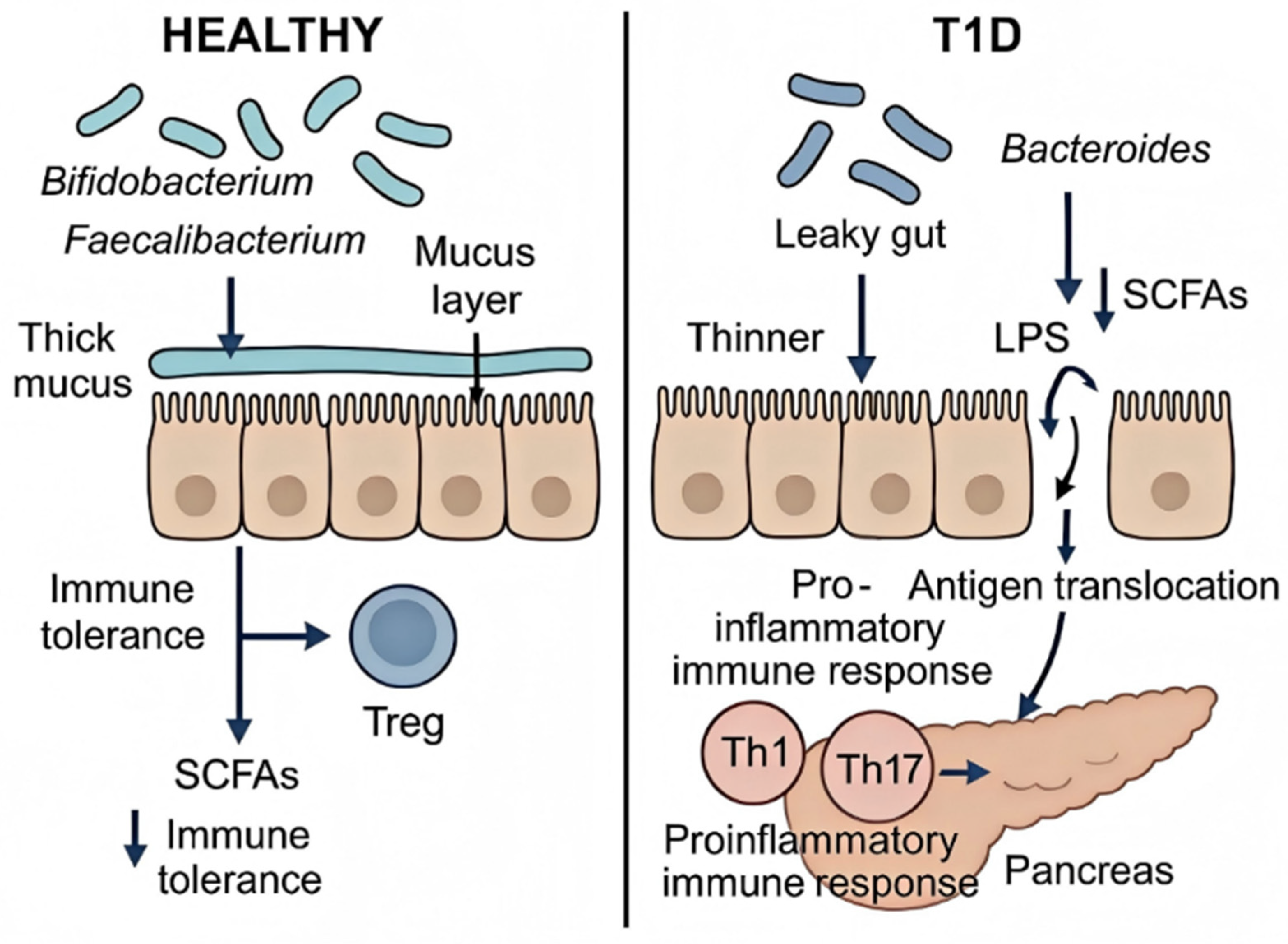
4. Gut Microenvironment in T1D Regulation
4.1. Gut Microbiota Dysbiosis and Metabolite Alterations
4.2. Breakdown of the Intestinal Barrier and Resultant Immune Dysregulation
4.2.1. Barrier Dysfunction and Antigen Translocation
4.2.2. Microbiota-Driven Immune Activation and Loss of Tolerance
5. Oxidative Stress, Mitochondrial Dysfunction, and Inflammatory Mechanisms
5.1. Oxidative Stress and Mitochondrial Dysfunction
5.2. Inflammatory Mechanisms
6. Epigenetic Modifications Mediated by Environmental Triggers
6.1. DNA Methylation: A Key Epigenetic Regulator in T1D
6.2. Histone Modifications and Non-Coding RNAs
6.3. Epigenetics in T1D Complications and Clinical Translation
7. Gene–Environment Interactions in T1D Pathogenesis
7.1. Genetic Susceptibility: Setting the Stage
7.2. Environmental Triggers: The External Catalysts
7.3. Mechanisms of Interaction: Bridging Genes and Environment
8. Conclusions and Future Perspectives
- From Association to Causation: While epidemiological studies robustly link environmental factors to T1D, establishing definitive causal relationships in humans is complex. Future research must move beyond correlation by leveraging longitudinal cohorts from birth, like the TEDDY study, refs. [6,118] with more frequent and precise exposure assessments. Integrating multi-omics approaches (genomics, epigenomics, metabolomics, metagenomics) from serial samples will be essential to delineate the temporal sequence of events from exposure to immune activation and β-cell damage [85,108].
- The Exposome and Multi-Factorial Interactions: Individuals are exposed to a mixture of environmental factors simultaneously, not in isolation. A major future direction is to characterize the “T1D exposome”—the totality of exposures throughout life—and understand how these factors interact synergistically or antagonistically [9,28]. Advanced statistical models and machine learning algorithms will be necessary to decipher these complex interactions and identify critical windows of exposure, particularly during gestation, early childhood, and puberty [26,27].
- Resolving Mechanistic Complexity: The precise molecular mechanisms linking specific exposures to the breakdown of immune tolerance require further elucidation. Key unanswered questions include the exact role of epigenetic modifications as a persistent memory of environmental insults and the relative contribution of direct β-cell toxicity versus immune dysregulation. Research should prioritize human-relevant models, such as humanized mice, stem-cell-derived islets, and sophisticated in vitro systems, to validate mechanisms identified in animal models [6,33,40,50,100].
- Towards Personalized Prevention and Therapy: The variable individual response to environmental triggers, influenced by genetics, epigenetics, and microbiome composition, calls for a personalized medicine approach. Future efforts should focus on developing integrated risk scores that combine genetic, epigenetic, and environmental data to identify individuals at the highest risk. This will enable targeted, cost-effective prevention trials. Moreover, the therapeutic potential of targeting environmental mechanisms, such as using short-chain fatty acid derivatives or engineered probiotics to restore immune tolerance, represents a promising frontier for intervention in pre-symptomatic or new-onset T1D [65,95,133].
- Interdisciplinary Collaboration: Addressing the multifaceted challenge of environment–T1D interactions demands unprecedented collaboration across disciplines, including epidemiology, immunology, toxicology, microbiology, bioinformatics, and public policy. Only through such integrated efforts can we translate mechanistic insights into tangible strategies to mitigate the global burden of T1D [107].
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | English Full Name |
| T1D | Type 1 Diabetes |
| HLA | Human Leukocyte Antigen |
| POPs | Persistent Organic Pollutants |
| BPA | Bisphenol A |
| PCBs | Polychlorinated Biphenyls |
| EDC | Endocrine-Disrupting Chemical |
| MLDSTZ | Multiple Low-Dose Streptozotocin |
| NOD | Non-Obese Diabetic (mouse model) |
| GMB | Gut Microbiome |
| SEARCH-CC | SEARCH for Diabetes in Youth Case Control Study |
| OR | Odds Ratio |
| CI | Confidence Interval |
| T1D/IS | Type 1 Diabetes with Normal Insulin Sensitivity |
| T1D/IR | Type 1 Diabetes with Insulin Resistance |
| INS-1E | Insulin-Secreting Cell Line 1-E |
| AhR | Aryl Hydrocarbon Receptor |
| ER | Endoplasmic Reticulum |
| Treg | Regulatory T-Cell |
| Th1 | T-Helper 1 Cell |
| Th17 | T-Helper 17 Cell |
| TIBC | Total Iron-Binding Capacity |
| HbA1c | Glycated Hemoglobin |
| Z-GCN | Zinc–Glucagon Complex |
| GNP | Gold Nanoparticle |
| PM | Particulate Matter |
| O3 | Ozone |
| ASD | Asian Sand Dust |
| CY | Cyclophosphamide |
| IFN-γ | Interferon-Gamma |
| CVB | Coxsackievirus B |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome Coronavirus 2 |
| ACE2 | Angiotensin-Converting Enzyme 2 |
| DKA | Diabetic Ketoacidosis |
| AGEs | Advanced Glycation End Products |
| PKC | Protein Kinase C |
| SCFA | Short-Chain Fatty Acid |
| GPR | G Protein-Coupled Receptor |
| PI3K-AKT | Phosphoinositide 3-Kinase-AKT Pathway |
| Foxp3 | Forkhead Box P3 |
| HDAC | Histone Deacetylase |
| LPS | Lipopolysaccharide |
| TLR4 | Toll-Like Receptor 4 |
| DC | Dendritic Cell |
| NF-κB | Nuclear Factor-Kappa B |
| FXR | Farnesoid X Receptor |
| EGFR | Epidermal Growth Factor Receptor |
| RhoA-Rock | RhoA-Rho-Associated Kinase Pathway |
| MUC2 | Mucin 2 |
| PRR | Pattern Recognition Receptor |
| NLRP3 | NOD-Like Receptor Pyrin Domain-Containing 3 |
| Breg | Regulatory B Cell |
| STAT3 | Signal Transducer and Activator of Transcription 3 |
| DNMT | DNA Methyltransferase |
| PDX1 | Pancreatic and Duodenal Homeobox 1 |
| MAFA | V-Maf Avian Musculoaponeurotic Fibrosarcoma Oncogene Homolog A |
| mtROS | Mitochondrial Reactive Oxygen Species |
| MnSOD | Manganese Superoxide Dismutase |
| NOX | NADPH Oxidase |
| mQTL | Methylation Quantitative Trait Locus |
| SNP | Single Nucleotide Polymorphism |
| PAD | Peptidylarginine Deiminase |
| miRNA | MicroRNA |
| circRNA | Circular RNA |
| DN | Diabetic Nephropathy |
| PDR | Proliferative Diabetic Retinopathy |
| DKD | Diabetic Kidney Disease |
| GWAS | Genome-Wide Association Study |
| TEDDY | The Environmental Determinants of Diabetes in the Young |
| HERV | Human Endogenous Retrovirus |
| CTSH | Cathepsin H |
| IL | Interleukin |
| TNF-α | Tumor Necrosis Factor-Alpha |
| VP1 | Viral Protein 1 |
| GAD65 | Glutamic Acid Decarboxylase 65 |
| CXCL10 | C-X-C Motif Chemokine Ligand 10 |
| iNKT | Invariant Natural Killer T Cell |
| pDC | Plasmacytoid Dendritic Cell |
| HSP60 | Heat Shock Protein 60 |
| DAMPs | Danger-Associated Molecular Patterns |
| PNPT1 | Polynucleotide Phosphorylase 1 |
| TET2 | Ten-Eleven Translocation 2 |
| GPER | G-Protein Coupled Estrogen Receptor |
| GapmeR | Gapmer Antisense Oligonucleotide |
| PAI-1 | Plasminogen Activator Inhibitor-1 |
| ET-1 | Endothelin-1 |
References
- Rewers, M.; Ludvigsson, J. Environmental risk factors for type 1 diabetes. Lancet 2016, 387, 2340–2348. [Google Scholar] [CrossRef] [PubMed]
- Bluestone, J.A.; Buckner, J.H.; Herold, K.C. Immunotherapy: Building a bridge to a cure for type 1 diabetes. Science 2021, 373, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Quattrin, T.; Mastrandrea, L.D.; Walker, L.S.K. Type 1 diabetes. Lancet 2023, 401, 2149–2162. [Google Scholar] [CrossRef]
- Keskesiadou, G.N.; Tsokkou, S.; Konstantinidis, I.; Georgaki, M.N.; Sioga, A.; Papamitsou, T.; Karachrysafi, S. Endocrine-Disrupting Chemicals and the Development of Diabetes Mellitus Type 1: A 5-Year Systematic Review. Int. J. Mol. Sci. 2024, 25, 10111. [Google Scholar] [CrossRef] [PubMed]
- Khalil, W.J.; Akeblersane, M.; Khan, A.S.; Moin, A.S.M.; Butler, A.E. Environmental Pollution and the Risk of Developing Metabolic Disorders: Obesity and Diabetes. Int. J. Mol. Sci. 2023, 24, 8870. [Google Scholar] [CrossRef]
- Cetkovic-Cvrlje, M.; Thinamany, S.; Bruner, K.A. Bisphenol A (BPA) aggravates multiple low-dose streptozotocin-induced Type 1 diabetes in C57BL/6 mice. J. Immunotoxicol. 2017, 14, 160–168. [Google Scholar] [CrossRef]
- Acconcia, F.; Pallottini, V.; Marino, M. Molecular Mechanisms of Action of BPA. Dose Response 2015, 13, 1559325815610582. [Google Scholar] [CrossRef]
- Xu, J.; Huang, G.; Nagy, T.; Teng, Q.; Guo, T.L. Sex-dependent effects of bisphenol A on type 1 diabetes development in non-obese diabetic (NOD) mice. Arch. Toxicol. 2019, 93, 997–1008. [Google Scholar] [CrossRef]
- Sinioja, T.; Bodin, J.; Duberg, D.; Dirven, H.; Berntsen, H.F.; Zimmer, K.; Nygaard, U.C.; Orešič, M.; Hyötyläinen, T. Exposure to persistent organic pollutants alters the serum metabolome in non-obese diabetic mice. Metabolomics 2022, 18, 87. [Google Scholar] [CrossRef]
- Bresson, S.E.; Isom, S.; Jensen, E.T.; Huber, S.; Oulhote, Y.; Rigdon, J.; Lovato, J.; Liese, A.D.; Pihoker, C.; Dabelea, D.; et al. Associations between persistent organic pollutants and type 1 diabetes in youth. Environ. Int. 2022, 163, 107175. [Google Scholar] [CrossRef]
- Johansen, V.B.I.; Josefsen, K.; Antvorskov, J.C. The Impact of Dietary Factors during Pregnancy on the Development of Islet Autoimmunity and Type 1 Diabetes: A Systematic Literature Review. Nutrients 2023, 15, 4333. [Google Scholar] [CrossRef]
- Størdal, K.; McArdle, H.J.; Hayes, H.; Tapia, G.; Viken, M.K.; Lund-Blix, N.A.; Haugen, M.; Joner, G.; Skrivarhaug, T.; Mårild, K.; et al. Prenatal iron exposure and childhood type 1 diabetes. Sci. Rep. 2018, 8, 9067. [Google Scholar] [CrossRef]
- Thorsen, S.U.; Liu, X.; Kataria, Y.; Mandrup-Poulsen, T.; Kaur, S.; Uusitalo, U.; Virtanen, S.M.; Norris, J.M.; Rewers, M.; Hagopian, W.; et al. Interaction Between Dietary Iron Intake and Genetically Determined Iron Overload: Risk of Islet Autoimmunity and Progression to Type 1 Diabetes in the TEDDY Study. Diabetes Care 2023, 46, 1014–1018. [Google Scholar] [CrossRef]
- Elhassan, S.; Dong, F.; Buckner, T.; Johnson, R.K.; Seifert, J.A.; Carry, P.M.; Vanderlinden, L.; Waugh, K.; Rewers, M.; Norris, J.M. Investigating iron intake in risk of progression from islet autoimmunity to type 1 diabetes: The diabetes autoimmunity study in the young. Front. Immunol. 2023, 14, 1124370. [Google Scholar] [CrossRef]
- Zhang, Y.; Peng, W.; Huang, J.; Zhang, W.; Jiang, P.; He, Y.; Wang, M. Causal Associations of Iron Status With the Renal Function and Diabetic Nephropathy in Patients With Diabetes Mellitus: A Two-Sample Mendelian Randomization Study. J. Diabetes Res. 2025, 2025, 6658794. [Google Scholar] [CrossRef] [PubMed]
- Moslhy, E.A.M.; Tadros, M.M.M.; Thabet, R.A.; Hemida, E.H.A.; Noureldeen, A.F.H. Impact of vitamin D deficiency on iron status in children with type I diabetes. Sci. Rep. 2024, 14, 12989. [Google Scholar] [CrossRef]
- Bjørklund, G.; Dadar, M.; Pivina, L.; Doşa, M.D.; Semenova, Y.; Aaseth, J. The Role of Zinc and Copper in Insulin Resistance and Diabetes Mellitus. Curr. Med. Chem. 2020, 27, 6643–6657. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, X.; Qi, X.; Bai, J.; Tong, B.; Zhang, D.; Yin, X.; Yu, P. The role of metal ion metabolism in the pathogenesis of diabetes and associated complications. Front. Endocrinol. 2025, 16, 1541809. [Google Scholar] [CrossRef] [PubMed]
- GhavamiNejad, A.; Liu, J.F.; Mirzaie, S.; Lu, B.; Samarikhalaj, M.; Giacca, A.; Wu, X.Y. Catechol-based chemistry for hypoglycemia-responsive delivery of zinc-glucagon via hydrogel-based microneedle patch technology. Nat. Commun. 2025, 16, 3124. [Google Scholar] [CrossRef]
- GhavamiNejad, A.; Li, J.; Lu, B.; Zhou, L.; Lam, L.; Giacca, A.; Wu, X.Y. Glucose-Responsive Composite Microneedle Patch for Hypoglycemia-Triggered Delivery of Native Glucagon. Adv. Mater. 2019, 31, e1901051. [Google Scholar] [CrossRef]
- da Silva, J.A.D.; Filetti, F.M.; da Silva, N.P.; Gomes, K.N.; Graceli, J.B.; Lopes, A.B.; Vassallo, D.V.; Nunes, K.Z. Copper exposure at a daily dose twice the recommended in diabetic rats induces oxidative stress, vascular dysfunction and perivascular adipose tissue inflammation in diabetic rats. Toxicol. Lett. 2025, 409, 97–108. [Google Scholar] [CrossRef]
- Atwa, H.A.; Hemeda, M.S.; Farh, R.M.; Khafagy, A.A.; Abdelaziz, B.M.; Ibrahim, M.A. Hair levels of mercury, lead, and cadmium and their association with glycemic control in Egyptian children with type 1 diabetes. BMC Pediatr. 2025, 25, 680. [Google Scholar] [CrossRef]
- Morsy, M.D.; Bin-Jaliah, I.; Bashir, S.O.; Shatoor, A.; Haidara, M.A. The impact of concomitant administration of vanadium and insulin on endothelial dysfunction markers (PAI-1 and ET-1) in type 1 diabetic rats. Arch. Physiol. Biochem. 2021, 127, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Ghalichi, F.; Ostadrahimi, A.; Saghafi-Asl, M. Vanadium and biomarkers of inflammation and oxidative stress in diabetes: A systematic review of animal studies. Health Promot. Perspect. 2022, 12, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Hanna, S.J.; Thayer, T.C.; Robinson, E.J.S.; Vinh, N.N.; Williams, N.; Landry, L.G.; Andrews, R.; Siah, Q.Z.; Leete, P.; Wyatt, R.; et al. Single-cell RNAseq identifies clonally expanded antigen-specific T-cells following intradermal injection of gold nanoparticles loaded with diabetes autoantigen in humans. Front. Immunol. 2023, 14, 1276255. [Google Scholar] [CrossRef]
- Taha-Khalde, A.; Haim, A.; Karakis, I.; Shashar, S.; Biederko, R.; Shtein, A.; Hershkovitz, E.; Novack, L. Air pollution and meteorological conditions during gestation and type 1 diabetes in offspring. Environ. Int. 2021, 154, 106546. [Google Scholar] [CrossRef]
- Badpa, M.; Wolf, K.; Schneider, A.; Winkler, C.; Haupt, F.; Peters, A.; Ziegler, A.G. Association of long-term environmental exposures in pregnancy and early life with islet autoimmunity development in children in Bavaria, Germany. Environ. Res. 2022, 212 Pt D, 113503, Erratum in Environ. Res. 2023, 231 Pt 2, 116179. [Google Scholar] [CrossRef] [PubMed]
- Lanzinger, S.; Altug, H.; Schikowski, T.; Khodaverdi, S.; Rosenbauer, J.; Rathmann, W.; Praedicow, K.; Schönau, E.; Holl, R.W. Longitudinal relationship of particulate matter and metabolic control and severe hypoglycaemia in children and adolescents with type 1 diabetes. Environ. Res. 2022, 203, 111859. [Google Scholar] [CrossRef]
- Chen, S.; Li, M.; Zhang, R.; Ye, L.; Jiang, Y.; Jiang, X.; Peng, H.; Wang, Z.; Guo, Z.; Chen, L.; et al. Type 1 diabetes and diet-induced obesity predispose C57BL/6J mice to PM(2.5)-induced lung injury: A comparative study. Part. Fibre Toxicol. 2023, 20, 10. [Google Scholar] [CrossRef]
- Liu, X.; Dos Santos, T.; Spigelman, A.F.; Duckett, S.; Smith, N.; Suzuki, K.; MacDonald, P.E. TMEM55A-mediated PI5P signalling regulates alpha cell actin depolymerisation and glucagon secretion. Diabetologia 2025, 68, 1509–1523. [Google Scholar] [CrossRef]
- Di Ciaula, A. Type I diabetes in paediatric age in Apulia (Italy): Incidence and associations with outdoor air pollutants. Diabetes Res. Clin. Pract. 2016, 111, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Morita, K.; Wang, D.; Baba, R.; Morimoto, H.; Song, Y.; Kanazawa, T.; Yoshida, Y. Particulate Matter, Asian Sand Dust Delays Cyclophosphamide-induced Type 1 Diabetes in NOD Mice. Immunol. Investig. 2020, 49, 698–710. [Google Scholar] [CrossRef]
- Eizirik, D.L.; Szymczak, F.; Mallone, R. Why does the immune system destroy pancreatic β-cells but not α-cells in type 1 diabetes? Nat. Rev. Endocrinol. 2023, 19, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, M.C.; Lambooij, J.M.; Pu, X.; Fagundes, R.R.; Enciso-Martinez, A.; Kats, K.; Giepmans, B.N.G.; Guigas, B.; Zaldumbide, A. Extracellular vesicles derived from stressed beta cells mediate monocyte activation and contribute to islet inflammation. Front. Immunol. 2024, 15, 1393248. [Google Scholar] [CrossRef] [PubMed]
- Dahl-Jørgensen, K. Virus as the cause of type 1 diabetes. Trends Mol. Med. 2024, 30, 1020–1027. [Google Scholar] [CrossRef]
- Dunne, J.L.; Richardson, S.J.; Atkinson, M.A.; Craig, M.E.; Dahl-Jørgensen, K.; Flodström-Tullberg, M.; Hyöty, H.; Insel, R.A.; Lernmark, Å.; Lloyd, R.E.; et al. Rationale for enteroviral vaccination and antiviral therapies in human type 1 diabetes. Diabetologia 2019, 62, 744–753. [Google Scholar] [CrossRef]
- Diana, J.; Brezar, V.; Beaudoin, L.; Dalod, M.; Mellor, A.; Tafuri, A.; von Herrath, M.; Boitard, C.; Mallone, R.; Lehuen, A. Viral infection prevents diabetes by inducing regulatory T cells through NKT cell-plasmacytoid dendritic cell interplay. J. Exp. Med. 2011, 208, 729–745. [Google Scholar] [CrossRef]
- Akimoto, H.; Fukuda-Kawaguchi, E.; Duramad, O.; Ishii, Y.; Tanabe, K. A Novel Liposome Formulation Carrying Both an Insulin Peptide and a Ligand for Invariant Natural Killer T Cells Induces Accumulation of Regulatory T Cells to Islets in Nonobese Diabetic Mice. J. Diabetes Res. 2019, 2019, 9430473. [Google Scholar] [CrossRef]
- Lombardi, A.; Tsomos, E.; Hammerstad, S.S.; Tomer, Y. Interferon alpha: The key trigger of type 1 diabetes. J. Autoimmun. 2018, 94, 7–15. [Google Scholar] [CrossRef]
- Rostron, C.R.; Evans-Molina, C. Molecular and Inflammatory Etiologies of β-Cell Dysfunction in Type 1 Diabetes. Physiology 2025, 40, 538–550. [Google Scholar] [CrossRef]
- Mauvais, F.X.; van Endert, P.M. Type 1 Diabetes: A Guide to Autoimmune Mechanisms for Clinicians. Diabetes Obes. Metab. 2025, 27 (Suppl. S6), 40–56. [Google Scholar] [CrossRef]
- Abdulreda, M.H.; Berggren, P.O. Islet inflammation in plain sight. Diabetes Obes. Metab. 2013, 15 (Suppl. S3), 105–116. [Google Scholar] [CrossRef]
- Afonso, G.; Mallone, R. Infectious triggers in type 1 diabetes: Is there a case for epitope mimicry? Diabetes Obes. Metab. 2013, 15 (Suppl. S3), 82–88. [Google Scholar] [CrossRef]
- Op de Beeck, A.; Eizirik, D.L. Viral infections in type 1 diabetes mellitus--why the β cells? Nat. Rev. Endocrinol. 2016, 12, 263–273. [Google Scholar] [CrossRef]
- Morse, Z.J.; Horwitz, M.S. Virus Infection Is an Instigator of Intestinal Dysbiosis Leading to Type 1 Diabetes. Front. Immunol. 2021, 12, 751337. [Google Scholar] [CrossRef]
- Morse, Z.J.; Simister, R.L.; Crowe, S.A.; Horwitz, M.S.; Osborne, L.C. Virus induced dysbiosis promotes type 1 diabetes onset. Front. Immunol. 2023, 14, 1096323. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, J.; Hadj Hassine, I.; Hassine, M.; Al-Malki, M.; Al-Yami, A.; Al-Bachir, A.; Ben M’hadheb, M. Viral Protein VP1 Virus-like Particles (VLP) of CVB4 Induces Protective Immunity against Lethal Challenges with Diabetogenic E2 and Wild Type JBV Strains in Mice Model. Viruses 2023, 15, 878. [Google Scholar] [CrossRef] [PubMed]
- Rasquinha, M.T.; Lasrado, N.; Sur, M.; Mone, K.; Qiu, H.; Riethoven, J.J.; Sobel, R.A.; Reddy, J. A Monovalent Mt10-CVB3 Vaccine Prevents CVB4-Accelerated Type 1 Diabetes in NOD Mice. Vaccines 2022, 11, 76. [Google Scholar] [CrossRef]
- Alhazmi, A.; Nekoua, M.P.; Michaux, H.; Sane, F.; Halouani, A.; Engelmann, I.; Alidjinou, E.K.; Martens, H.; Jaidane, H.; Geenen, V.; et al. Effect of Coxsackievirus B4 Infection on the Thymus: Elucidating Its Role in the Pathogenesis of Type 1 Diabetes. Microorganisms 2021, 9, 1177. [Google Scholar] [CrossRef] [PubMed]
- Di Vincenzo, F.; Del Gaudio, A.; Petito, V.; Lopetuso, L.R.; Scaldaferri, F. Gut microbiota, intestinal permeability, and systemic inflammation: A narrative review. Intern. Emerg. Med. 2024, 19, 275–293. [Google Scholar] [CrossRef]
- Mouries, J.; Brescia, P.; Silvestri, A.; Spadoni, I.; Sorribas, M.; Wiest, R.; Mileti, E.; Galbiati, M.; Invernizzi, P.; Adorini, L.; et al. Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J. Hepatol. 2019, 71, 1216–1228. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, H.; Wang, G.; Zhai, J.; Du, B. COVID-19 as a Trigger for Type 1 Diabetes. J. Clin. Endocrinol. Metab. 2023, 108, 2176–2183. [Google Scholar] [CrossRef]
- Tang, X.; He, B.; Liu, Z.; Zhou, Z.; Li, X. Fulminant type 1 diabetes after COVID-19 vaccination. Diabetes Metab. 2022, 48, 101324. [Google Scholar] [CrossRef]
- Rahmati, M.; Keshvari, M.; Mirnasuri, S.; Yon, D.K.; Lee, S.W.; Il Shin, J.; Smith, L. The global impact of COVID-19 pandemic on the incidence of pediatric new-onset type 1 diabetes and ketoacidosis: A systematic review and meta-analysis. J. Med. Virol. 2022, 94, 5112–5127. [Google Scholar] [CrossRef]
- DiMeglio, L.A. COVID-19 and Type 1 Diabetes: Addressing Concerns and Maintaining Control. Diabetes Care 2021, 44, 1924–1928. [Google Scholar] [CrossRef] [PubMed]
- Debuysschere, C.; Nekoua, M.P.; Alidjinou, E.K.; Hober, D. The relationship between SARS-CoV-2 infection and type 1 diabetes mellitus. Nat. Rev. Endocrinol. 2024, 20, 588–599. [Google Scholar] [CrossRef]
- Knip, M.; Parviainen, A.; Turtinen, M.; But, A.; Härkönen, T.; Hepojoki, J.; Sironen, T.; Iheozor-Ejiofor, R.; Uğurlu, H.; Saksela, K.; et al. SARS-CoV-2 and type 1 diabetes in children in Finland: An observational study. Lancet Diabetes Endocrinol. 2023, 11, 251–260. [Google Scholar] [CrossRef]
- Kountouri, A.; Korakas, E.; Ikonomidis, I.; Raptis, A.; Tentolouris, N.; Dimitriadis, G.; Lambadiari, V. Type 1 Diabetes Mellitus in the SARS-CoV-2 Pandemic: Oxidative Stress as a Major Pathophysiological Mechanism Linked to Adverse Clinical Outcomes. Antioxidants 2021, 10, 752. [Google Scholar] [CrossRef] [PubMed]
- Rahmati, M.; Yon, D.K.; Lee, S.W.; Udeh, R.; Mc, E.M.; Kim, M.S.; Gyasi, R.M.; Oh, H.; López Sánchez, G.F.; Jacob, L.; et al. New-onset type 1 diabetes in children and adolescents as postacute sequelae of SARS-CoV-2 infection: A systematic review and meta-analysis of cohort studies. J. Med. Virol. 2023, 95, e28833. [Google Scholar] [CrossRef] [PubMed]
- Levet, S.; Charvet, B.; Bertin, A.; Deschaumes, A.; Perron, H.; Hober, D. Human Endogenous Retroviruses and Type 1 Diabetes. Curr. Diab Rep. 2019, 19, 141. [Google Scholar] [CrossRef]
- Del Chierico, F.; Rapini, N.; Deodati, A.; Matteoli, M.C.; Cianfarani, S.; Putignani, L. Pathophysiology of Type 1 Diabetes and Gut Microbiota Role. Int. J. Mol. Sci. 2022, 23, 14650. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, R.; Han, B.; Sun, C.; Chen, R.; Wei, H.; Chen, L.; Du, H.; Li, G.; Yang, Y.; et al. Functional and metabolic alterations of gut microbiota in children with new-onset type 1 diabetes. Nat. Commun. 2022, 13, 6356. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, C.S.; Hesse, D.; Fernandes, G.R.; Hansen, T.H.; Kern, T.; Linneberg, A.; Van Espen, L.; Jørgensen, T.; Nielsen, T.; Alibegovic, A.C.; et al. Characterization of the gut bacterial and viral microbiota in latent autoimmune diabetes in adults. Sci. Rep. 2024, 14, 8315. [Google Scholar] [CrossRef]
- Wook Kim, K.; Allen, D.W.; Briese, T.; Couper, J.J.; Barry, S.C.; Colman, P.G.; Cotterill, A.M.; Davis, E.A.; Giles, L.C.; Harrison, L.C.; et al. Distinct Gut Virome Profile of Pregnant Women With Type 1 Diabetes in the ENDIA Study. Open Forum Infect. Dis. 2019, 6, ofz025. [Google Scholar] [CrossRef] [PubMed]
- Rosell-Mases, E.; Santiago, A.; Corral-Pujol, M.; Yáñez, F.; Varela, E.; Egia-Mendikute, L.; Arpa, B.; Cosovanu, C.; Panosa, A.; Serrano-Gómez, G.; et al. Mutual modulation of gut microbiota and the immune system in type 1 diabetes models. Nat. Commun. 2023, 14, 7770. [Google Scholar] [CrossRef] [PubMed]
- Kinashi, Y.; Hase, K. Partners in Leaky Gut Syndrome: Intestinal Dysbiosis and Autoimmunity. Front. Immunol. 2021, 12, 673708. [Google Scholar] [CrossRef]
- Luo, S.; Yue, T.; Liu, Z.; Yang, D.; Xu, M.; Ding, Y.; Jiang, W.; Xu, W.; Yan, J.; Weng, J.; et al. Gut microbiome and metabolic activity in type 1 diabetes: An analysis based on the presence of GADA. Front. Endocrinol. 2022, 13, 938358. [Google Scholar] [CrossRef]
- Belvoncikova, P.; Maronek, M.; Gardlik, R. Gut Dysbiosis and Fecal Microbiota Transplantation in Autoimmune Diseases. Int. J. Mol. Sci. 2022, 23, 10729. [Google Scholar] [CrossRef]
- Fuhri Snethlage, C.M.; Nieuwdorp, M.; van Raalte, D.H.; Rampanelli, E.; Verchere, B.C.; Hanssen, N.M.J. Auto-immunity and the gut microbiome in type 1 diabetes: Lessons from rodent and human studies. Best. Pract. Res. Clin. Endocrinol. Metab. 2021, 35, 101544. [Google Scholar] [CrossRef]
- Mishra, S.; Jain, S.; Agadzi, B.; Yadav, H. A Cascade of Microbiota-Leaky Gut-Inflammation- Is it a Key Player in Metabolic Disorders? Curr. Obes. Rep. 2025, 14, 32. [Google Scholar] [CrossRef]
- Li, X.; Atkinson, M.A. The role for gut permeability in the pathogenesis of type 1 diabetes—A solid or leaky concept? Pediatr. Diabetes 2015, 16, 485–492. [Google Scholar] [CrossRef]
- de Groot, P.; Nikolic, T.; Pellegrini, S.; Sordi, V.; Imangaliyev, S.; Rampanelli, E.; Hanssen, N.; Attaye, I.; Bakker, G.; Duinkerken, G.; et al. Faecal microbiota transplantation halts progression of human new-onset type 1 diabetes in a randomised controlled trial. Gut 2021, 70, 92–105. [Google Scholar] [CrossRef]
- Paun, A.; Yau, C.; Danska, J.S. Immune recognition and response to the intestinal microbiome in type 1 diabetes. J. Autoimmun. 2016, 71, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, S.M.; Henschel, A.M.; Hessner, M.J. Innate inflammation in type 1 diabetes. Transl. Res. 2016, 167, 214–227. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, L.M.; Newby, B.N.; Perry, D.J.; Posgai, A.L.; Haller, M.J.; Brusko, T.M. Immune Mechanisms and Pathways Targeted in Type 1 Diabetes. Curr. Diab Rep. 2018, 18, 90. [Google Scholar] [CrossRef]
- Ferretti, C.; La Cava, A. Adaptive immune regulation in autoimmune diabetes. Autoimmun. Rev. 2016, 15, 236–241. [Google Scholar] [CrossRef]
- Girdhar, K.; Huang, Q.; Chow, I.T.; Vatanen, T.; Brady, C.; Raisingani, A.; Autissier, P.; Atkinson, M.A.; Kwok, W.W.; Kahn, C.R.; et al. A gut microbial peptide and molecular mimicry in the pathogenesis of type 1 diabetes. Proc. Natl. Acad. Sci. USA 2022, 119, e2120028119, Erratum in Proc. Natl. Acad. Sci. USA 2023, 120, e2309963120. [Google Scholar] [CrossRef]
- Mokhtari, P.; Metos, J.; Anandh Babu, P.V. Impact of type 1 diabetes on the composition and functional potential of gut microbiome in children and adolescents: Possible mechanisms, current knowledge, and challenges. Gut Microbes 2021, 13, 1–18. [Google Scholar] [CrossRef]
- Abuqwider, J.; Corrado, A.; Scidà, G.; Lupoli, R.; Costabile, G.; Mauriello, G.; Bozzetto, L. Gut microbiome and blood glucose control in type 1 diabetes: A systematic review. Front. Endocrinol. 2023, 14, 1265696. [Google Scholar] [CrossRef]
- Wang, Z.; Xie, Z.; Lu, Q.; Chang, C.; Zhou, Z. Beyond Genetics: What Causes Type 1 Diabetes. Clin. Rev. Allergy Immunol. 2017, 52, 273–286. [Google Scholar] [CrossRef]
- Wagner, L.E.; Melnyk, O.; Turner, A.; Duffett, B.E.; Muralidharan, C.; Martinez-Irizarry, M.M.; Arvin, M.C.; Orr, K.S.; Manduchi, E.; Kaestner, K.H.; et al. IFN-α Induces Heterogenous ROS Production in Human β-Cells. bioRxiv, 2025; preprint. [Google Scholar] [CrossRef] [PubMed]
- Coughlan, M.T.; Yap, F.Y.; Tong, D.C.; Andrikopoulos, S.; Gasser, A.; Thallas-Bonke, V.; Webster, D.E.; Miyazaki, J.; Kay, T.W.; Slattery, R.M.; et al. Advanced glycation end products are direct modulators of β-cell function. Diabetes 2011, 60, 2523–2532. [Google Scholar] [CrossRef]
- Lorenzo, O.; Ramírez, E.; Picatoste, B.; Egido, J.; Tuñón, J. Alteration of energy substrates and ROS production in diabetic cardiomyopathy. Mediat. Inflamm. 2013, 2013, 461967. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Sussel, L.; Davidson, H.W. Inherent Beta Cell Dysfunction Contributes to Autoimmune Susceptibility. Biomolecules 2021, 11, 512. [Google Scholar] [CrossRef] [PubMed]
- Balzano-Nogueira, L.; Ramirez, R.; Zamkovaya, T.; Dailey, J.; Ardissone, A.N.; Chamala, S.; Serrano-Quílez, J.; Rubio, T.; Haller, M.J.; Concannon, P.; et al. Integrative analyses of TEDDY Omics data reveal lipid metabolism abnormalities, increased intracellular ROS and heightened inflammation prior to autoimmunity for type 1 diabetes. Genome Biol. 2021, 22, 39. [Google Scholar] [CrossRef]
- Padgett, L.E.; Burg, A.R.; Lei, W.; Tse, H.M. Loss of NADPH oxidase-derived superoxide skews macrophage phenotypes to delay type 1 diabetes. Diabetes 2015, 64, 937–946. [Google Scholar] [CrossRef]
- Feduska, J.M.; Tse, H.M. The proinflammatory effects of macrophage-derived NADPH oxidase function in autoimmune diabetes. Free Radic. Biol. Med. 2018, 125, 81–89. [Google Scholar] [CrossRef]
- Chávez, M.D.; Tse, H.M. Targeting Mitochondrial-Derived Reactive Oxygen Species in T Cell-Mediated Autoimmune Diseases. Front. Immunol. 2021, 12, 703972. [Google Scholar] [CrossRef]
- Padgett, L.E.; Anderson, B.; Liu, C.; Ganini, D.; Mason, R.P.; Piganelli, J.D.; Mathews, C.E.; Tse, H.M. Loss of NOX-Derived Superoxide Exacerbates Diabetogenic CD4 T-Cell Effector Responses in Type 1 Diabetes. Diabetes 2015, 64, 4171–4183. [Google Scholar] [CrossRef]
- Ribatti, D. Gut microbiota, intestinal permeability, and systemic inflammation: A narrative review. Comment. Intern. Emerg. Med. 2024, 19, 1515–1516. [Google Scholar] [CrossRef]
- Usuda, H.; Okamoto, T.; Wada, K. Leaky Gut: Effect of Dietary Fiber and Fats on Microbiome and Intestinal Barrier. Int. J. Mol. Sci. 2021, 22, 7613. [Google Scholar] [CrossRef]
- Bertrand, L.; Chervonsky, A.V.; Lehuen, A. Innate Immunity in Type 1 Diabetes. Cold Spring Harb. Perspect. Med. 2025, 15, a041595. [Google Scholar] [CrossRef]
- Clark, M.; Kroger, C.J.; Tisch, R.M. Type 1 Diabetes: A Chronic Anti-Self-Inflammatory Response. Front. Immunol. 2017, 8, 1898. [Google Scholar] [CrossRef]
- Zipris, D. Visceral Adipose Tissue: A New Target Organ in Virus-Induced Type 1 Diabetes. Front. Immunol. 2021, 12, 702506. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Yip, L.; Wang, F.; Marty, S.E.; Fathman, C.G. Autoimmune disease: Genetic susceptibility, environmental triggers, and immune dysregulation. Where can we develop therapies? Front. Immunol. 2025, 16, 1626082. [Google Scholar] [CrossRef] [PubMed]
- Christen, U.; Kimmel, R. Chemokines as Drivers of the Autoimmune Destruction in Type 1 Diabetes: Opportunity for Therapeutic Intervention in Consideration of an Optimal Treatment Schedule. Front. Endocrinol. 2020, 11, 591083. [Google Scholar] [CrossRef] [PubMed]
- Frazzei, G.; van Vollenhoven, R.F.; de Jong, B.A.; Siegelaar, S.E.; van Schaardenburg, D. Preclinical Autoimmune Disease: A Comparison of Rheumatoid Arthritis, Systemic Lupus Erythematosus, Multiple Sclerosis and Type 1 Diabetes. Front. Immunol. 2022, 13, 899372. [Google Scholar] [CrossRef]
- Qu, H.Q.; Hakonarson, H. Sex as a modifier of genetic risk for type 1 diabetes. Diabetes Obes. Metab. 2025, 27, 6857–6868. [Google Scholar] [CrossRef]
- Knip, M.; Simell, O. Environmental triggers of type 1 diabetes. Cold Spring Harb. Perspect. Med. 2012, 2, a007690. [Google Scholar] [CrossRef]
- Stefan, M.; Zhang, W.; Concepcion, E.; Yi, Z.; Tomer, Y. DNA methylation profiles in type 1 diabetes twins point to strong epigenetic effects on etiology. J. Autoimmun. 2014, 50, 33–37. [Google Scholar] [CrossRef]
- Wu, H.; Chen, Y.; Zhu, H.; Zhao, M.; Lu, Q. The Pathogenic Role of Dysregulated Epigenetic Modifications in Autoimmune Diseases. Front. Immunol. 2019, 10, 2305. [Google Scholar] [CrossRef]
- Agardh, E.; Lundstig, A.; Perfilyev, A.; Volkov, P.; Freiburghaus, T.; Lindholm, E.; Rönn, T.; Agardh, C.D.; Ling, C. Genome-wide analysis of DNA methylation in subjects with type 1 diabetes identifies epigenetic modifications associated with proliferative diabetic retinopathy. BMC Med. 2015, 13, 182. [Google Scholar] [CrossRef]
- Paul, D.S.; Teschendorff, A.E.; Dang, M.A.; Lowe, R.; Hawa, M.I.; Ecker, S.; Beyan, H.; Cunningham, S.; Fouts, A.R.; Ramelius, A.; et al. Increased DNA methylation variability in type 1 diabetes across three immune effector cell types. Nat. Commun. 2016, 7, 13555. [Google Scholar] [CrossRef]
- Ye, J.; Richardson, T.G.; McArdle, W.L.; Relton, C.L.; Gillespie, K.M.; Suderman, M.; Hemani, G. Identification of loci where DNA methylation potentially mediates genetic risk of type 1 diabetes. J. Autoimmun. 2018, 93, 66–75. [Google Scholar] [CrossRef]
- Stefan-Lifshitz, M.; Karakose, E.; Cui, L.; Ettela, A.; Yi, Z.; Zhang, W.; Tomer, Y. Epigenetic modulation of β cells by interferon-α via PNPT1/mir-26a/TET2 triggers autoimmune diabetes. JCI Insight 2019, 4, e126663. [Google Scholar] [CrossRef]
- Vanderlinden, L.A.; Wong, E.; Johnson, R.K.; Carry, P.; Dong, F.; Kechris, K.; Rewers, M.; Norris, J.M. DNA Methylation Smoking Scores and Risk of Islet Autoimmunity and Type 1 Diabetes. Diabetes Care 2025, 48, 1628–1636. [Google Scholar] [CrossRef]
- Tobias, D.K.; Merino, J.; Ahmad, A.; Aiken, C.; Benham, J.L.; Bodhini, D.; Clark, A.L.; Colclough, K.; Corcoy, R.; Cromer, S.J.; et al. Second international consensus report on gaps and opportunities for the clinical translation of precision diabetes medicine. Nat. Med. 2023, 29, 2438–2457. [Google Scholar] [CrossRef]
- Chen, Z.; Satake, E.; Pezzolesi, M.G.; Md Dom, Z.I.; Stucki, D.; Kobayashi, H.; Syreeni, A.; Johnson, A.T.; Wu, X.; Dahlström, E.H.; et al. Integrated analysis of blood DNA methylation, genetic variants, circulating proteins, microRNAs, and kidney failure in type 1 diabetes. Sci. Transl. Med. 2024, 16, eadj3385. [Google Scholar] [CrossRef] [PubMed]
- Hixon, J.C.; Rivas Zarete, J.I.; White, J.; Hilaire, M.; Muhammad, A.; Yusuf, A.P.; Adu-Addai, B.; Yates, C.C.; Mahavadi, S. Epigenetic Modulation of GPER Expression in Gastric and Colonic Smooth Muscle of Male and Female Non-Obese Diabetic (NOD) Mice: Insights into H3K4me3 and H3K27ac Modifications. Int. J. Mol. Sci. 2024, 25, 5260. [Google Scholar] [CrossRef]
- Yang, M.L.; Sodré, F.M.C.; Mamula, M.J.; Overbergh, L. Citrullination and PAD Enzyme Biology in Type 1 Diabetes—Regulators of Inflammation, Autoimmunity, and Pathology. Front. Immunol. 2021, 12, 678953. [Google Scholar] [CrossRef] [PubMed]
- Roshandel, D.; Chen, Z.; Canty, A.J.; Bull, S.B.; Natarajan, R.; Paterson, A.D. DNA methylation age calculators reveal association with diabetic neuropathy in type 1 diabetes. Clin. Epigenet. 2020, 12, 52. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, L.; Zeng, H.; Gao, L.; Guo, S.; Chen, C.; Liu, X.; Zhang, M.; Ma, L.; Li, Y.; et al. Circ-0000953 deficiency exacerbates podocyte injury and autophagy disorder by targeting Mir665-3p-Atg4b in diabetic nephropathy. Autophagy 2024, 20, 1072–1097. [Google Scholar] [CrossRef]
- Kato, M.; Abdollahi, M.; Omori, K.; Malek, V.; Lanting, L.; Kandeel, F.; Rawson, J.; Tsark, W.; Zhang, L.; Wang, M.; et al. Lowering an ER stress-regulated long noncoding RNA protects mice from diabetes and isolated pancreatic islets from cell death. Mol. Ther. Nucleic Acids 2024, 35, 102252. [Google Scholar] [CrossRef]
- Mittal, R.; Camick, N.; Lemos, J.R.N.; Hirani, K. Gene-environment interaction in the pathophysiology of type 1 diabetes. Front Endocrinol 2024, 15, 1335435. [Google Scholar] [CrossRef]
- Pang, H.; Lin, J.; Luo, S.; Huang, G.; Li, X.; Xie, Z.; Zhou, Z. The missing heritability in type 1 diabetes. Diabetes Obes. Metab. 2022, 24, 1901–1911. [Google Scholar] [CrossRef] [PubMed]
- Yau, C.; Danska, J.S. Cracking the type 1 diabetes code: Genes, microbes, immunity, and the early life environment. Immunol. Rev. 2024, 325, 23–45. [Google Scholar] [CrossRef] [PubMed]
- Chia, J.S.J.; McRae, J.L.; Enjapoori, A.K.; Lefèvre, C.M.; Kukuljan, S.; Dwyer, K.M. Dietary Cows’ Milk Protein A1 Beta-Casein Increases the Incidence of T1D in NOD Mice. Nutrients 2018, 10, 1291. [Google Scholar] [CrossRef]
- Rewers, M.; Hyöty, H.; Lernmark, Å.; Hagopian, W.; She, J.X.; Schatz, D.; Ziegler, A.G.; Toppari, J.; Akolkar, B.; Krischer, J. The Environmental Determinants of Diabetes in the Young (TEDDY) Study: 2018 Update. Curr. Diab Rep. 2018, 18, 136. [Google Scholar] [CrossRef]
- Blanter, M.; Sork, H.; Tuomela, S.; Flodström-Tullberg, M. Genetic and Environmental Interaction in Type 1 Diabetes: A Relationship Between Genetic Risk Alleles and Molecular Traits of Enterovirus Infection? Curr. Diab Rep. 2019, 19, 82. [Google Scholar] [CrossRef]
- Uusitalo, U.; Lee, H.S.; Andrén Aronsson, C.; Vehik, K.; Yang, J.; Hummel, S.; Silvis, K.; Lernmark, Å.; Rewers, M.; Hagopian, W.; et al. Early Infant Diet and Islet Autoimmunity in the TEDDY Study. Diabetes Care 2018, 41, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Rønningen, K.S. Type 1 diabetes: Prospective cohort studies for identification of the environmental trigger. Arch. Immunol. Ther. Exp. 2013, 61, 459–468. [Google Scholar] [CrossRef]
- Ye, J.; Stefan-Lifshitz, M.; Tomer, Y. Genetic and environmental factors regulate the type 1 diabetes gene CTSH via differential DNA methylation. J. Biol. Chem. 2021, 296, 100774. [Google Scholar] [CrossRef]
- Badenhoop, K.; Kahles, H.; Seidl, C.; Kordonouri, O.; Lopez, E.R.; Walter, M.; Rosinger, S.; Ziegler, A.; Böhm, B.O. MHC-environment interactions leading to type 1 diabetes: Feasibility of an analysis of HLA DR-DQ alleles in relation to manifestation periods and dates of birth. Diabetes Obes. Metab. 2009, 11 (Suppl. S1), 88–91. [Google Scholar] [CrossRef]
- Hajat, A.; Hsia, C.; O’Neill, M.S. Socioeconomic Disparities and Air Pollution Exposure: A Global Review. Curr. Environ. Health Rep. 2015, 2, 440–450. [Google Scholar] [CrossRef]
- Howard, S.G. Exposure to environmental chemicals and type 1 diabetes: An update. J. Epidemiol. Community Health 2019, 73, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Di Ciaula, A.; Portincasa, P. Relationships between emissions of toxic airborne molecules and type 1 diabetes incidence in children: An ecologic study. World J. Diabetes 2021, 12, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Beyerlein, A.; Donnachie, E.; Jergens, S.; Ziegler, A.G. Infections in Early Life and Development of Type 1 Diabetes. Jama 2016, 315, 1899–1901. [Google Scholar] [CrossRef] [PubMed]
- Verduci, E.; Mameli, C.; Amatruda, M.; Petitti, A.; Vizzuso, S.; El Assadi, F.; Zuccotti, G.; Alabduljabbar, S.; Terranegra, A. Early Nutrition and Risk of Type 1 Diabetes: The Role of Gut Microbiota. Front. Nutr. 2020, 7, 612377. [Google Scholar] [CrossRef]
- Dedrick, S.; Sundaresh, B.; Huang, Q.; Brady, C.; Yoo, T.; Cronin, C.; Rudnicki, C.; Flood, M.; Momeni, B.; Ludvigsson, J.; et al. The Role of Gut Microbiota and Environmental Factors in Type 1 Diabetes Pathogenesis. Front. Endocrinol. 2020, 11, 78. [Google Scholar] [CrossRef]
- Hou, Y.; Song, A.; Jin, Y.; Xia, Q.; Song, G.; Xing, X. A dose-response meta-analysis between serum concentration of 25-hydroxy vitamin D and risk of type 1 diabetes mellitus. Eur. J. Clin. Nutr. 2021, 75, 1010–1023. [Google Scholar] [CrossRef]
- Luppi, S.; Aldegheri, L.; Azzalini, E.; Pacetti, E.; Barucca Sebastiani, G.; Fabiani, C.; Robino, A.; Comar, M. Unravelling the Role of Gut and Oral Microbiota in the Pediatric Population with Type 1 Diabetes Mellitus. Int. J. Mol. Sci. 2024, 25, 10611. [Google Scholar] [CrossRef] [PubMed]
- Nurminen, N.; Cerrone, D.; Lehtonen, J.; Parajuli, A.; Roslund, M.; Lönnrot, M.; Ilonen, J.; Toppari, J.; Veijola, R.; Knip, M.; et al. Land Cover of Early-Life Environment Modulates the Risk of Type 1 Diabetes. Diabetes Care 2021, 44, 1506–1514. [Google Scholar] [CrossRef] [PubMed]
- Herold, K.C.; Delong, T.; Perdigoto, A.L.; Biru, N.; Brusko, T.M.; Walker, L.S.K. The immunology of type 1 diabetes. Nat. Rev. Immunol. 2024, 24, 435–451. [Google Scholar] [CrossRef] [PubMed]
| Pollutant Class | Representative Agent(s) | Proposed Primary Mechanism(s) | Association with T1D |
|---|---|---|---|
| Persistent Organic Pollutants (POPs) | Bisphenol A (BPA) | Endocrine disruption; Immune dysregulation; Metabolic alteration; AhR pathway activation inducing oxidative stress & inflammation | Increased risk |
| Polychlorinated Biphenyls (PCBs), Organochlorine Pesticides (e.g., p,p′-DDE) | Direct β-cell toxicity (impaired insulin secretion); AhR pathway activation; Induction of oxidative stress & β-cell apoptosis | Increased risk | |
| Heavy Metals | Lead (Pb) | Mitochondrial dysfunction; Disruption of insulin signaling; Promotion of oxidative stress | Increased risk (associated with poorer glycemic control) |
| Cadmium (Cd) | Tissue accumulation; Direct pancreatic β-cell damage | Increased risk | |
| Air Particulate Matter | PM2.5/PM10 | Systemic inflammation; Oxidative stress; Impaired insulin sensitivity; Activation of pro-inflammatory pathways | Increased risk (associated with elevated HbA1c & hypoglycemia) |
| Ozone (O3) | Prenatal immune disruption; Oxidative stress; Impaired fetal immune cell differentiation and β-cell development; Indirect effects via maternal metabolic alterations (e.g., gestational diabetes) | Increased risk (maternal exposure associated with higher T1D risk in offspring) | |
| Trace Elements | Iron (Fe) | Oxidative stress (U-shaped response); Gene–environment interactions | Complex (U-shaped association) |
| Zinc (Zn) | Immune system integrity; Insulin metabolism; Antioxidant defense | Deficiency linked to increased risk | |
| Copper (Cu) | Disruption of metabolic homeostasis; Promotion of oxidative stress (Fenton-like reactions) | Dysregulation observed (elevated in new-onset T1D) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Y.; Wang, W.; Huang, Z.; Zhang, C.; Zhang, J.; Pang, Y.; Li, S. Environmental Mechanisms Influencing the Pathogenesis and Progression of Type 1 Diabetes. Int. J. Mol. Sci. 2025, 26, 11613. https://doi.org/10.3390/ijms262311613
Tang Y, Wang W, Huang Z, Zhang C, Zhang J, Pang Y, Li S. Environmental Mechanisms Influencing the Pathogenesis and Progression of Type 1 Diabetes. International Journal of Molecular Sciences. 2025; 26(23):11613. https://doi.org/10.3390/ijms262311613
Chicago/Turabian StyleTang, Yuntao, Weizhou Wang, Zhengsha Huang, Chenxi Zhang, Jia Zhang, Yafang Pang, and Shangze Li. 2025. "Environmental Mechanisms Influencing the Pathogenesis and Progression of Type 1 Diabetes" International Journal of Molecular Sciences 26, no. 23: 11613. https://doi.org/10.3390/ijms262311613
APA StyleTang, Y., Wang, W., Huang, Z., Zhang, C., Zhang, J., Pang, Y., & Li, S. (2025). Environmental Mechanisms Influencing the Pathogenesis and Progression of Type 1 Diabetes. International Journal of Molecular Sciences, 26(23), 11613. https://doi.org/10.3390/ijms262311613






