Integrated DNA Methylation and Transcriptome Analysis Reveals Epigenetic Mechanisms of Lactation Performance Differences in Cloned Buffalo
Abstract
1. Introduction
2. Result
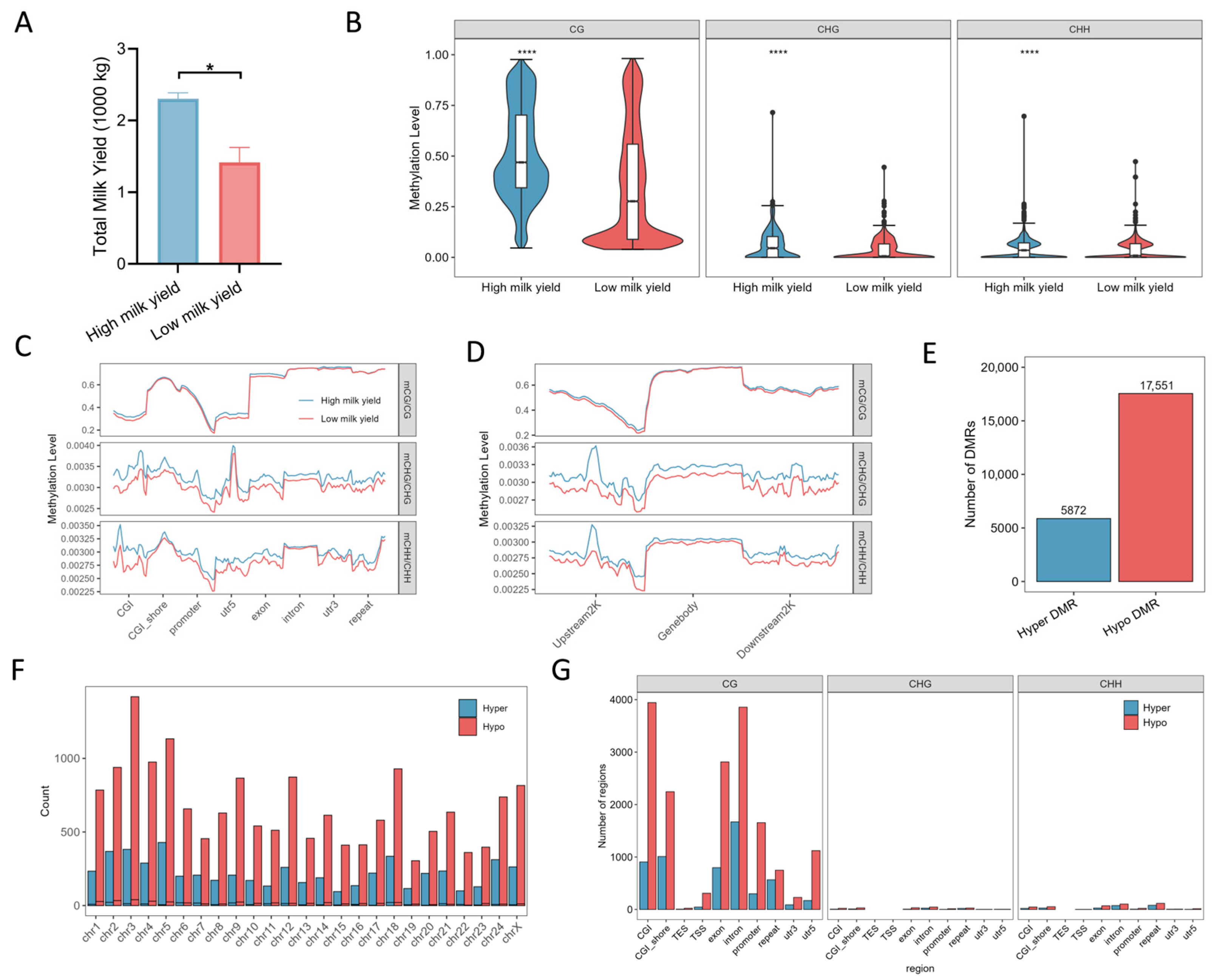
2.1. Genome-Wide DNA Methylation Landscapes in Mammary Tissues of Cloned Buffalo
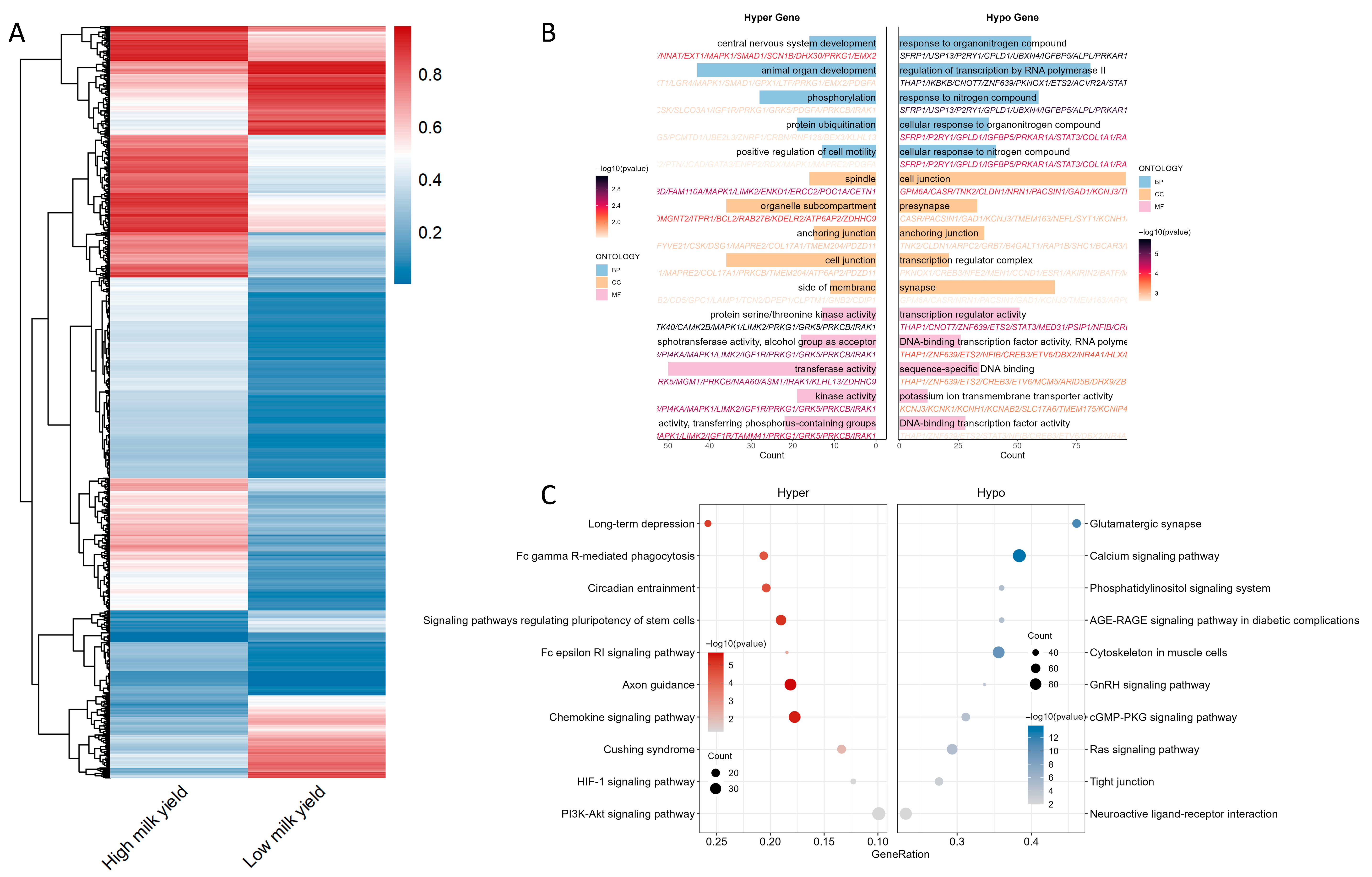
2.2. Functional Annotation and Pathway Enrichment of Differentially Methylated Genes
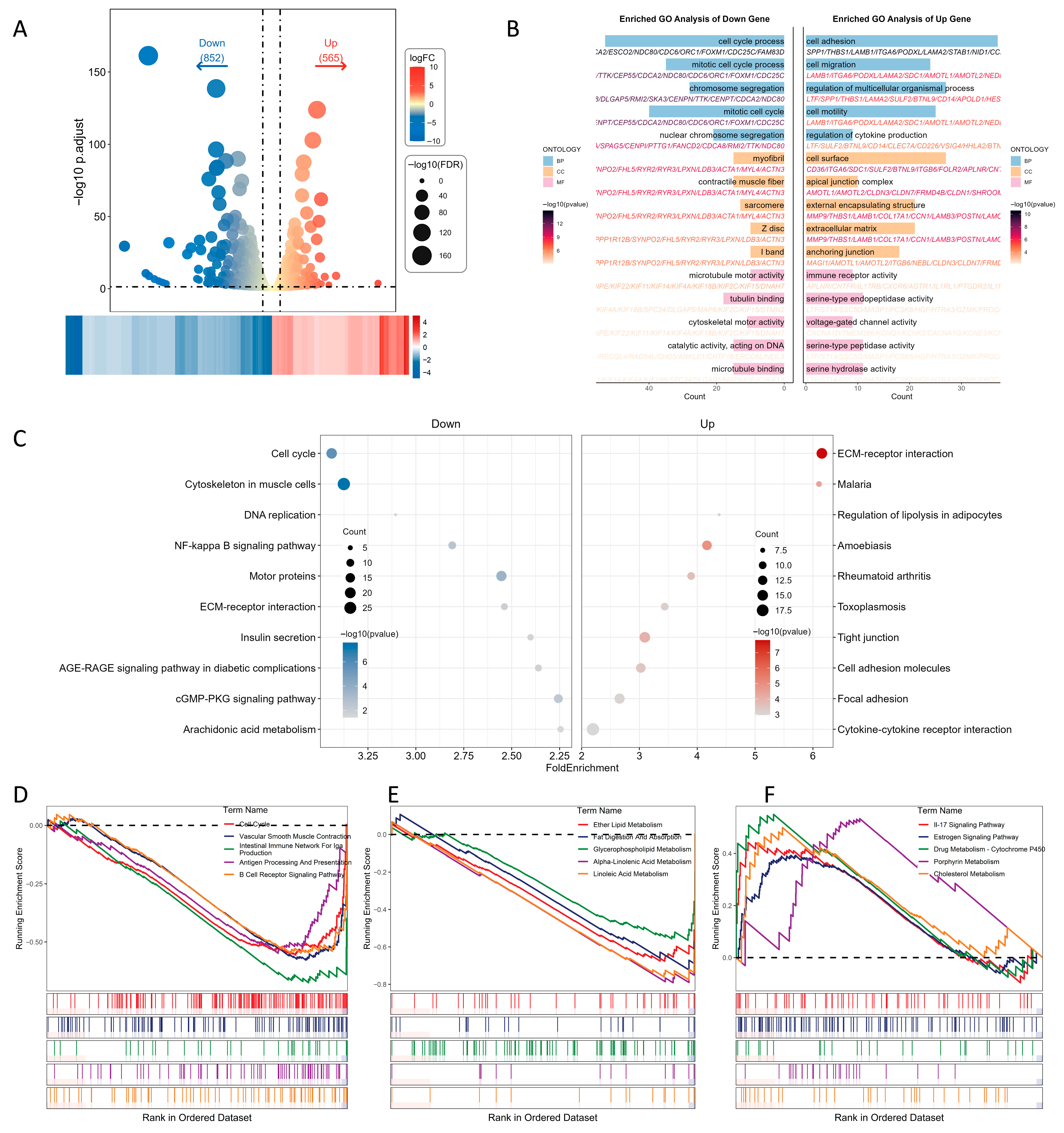
2.3. Transcriptomic Profiling Reveals Differential Gene Expression Associated with Milk Yield
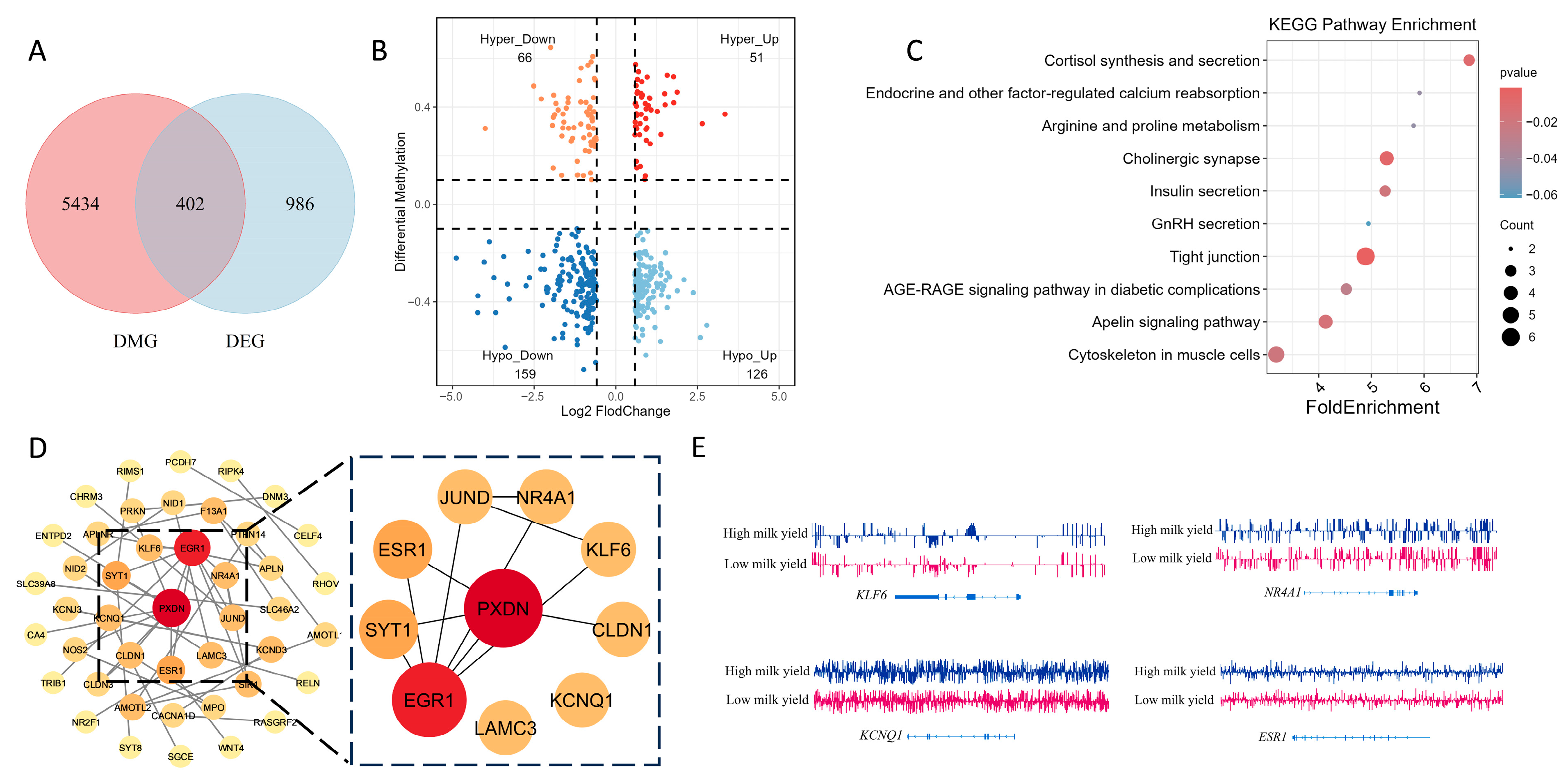
2.4. Integrated Methylome–Transcriptome Analysis Identifies Hypo-Up Genes and Hub Regulators
3. Discussion
4. Materials and Methods
4.1. Tissue Sampling
4.2. DNA and RNA Extraction
4.3. RNA-Sequencing
4.4. Whole-Genome Bisulfite Sequencing
4.5. GO and KEGG Enrichment Analysis of DEGs and DMGs
4.6. Protein–Protein Interaction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Y.; Colli, L.; Barker, J.S.F. Asian water buffalo: Domestication, history and genetics. Anim. Genet. 2020, 51, 177–191. [Google Scholar] [CrossRef]
- Zicarelli, L. Buffalo milk: Its properties, dairy yield and mozzarella production. Vet. Res. Commun. 2004, 28 (Suppl. S1), 127–135. [Google Scholar] [CrossRef]
- Du, C.; Deng, T.; Zhou, Y.; Ye, T.; Zhou, Z.; Zhang, S.; Shao, B.; Wei, P.; Sun, H.; Khan, F.A.; et al. Systematic analyses for candidate genes of milk production traits in water buffalo (Bubalus bubalis). Anim. Genet. 2019, 50, 207–216. [Google Scholar] [CrossRef]
- Xue, Q.; Huang, Y.; Cheng, C.; Wang, Y.; Liao, F.; Duan, Q.; Wang, X.; Miao, C. Progress in epigenetic regulation of milk synthesis, with particular emphasis on mRNA regulation and DNA methylation. Cell Cycle 2023, 22, 1675–1693. [Google Scholar] [CrossRef] [PubMed]
- Ju, Z.; Jiang, Q.; Wang, J.; Wang, X.; Yang, C.; Sun, Y.; Zhang, Y.; Wang, C.; Gao, Y.; Wei, X.; et al. Genome-wide methylation and transcriptome of blood neutrophils reveal the roles of DNA methylation in affecting transcription of protein-coding genes and miRNAs in E. coli-infected mastitis cows. BMC Genom. 2020, 21, 102. [Google Scholar] [CrossRef]
- Deng, T.; Liang, A.; Liang, S.; Ma, X.; Lu, X.; Duan, A.; Pang, C.; Hua, G.; Liu, S.; Campanile, G.; et al. Integrative Analysis of Transcriptome and GWAS Data to Identify the Hub Genes Associated with Milk Yield Trait in Buffalo. Front. Genet. 2019, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Zhu La, A.T.; Gao, S.; Gao, W.; Ma, L.; Bu, D.; Zhang, W. Hepatic Transcriptome Reveals Potential Key Genes Contributing to Differential Milk Production. Genes 2024, 15, 1229. [Google Scholar] [CrossRef]
- Hao, M.; Jiang, J.; Zhang, Y.; Wang, S.; Fu, G.; Zou, F.; Xie, Y.; Zhao, S.; Li, W. Transcriptional profiling of buffalo mammary gland with different milk fat contents. Gene 2021, 802, 145864. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Zheng, Z.; Liu, B.; Ji, X.; Bai, Y.; Zhang, W. Whole blood transcriptional profiling comparison between different milk yield of Chinese Holstein cows using RNA-seq data. BMC Genom. 2016, 17 (Suppl. S7), 512. [Google Scholar] [CrossRef]
- Cui, X.; Hou, Y.; Yang, S.; Xie, Y.; Zhang, S.; Zhang, Y.; Zhang, Q.; Lu, X.; Liu, G.E.; Sun, D. Transcriptional profiling of mammary gland in Holstein cows with extremely different milk protein and fat percentage using RNA sequencing. BMC Genom. 2014, 15, 226. [Google Scholar] [CrossRef]
- Tian, P.; Luo, Y.; Li, X.; Tian, J.; Tao, S.; Hua, C.; Geng, Y.; Ni, Y.; Zhao, R. Negative effects of long-term feeding of high-grain diets to lactating goats on milk fat production and composition by regulating gene expression and DNA methylation in the mammary gland. J. Anim. Sci. Biotechnol. 2017, 8, 74. [Google Scholar] [CrossRef]
- Ahmad, S.M.; Bhat, B.; Bhat, S.A.; Yaseen, M.; Mir, S.; Raza, M.; Iquebal, M.A.; Shah, R.A.; Ganai, N.A. SNPs in Mammary Gland Epithelial Cells Unraveling Potential Difference in Milk Production Between Jersey and Kashmiri Cattle Using RNA Sequencing. Front. Genet. 2021, 12, 666015. [Google Scholar] [CrossRef]
- Matoba, S.; Zhang, Y. Somatic Cell Nuclear Transfer Reprogramming: Mechanisms and Applications. Cell Stem Cell 2018, 23, 471–485. [Google Scholar] [CrossRef]
- Cao, P.; Li, H.; Zuo, Y.; Nashun, B. Characterization of DNA Methylation Patterns and Mining of Epigenetic Markers During Genomic Reprogramming in SCNT Embryos. Front. Cell Dev. Biol. 2020, 8, 570107. [Google Scholar] [CrossRef] [PubMed]
- Montazer-Torbati, F.; Boutinaud, M.; Brun, N.; Richard, C.; Neveu, A.; Jaffrezic, F.; Laloe, D.; LeBourhis, D.; Nguyen, M.; Chadi, S.; et al. Differences during the first lactation between cows cloned by somatic cell nuclear transfer and noncloned cows. J. Dairy Sci. 2016, 99, 4778–4794. [Google Scholar] [CrossRef]
- Matoba, S.; Wang, H.; Jiang, L.; Lu, F.; Iwabuchi, K.A.; Wu, X.; Inoue, K.; Yang, L.; Press, W.; Lee, J.T.; et al. Loss of H3K27me3 Imprinting in Somatic Cell Nuclear Transfer Embryos Disrupts Post-Implantation Development. Cell Stem Cell 2018, 23, 343–354.e345. [Google Scholar] [CrossRef] [PubMed]
- Wijenayake, S.; Eisha, S.; Purohit, M.K.; McGowan, P.O. Milk derived extracellular vesicle uptake in human microglia regulates the DNA methylation machinery: Short title: Milk-derived extracellular vesicles and the epigenetic machinery. Sci. Rep. 2024, 14, 28630. [Google Scholar] [CrossRef]
- Dechow, C.D.; Liu, W.S. DNA methylation patterns in peripheral blood mononuclear cells from Holstein cattle with variable milk yield. BMC Genom. 2018, 19, 744. [Google Scholar] [CrossRef]
- Nayan, V.; Singh, K.; Iquebal, M.A.; Jaiswal, S.; Bhardwaj, A.; Singh, C.; Bhatia, T.; Kumar, S.; Singh, R.; Swaroop, M.N.; et al. Genome-Wide DNA Methylation and Its Effect on Gene Expression During Subclinical Mastitis in Water Buffalo. Front. Genet. 2022, 13, 828292. [Google Scholar] [CrossRef]
- Punetha, M.; Kumar, D.; Kumar, S.; Maggo, B.; Dahiya, P.; Kumar, P.; Sharma, R.K.; Pal, Y.; Yadav, P.S. Unravelling High Nuclear Genomic Similarity and Mitochondria Linked Epigenetic Divergence in SCNT Derived Buffalo Clones via Long-Read Nanopore Genome Sequencing. Int. J. Mol. Sci. 2025, 26, 8836. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Bissonnette, N.; Dudemaine, P.L.; Zhao, X.; Ibeagha-Awemu, E.M. Whole Genome DNA Methylation Variations in Mammary Gland Tissues from Holstein Cattle Producing Milk with Various Fat and Protein Contents. Genes 2021, 12, 1727. [Google Scholar] [CrossRef]
- Wang, M.; Liang, Y.; Ibeagha-Awemu, E.M.; Li, M.; Zhang, H.; Chen, Z.; Sun, Y.; Karrow, N.A.; Yang, Z.; Mao, Y. Genome-Wide DNA Methylation Analysis of Mammary Gland Tissues from Chinese Holstein Cows with Staphylococcus aureus Induced Mastitis. Front. Genet. 2020, 11, 550515. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, G.E.; Ma, L.; Fang, L.; Li, C.J.; Baldwin, R.L.T. Transcriptomic profiling of gastrointestinal tracts in dairy cattle during lactation reveals molecular adaptations for milk synthesis. J. Adv. Res. 2025, 71, 67–80. [Google Scholar] [CrossRef]
- Sengupta, T.; Abraham, G.; Xu, Y.; Clurman, B.E.; Minella, A.C. Hypoxia-inducible factor 1 is activated by dysregulated cyclin E during mammary epithelial morphogenesis. Mol. Cell Biol. 2011, 31, 3885–3895. [Google Scholar] [CrossRef]
- Xu, Q.; Li, X.; Ma, L.; Loor, J.J.; Coleman, D.N.; Jia, H.; Liu, G.; Xu, C.; Wang, Y.; Li, X. Adipose tissue proteomic analysis in ketotic or healthy Holstein cows in early lactation1. J. Anim. Sci. 2019, 97, 2837–2849. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Liu, X.; Li, H.; Lin, X.; Yan, Z.; Cao, Q.; Zhao, M.; Xu, Z.; Wang, Z. Menin Modulates Mammary Epithelial Cell Numbers in Bovine Mammary Glands Through Cyclin D1. J. Mammary Gland. Biol. Neoplasia 2017, 22, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Buitenhuis, B.; Janss, L.L.; Poulsen, N.A.; Larsen, L.B.; Larsen, M.K.; Sorensen, P. Genome-wide association and biological pathway analysis for milk-fat composition in Danish Holstein and Danish Jersey cattle. BMC Genom. 2014, 15, 1112. [Google Scholar] [CrossRef]
- Khan, M.Z.; Khan, A.; Xiao, J.; Ma, Y.; Ma, J.; Gao, J.; Cao, Z. Role of the JAK-STAT Pathway in Bovine Mastitis and Milk Production. Animals 2020, 10, 2107. [Google Scholar] [CrossRef]
- Rainard, P.; Cunha, P.; Bougarn, S.; Fromageau, A.; Rossignol, C.; Gilbert, F.B.; Berthon, P. T helper 17-associated cytokines are produced during antigen-specific inflammation in the mammary gland. PLoS ONE 2013, 8, e63471. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, M.; Su, H.; Zhao, F.; Wang, D.; Zhang, Y.; Cao, G.; Zhang, Y. Regulation of Inflammatory Responses of Cow Mammary Epithelial Cells through MAPK Signaling Pathways of IL-17A Cytokines. Animals 2024, 14, 1572. [Google Scholar] [CrossRef] [PubMed]
- Hirosawa, T.; Kawamoto, S.; Shimizu, T. SAPHO syndrome. BMJ Case Rep. 2019, 12, e233221. [Google Scholar] [CrossRef]
- Wei, J.L.; Villwock, J.A. Balance Versus Integration: Work-Life Considerations. Otolaryngol. Clin. N. Am. 2021, 54, 823–837. [Google Scholar] [CrossRef]
- Hu, Z.L.; Reecy, J.M. Animal QTLdb: Beyond a repository. A public platform for QTL comparisons and integration with diverse types of structural genomic information. Mamm. Genome 2007, 18, 1–4. [Google Scholar] [CrossRef]
- Liu, Y.; Han, B.; Zheng, W.; Peng, P.; Yang, C.; Jiang, G.; Ma, Y.; Li, J.; Ni, J.; Sun, D. Identification of genetic associations and functional SNPs of bovine KLF6 gene on milk production traits in Chinese holstein. BMC Genom. Data 2023, 24, 72. [Google Scholar] [CrossRef]
- Iqbal, A.; Yu, H.; Jiang, P.; Zhao, Z. Deciphering the Key Regulatory Roles of KLF6 and Bta-miR-148a on Milk Fat Metabolism in Bovine Mammary Epithelial Cells. Genes 2022, 13, 1828. [Google Scholar] [CrossRef]
- Tarcic, G.; Avraham, R.; Pines, G.; Amit, I.; Shay, T.; Lu, Y.; Zwang, Y.; Katz, M.; Ben-Chetrit, N.; Jacob-Hirsch, J.; et al. EGR1 and the ERK-ERF axis drive mammary cell migration in response to EGF. FASEB J. 2012, 26, 1582–1592. [Google Scholar] [CrossRef]
- Rusidze, M.; Adlanmerini, M.; Chantalat, E.; Raymond-Letron, I.; Cayre, S.; Arnal, J.F.; Deugnier, M.A.; Lenfant, F. Estrogen receptor-alpha signaling in post-natal mammary development and breast cancers. Cell Mol. Life Sci. 2021, 78, 5681–5705. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Han, B.; Liu, L.; Zhao, F.; Liang, W.; Jiang, J.; Yang, Y.; Ma, Z.; Sun, D. Genetic association of DDIT3, RPL23A, SESN2 and NR4A1 genes with milk yield and composition in dairy cattle. Anim. Genet. 2019, 50, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Mi, S.; Xing, Y.; An, S.; Chen, S.; Tang, Y.; Wang, Y.; Yu, Y. Transcriptome analysis identifies the NR4A subfamily involved in the alleviating effect of folic acid on mastitis induced by high concentration of Staphylococcus aureus lipoteichoic acid. BMC Genom. 2024, 25, 1051. [Google Scholar] [CrossRef]
- Tribout, T.; Croiseau, P.; Lefebvre, R.; Barbat, A.; Boussaha, M.; Fritz, S.; Boichard, D.; Hoze, C.; Sanchez, M.P. Confirmed effects of candidate variants for milk production, udder health, and udder morphology in dairy cattle. Genet. Sel. Evol. 2020, 52, 55. [Google Scholar] [CrossRef]
- Berenguier, C.; Chen, X.; Allegrini, B.; Guizouarn, H.; Borgese, F.; Etchebest, C.; Soriani, O.; Rapetti-Mauss, R. Cancer-associated loss-of-function mutations in KCNQ1 enhance Wnt/beta-catenin signalling disrupting epithelial homeostasis. Oncogene 2025, 44, 2715–2729. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, L.; Lun, A.T.L.; Baldoni, P.L.; Smyth, G.K. edgeR v4: Powerful differential analysis of sequencing data with expanded functionality and improved support for small counts and larger datasets. Nucleic Acids Res. 2025, 53, gkaf018. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Wu, H. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics 2016, 32, 1446–1453. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, J.-H.; Zheng, H.-Y.; Yang, C.-Y.; Shang, J.-H. Integrated DNA Methylation and Transcriptome Analysis Reveals Epigenetic Mechanisms of Lactation Performance Differences in Cloned Buffalo. Int. J. Mol. Sci. 2025, 26, 11585. https://doi.org/10.3390/ijms262311585
Hu J-H, Zheng H-Y, Yang C-Y, Shang J-H. Integrated DNA Methylation and Transcriptome Analysis Reveals Epigenetic Mechanisms of Lactation Performance Differences in Cloned Buffalo. International Journal of Molecular Sciences. 2025; 26(23):11585. https://doi.org/10.3390/ijms262311585
Chicago/Turabian StyleHu, Jia-Hao, Hai-Ying Zheng, Chun-Yan Yang, and Jiang-Hua Shang. 2025. "Integrated DNA Methylation and Transcriptome Analysis Reveals Epigenetic Mechanisms of Lactation Performance Differences in Cloned Buffalo" International Journal of Molecular Sciences 26, no. 23: 11585. https://doi.org/10.3390/ijms262311585
APA StyleHu, J.-H., Zheng, H.-Y., Yang, C.-Y., & Shang, J.-H. (2025). Integrated DNA Methylation and Transcriptome Analysis Reveals Epigenetic Mechanisms of Lactation Performance Differences in Cloned Buffalo. International Journal of Molecular Sciences, 26(23), 11585. https://doi.org/10.3390/ijms262311585





