Integrating Bioprinting and Increased Throughput: Next-Generation Models for Cardiac Research
Abstract
1. Introduction
2. Technologies Driving High-Throughput Engineered Cardiac Tissue Generation
2.1. Types of Bioprinting
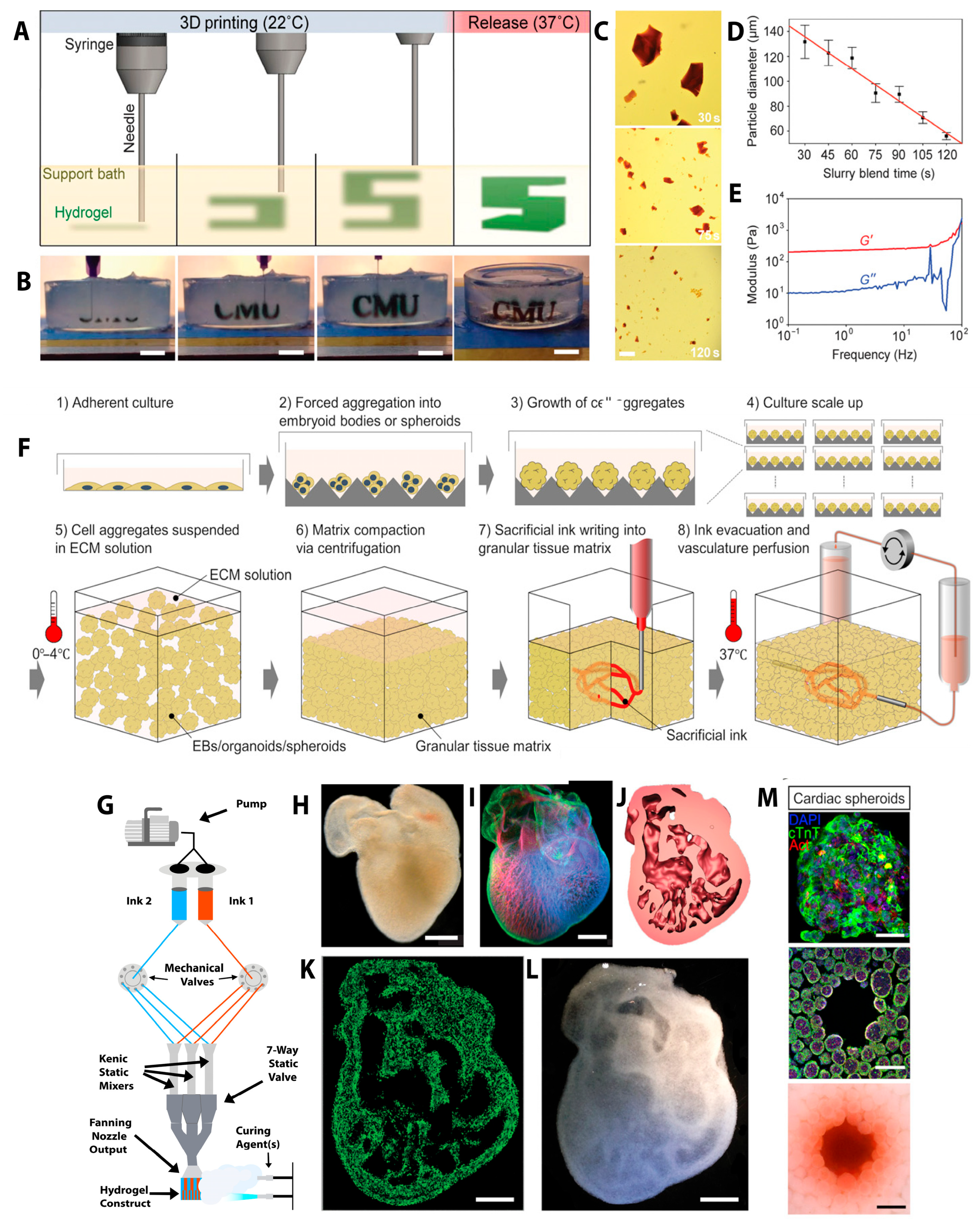
2.2. Three-Dimensional Bioprinting of the Heart
2.3. Three-Dimensional Bioprinting Strategies for Next-Generation Tissues
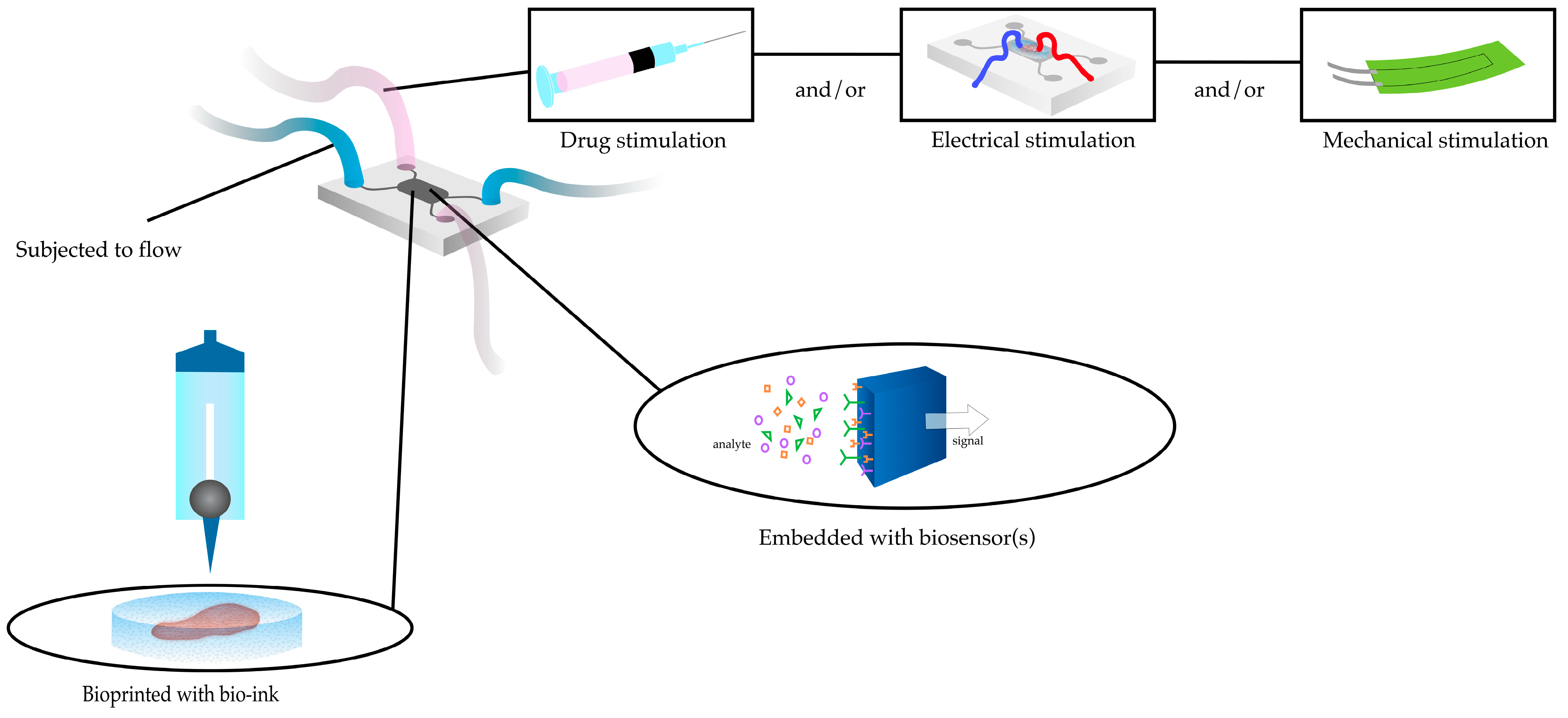
2.4. Heart-on-a-Chip
2.5. Challenges in Integrating 3DBP with HOC
3. Next-Generation Cardiac Model Systems
3.1. Organ Building Blocks
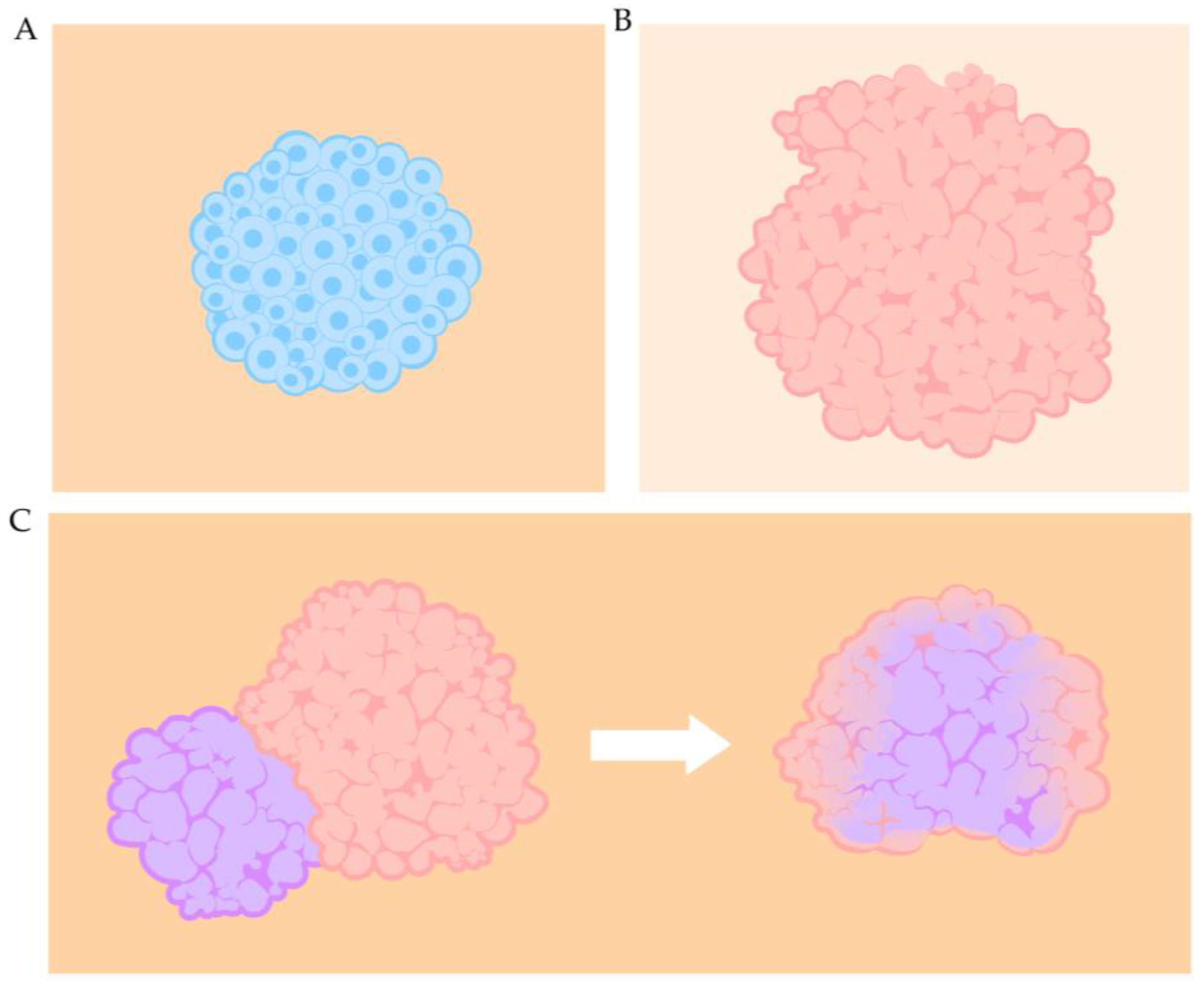
3.2. Spheroids
3.3. Organoids
3.4. Assembloids
3.5. Challenges and Strategies of Organ Building Block Systems
4. High-Throughput Screening of the Bioprinted Cardiac Tissue Models
5. Transcriptomics for High-Throughput Improvement in Analysis of 3D Cardiac Tissue
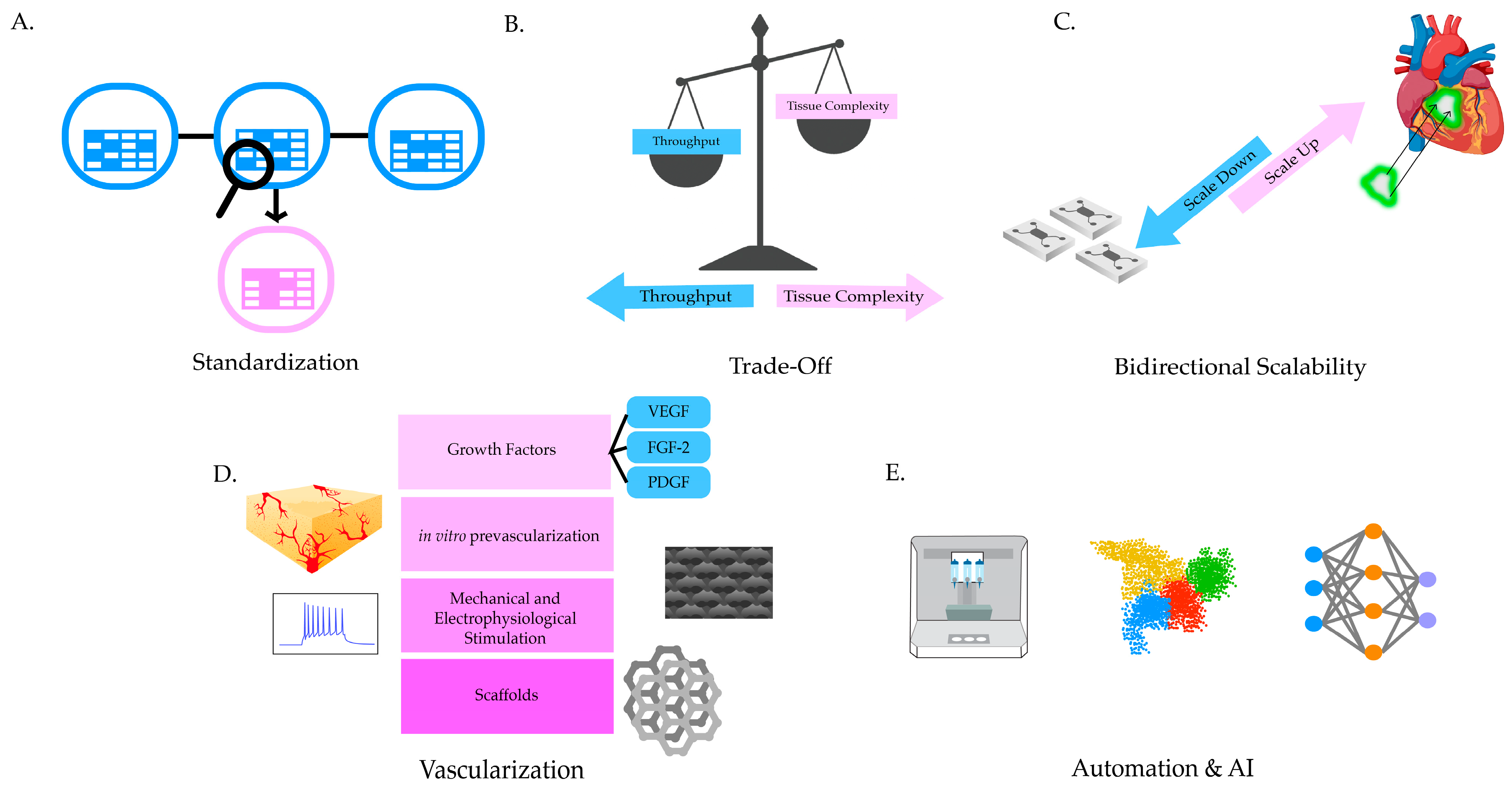
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
Abbreviations
| 3DCC | 3-Dimensional Cell Culture |
| 3DBP | 3-Dimensional Bioprinting |
| BAB400 | BioAssemblyBot®400 |
| HTS | High-Throughput Screening |
| CVD | Cardiovascular Diseases |
| dECM | Decellularized Extracellular Matrix |
| CAD | Computer-aided Design |
| CEVIC | Continuously Extruded Variable Internal Channeling |
| DLP | Digital Light Processing |
| EBB | Extrusion-Based Bioprinting |
| EHT | Engineered Heart Tissue |
| FRESH | Freeform Reversible Embedding of Suspended Hydrogels |
| HoC | Heart-on-a-Chip |
| CM | Cardiomyocytes |
| iPSC | Induced Pluripotent Stem Cell |
| hiPSC | Human Induced Pluripotent Stem Cell |
| CF | Cardiofibroblast |
| EC | Endothelial Cell |
| ECM | Extracellular Matrix |
| LBB | Laser-Assisted Bioprinting |
| OBB | Organ Building Block |
| NCC | Neural Crest Cells |
| OCT | Optical Coherence Tomography |
| PEDOT:PSS | Poly(3,4-ethylenedioxythiophene) polystyrene sulfonate |
| PEG | Poly(ethylene) Glycol |
| rGO | Reduced Graphene Oxide |
| sc-RNA Seq | Single-Cell Ribonucleic Acid Sequencing |
| SLA | Stereolithography |
| Sn-RNA seq | Single-Nuclei Ribonucleic Acid Sequencing |
| ST | Spatial transcriptomics |
| FISH | Fluorescence In Situ Hybridization |
| smFISH | Multiplex Single-Molecule Fluorescence In Situ Hybridization |
| HDST | High-Density Spatial Transcriptomics |
| UV | Ultraviolet |
References
- Di Cesare, M.; Perel, P.; Taylor, S.; Kabudula, C.; Bixby, H.; Gaziano, T.A.; McGhie, D.V.; Mwangi, J.; Pervan, B.; Narula, J.; et al. The Heart of the World. Glob. Heart 2024, 19, 11. [Google Scholar] [CrossRef]
- Chong, B.; Jayabaskaran, J.; Jauhari, S.M.; Chan, S.P.; Goh, R.; Kueh, M.T.W.; Li, H.; Chin, Y.H.; Kong, G.; Anand, V.V.; et al. Global burden of cardiovascular diseases: Projections from 2025 to 2050. Eur. J. Prev. Cardiol. 2024, 32, 1001–1015. [Google Scholar] [CrossRef]
- Cohn, J.N. Structural changes in cardiovascular disease. Am. J. Cardiol. 1995, 76, 34E–37E. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhang, Z.; Sun, Z. The potential and challenges of using stem cells for cardiovascular repair and regeneration. Genes Dis. 2014, 1, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Olvera Lopez, E.; Ballard, B.D.; Jan, A. Cardiovascular Disease. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. Available online: http://www.ncbi.nlm.nih.gov/books/NBK535419/ (accessed on 4 August 2025).
- Hernandez, G.A.; Lemor, A.; Clark, D.; Blumer, V.; Burstein, D.; Byrne, R.; Fowler, R.; Frischhertz, B.; Sandhaus, E.; Schlendorf, K.; et al. Heart transplantation and in-hospital outcomes in adult congenital heart disease patients with Fontan: A decade nationwide analysis from 2004 to 2014. J. Card. Surg. 2020, 35, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Jain, A. Heart Transplantation. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK557571/ (accessed on 17 August 2025).
- Goldfracht, I.; Efraim, Y.; Shinnawi, R.; Kovalev, E.; Huber, I.; Gepstein, A.; Arbel, G.; Shaheen, N.; Tiburcy, M.; Zimmermann, W.H.; et al. Engineered heart tissue models from hiPSC-derived cardiomyocytes and cardiac ECM for disease modeling and drug testing applications. Acta Biomater. 2019, 92, 145–159. [Google Scholar] [CrossRef]
- Zimmermann, W.-H.; Melnychenko, I.; Wasmeier, G.; Didié, M.; Naito, H.; Nixdorff, U.; Hess, A.; Budinsky, L.; Brune, K.; Michaelis, B.; et al. Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nat. Med. 2006, 12, 452–458. [Google Scholar] [CrossRef]
- Polonchuk, L.; Gentile, C. Current state and future of 3D bioprinted models for cardiovascular research and drug development. ADMET DMPK 2021, 9, 231–242. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Ma, S.P.; Yeager, K.; Chen, T.; Song, L.; Sirabella, D.; Morikawa, K.; Teles, D.; Yazawa, M.; Vunjak-Novakovic, G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 2019, 556, 239–243, Correction in Nature 2018, 572, E16–E17 . [Google Scholar] [CrossRef]
- Min, S.; Lee, H.-J.; Jin, Y.; Kim, Y.H.; Sung, J.; Choi, H.-J.; Cho, S.-W. Biphasic Electrical Pulse by a Micropillar Electrode Array Enhances Maturation and Drug Response of Reprogrammed Cardiac Spheroids. Nano Lett. 2020, 20, 6947–6956. [Google Scholar] [CrossRef]
- Lind, J.U.; Busbee, T.A.; Valentine, A.D.; Pasqualini, F.S.; Yuan, H.; Yadid, M.; Park, S.-J.; Kotikian, A.; Nesmith, A.P.; Campbell, P.H.; et al. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat. Mater. 2017, 16, 303–308. [Google Scholar] [CrossRef]
- Lu, K.; Seidel, T.; Cao-Ehlker, X.; Dorn, T.; Batcha, A.M.N.; Schneider, C.M.; Semmler, M.; Volk, T.; Moretti, A.; Dendorfer, A.; et al. Progressive stretch enhances growth and maturation of 3D stem-cell-derived myocardium. Theranostics 2021, 11, 6138–6153. [Google Scholar] [CrossRef]
- Imboden, M.; de Coulon, E.; Poulin, A.; Dellenbach, C.; Rosset, S.; Shea, H.; Rohr, S. High-speed mechano-active multielectrode array for investigating rapid stretch effects on cardiac tissue. Nat. Commun. 2019, 10, 834. [Google Scholar] [CrossRef] [PubMed]
- Lewis-Israeli, Y.R.; Wasserman, A.H.; Gabalski, M.A.; Volmert, B.D.; Ming, Y.; Ball, K.A.; Yang, W.; Zou, J.; Ni, G.; Pajares, N.; et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat. Commun. 2021, 12, 5142. [Google Scholar] [CrossRef] [PubMed]
- Hofbauer, P.; Jahnel, S.M.; Papai, N.; Giesshammer, M.; Deyett, A.; Schmidt, C.; Penc, M.; Tavernini, K.; Grdseloff, N.; Meledeth, C.; et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell 2021, 184, 3299–3317.e22. [Google Scholar] [CrossRef]
- Bremner, S.B.; Gaffney, K.S.; Sniadecki, N.J.; Mack, D.L. A Change of Heart: Human Cardiac Tissue Engineering as a Platform for Drug Development. Curr. Cardiol. Rep. 2022, 24, 473–486. [Google Scholar] [CrossRef]
- Agarwal, T.; Fortunato, G.M.; Hann, S.Y.; Ayan, B.; Vajanthri, K.Y.; Presutti, D.; Cui, H.; Chan, A.H.-P.; Costantini, M.; Onesto, V.; et al. Recent Advances in Bioprinting Technologies for Engineering Cardiac Tissue. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 124, 112057. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.A.; Zhu, Y.; Woo, Y.J. Advances in 3D Bioprinting: Techniques, Applications, and Future Directions for Cardiac Tissue Engineering. Bioengineering 2023, 10, 842. [Google Scholar] [CrossRef]
- Cui, X.; Boland, T.; D’Lima, D.D.; Lotz, M.K. Thermal Inkjet Printing in Tissue Engineering and Regenerative Medicine. Recent Pat. Drug Deliv. Formul. 2012, 6, 149–155. [Google Scholar] [CrossRef]
- Gu, Z.; Fu, J.; Lin, H.; He, Y. Development of 3D bioprinting: From printing methods to biomedical applications. Asian J. Pharm. Sci. 2020, 15, 529–557. [Google Scholar] [CrossRef]
- Takagi, D.; Lin, W.; Matsumoto, T.; Yaginuma, H.; Hemmi, N.; Hatada, S.; Seo, M. High-precision three-dimensional inkjet technology for live cell bioprinting. Int. J. Bioprint. 2019, 5, 208. [Google Scholar] [CrossRef]
- Cui, X.; Dean, D.; Ruggeri, Z.M.; Boland, T. Cell damage evaluation of thermal inkjet printed Chinese hamster ovary cells. Biotechnol. Bioeng. 2010, 106, 963–969. [Google Scholar] [CrossRef]
- Liu, N.; Ye, X.; Yao, B.; Zhao, M.; Wu, P.; Liu, G.; Zhuang, D.; Jiang, H.; Chen, X.; He, Y.; et al. Advances in 3D bioprinting technology for cardiac tissue engineering and regeneration. Bioact. Mater. 2021, 6, 1388–1401. [Google Scholar] [CrossRef] [PubMed]
- de Gans, B.-J.; Duineveld, P.C.; Schubert, U.S. Inkjet Printing of Polymers: State of the Art and Future Developments. Adv. Mater. 2004, 16, 203–213. [Google Scholar] [CrossRef]
- Derakhshanfar, S.; Mbeleck, R.; Xu, K.; Zhang, X.; Zhong, W.; Xing, M. 3D bioprinting for biomedical devices and tissue engineering: A review of recent trends and advances. Bioact. Mater. 2018, 3, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Manji, R.A.; Menkis, A.H.; Ekser, B.; Cooper, D.K.C. Porcine bioprosthetic heart valves: The next generation. Am. Heart J. 2012, 164, 177–185. [Google Scholar] [CrossRef]
- Desai, M.; Seifalian, A.M.; Hamilton, G. Role of prosthetic conduits in coronary artery bypass grafting. Eur. J. Cardio-Thorac. Surg. 2011, 40, 394–398. [Google Scholar] [CrossRef]
- Alonzo, M.; AnilKumar, S.; Roman, B.; Tasnim, N.; Joddar, B. 3D Bioprinting of cardiac tissue and cardiac stem cell therapy. Transl. Res. 2019, 211, 64–83. [Google Scholar] [CrossRef]
- Chingale, M.; Cheng, K.; Huang, K. 3D Bioprinting Technology—One Step Closer Towards Cardiac Tissue Regeneration. Front. Mater. 2022, 8, 804134. [Google Scholar] [CrossRef]
- You, S.; Xiang, Y.; Hwang, H.H.; Berry, D.B.; Kiratitanaporn, W.; Guan, J.; Yao, E.; Tang, M.; Zhong, Z.; Ma, X.; et al. High cell density and high-resolution 3D bioprinting for fabricating vascularized tissues. Sci. Adv. 2023, 9, eade7923. [Google Scholar] [CrossRef]
- Dababneh, A.B.; Ozbolat, I.T. Bioprinting Technology: A Current State-of-the-Art Review. J. Manuf. Sci. Eng. 2014, 136, 061016. [Google Scholar] [CrossRef]
- Prendergast, M.E.; Davidson, M.D.; Burdick, J.A. A Biofabrication Method to Align Cells within Bioprinted Photocrosslinkable and Cell-degradable Hydrogel Constructs via Embedded Fibers. Biofabrication 2021, 13, 044108. [Google Scholar] [CrossRef]
- Ozbolat, I.T.; Hospodiuk, M. Current advances and future perspectives in extrusion-based bioprinting. Biomaterials 2016, 76, 321–343. [Google Scholar] [CrossRef]
- Gillispie, G.; Prim, P.; Copus, J.; Fisher, J.; Mikos, A.G.; Yoo, J.J.; Atala, A.; Lee, S.J. Assessment Methodologies for Extrusion-Based Bioink Printability. Biofabrication 2020, 12, 022003. [Google Scholar] [CrossRef] [PubMed]
- Arslan-Yildiz, A.; Assal, R.E.; Chen, P.; Guven, S.; Inci, F.; Demirci, U. Towards artificial tissue models: Past, present, and future of 3D bioprinting. Biofabrication 2016, 8, 014103. [Google Scholar] [CrossRef] [PubMed]
- Budharaju, H.; Sundaramurthi, D.; Sethuraman, S. Embedded 3D bioprinting—An emerging strategy to fabricate biomimetic & large vascularized tissue constructs. Bioact. Mater. 2024, 32, 356–384. [Google Scholar] [CrossRef] [PubMed]
- Zennifer, A.; Subramanian, A.; Sethuraman, S. Design considerations of bioinks for laser bioprinting technique towards tissue regenerative applications. Bioprinting 2022, 27, e00205. [Google Scholar] [CrossRef]
- Kim, M.H.; Singh, Y.P.; Celik, N.; Yeo, M.; Rizk, E.; Hayes, D.J.; Ozbolat, I.T. High-throughput bioprinting of spheroids for scalable tissue fabrication. Nat. Commun. 2024, 15, 10083. [Google Scholar] [CrossRef]
- Mobaraki, M.; Ghaffari, M.; Yazdanpanah, A.; Luo, Y.; Mills, D.K. Bioinks and bioprinting: A focused review. Bioprinting 2020, 18, e00080. [Google Scholar] [CrossRef]
- Hwang, H.H.; Zhu, W.; Victorine, G.; Lawrence, N.; Chen, S. 3D-Printing of Functional Biomedical Microdevices via Light- and Extrusion-Based Approaches. Small Methods 2018, 2, 1700277. [Google Scholar] [CrossRef]
- Alparslan, C.; Bayraktar, Ş. Advances in Digital Light Processing (DLP) Bioprinting: A Review of Biomaterials and Its Applications, Innovations, Challenges, and Future Perspectives. Polymers 2025, 17, 1287. [Google Scholar] [CrossRef]
- Ke, D.; Niu, C.; Yang, X. Evolution of 3D bioprinting-from the perspectives of bioprinting companies. Bioprinting 2022, 25, e00193. [Google Scholar] [CrossRef]
- Roy, A.; Saxena, V.; Pandey, L.M. 3D printing for cardiovascular tissue engineering: A review. Mater. Technol. 2018, 33, 433–442. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, L.; Li, T.; Liu, S.; Guo, B.; Huang, W.; Wu, Y. 3D bioprinting in cardiac tissue engineering. Theranostics 2021, 11, 7948–7969. [Google Scholar] [CrossRef]
- Pati, F.; Jang, J.; Ha, D.-H.; Won Kim, S.; Rhie, J.-W.; Shim, J.-H.; Kim, D.-H.; Cho, D.-W. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat. Commun. 2014, 5, 3935. [Google Scholar] [CrossRef]
- Ainsworth, M.J.; Chirico, N.; de Ruijter, M.; Hrynevich, A.; Dokter, I.; Sluijter, J.P.G.; Malda, J.; van Mil, A.; Castilho, M. Convergence of melt electrowriting and extrusion-based bioprinting for vascular patterning of a myocardial construct. Biofabrication 2023, 15, 035025. [Google Scholar] [CrossRef] [PubMed]
- Bera, A.K.; Rizvi, M.S.; KN, V.; Pati, F. Engineering anisotropic tissue analogues: Harnessing synergistic potential of extrusion-based bioprinting and extracellular matrix-based bioink. Biofabrication 2024, 17, 015003. [Google Scholar] [CrossRef] [PubMed]
- Duan, B. State-of-the-Art Review of 3D Bioprinting for Cardiovascular Tissue Engineering. Ann. Biomed. Eng. 2017, 45, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Kim, K. Stereolithography 3D Bioprinting. In 3D Bioprinting: Principles and Protocols; Crook, J.M., Ed.; Springer: New York, NY, USA, 2020; pp. 93–108. ISBN 978-1-0716-0520-2. [Google Scholar] [CrossRef]
- O’Connell, C.D.; Zhang, B.; Onofrillo, C.; Duchi, S.; Blanchard, R.; Quigley, A.; Bourke, J.; Gambhir, S.; Kapsa, R.; Di Bella, C.; et al. Tailoring the mechanical properties of gelatin methacryloyl hydrogels through manipulation of the photocrosslinking conditions. Soft Matter 2018, 14, 2142–2151. [Google Scholar] [CrossRef]
- Borovjagin, A.V.; Ogle, B.M.; Berry, J.L.; Zhang, J. From Microscale Devices to 3D Printing. Circ. Res. 2017, 120, 150–165. [Google Scholar] [CrossRef]
- Rider, P.; Kačarević, Ž.P.; Alkildani, S.; Retnasingh, S.; Barbeck, M. Bioprinting of tissue engineering scaffolds. J. Tissue Eng. 2018, 9, 2041731418802090. [Google Scholar] [CrossRef]
- Lim, K.S.; Schon, B.S.; Mekhileri, N.V.; Brown, G.C.J.; Chia, C.M.; Prabakar, S.; Hooper, G.J.; Woodfield, T.B.F. New Visible-Light Photoinitiating System for Improved Print Fidelity in Gelatin-Based Bioinks. ACS Biomater. Sci. Eng. 2016, 2, 1752–1762. [Google Scholar] [CrossRef]
- Sharma, P.; Wang, X.; Ming, C.L.C.; Vettori, L.; Figtree, G.; Boyle, A.; Gentile, C. Considerations for the Bioengineering of Advanced Cardiac In Vitro Models of Myocardial Infarction. Small 2021, 17, 2003765. [Google Scholar] [CrossRef] [PubMed]
- Roche, C.D.; Sharma, P.; Ashton, A.W.; Jackson, C.; Xue, M.; Gentile, C. Printability, Durability, Contractility and Vascular Network Formation in 3D Bioprinted Cardiac Endothelial Cells Using Alginate–Gelatin Hydrogels. Front. Bioeng. Biotechnol. 2021, 9, 636257. [Google Scholar] [CrossRef]
- Zhou, P.; Pu, W.T. Recounting cardiac cellular composition. Circ. Res. 2016, 118, 368–370. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.P.; Squirrell, J.M.; Lyons, G.E.; Eliceiri, K.W.; Ogle, B.M. Imaging cardiac extracellular matrices: A blueprint for regeneration. Trends Biotechnol. 2012, 30, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Lesman, A.; Gepstein, L.; Levenberg, S. Vascularization shaping the heart. Ann. N. Y. Acad. Sci. 2010, 1188, 46–51. [Google Scholar] [CrossRef]
- Khan, A.; Kumari, P.; Kumari, N.; Shaikh, U.; Ekhator, C.; Halappa Nagaraj, R.; Yadav, V.; Khan, A.W.; Lazarevic, S.; Bharati, B.; et al. Biomimetic Approaches in Cardiac Tissue Engineering: Replicating the Native Heart Microenvironment. Cureus 2023, 15, e43431. [Google Scholar] [CrossRef]
- Elson, E.L.; Genin, G.M. Tissue constructs: Platforms for basic research and drug discovery. Interface Focus 2016, 6, 20150095. [Google Scholar] [CrossRef]
- Gaebel, R.; Ma, N.; Liu, J.; Guan, J.; Koch, L.; Klopsch, C.; Gruene, M.; Toelk, A.; Wang, W.; Mark, P.; et al. Patterning human stem cells and endothelial cells with laser printing for cardiac regeneration. Biomaterials 2011, 32, 9218–9230. [Google Scholar] [CrossRef]
- Wissing, T.B.; Bonito, V.; Bouten, C.V.C.; Smits, A.I.P.M. Biomaterial-driven in situ cardiovascular tissue engineering—A multi-disciplinary perspective. npj Regen. Med. 2017, 2, 18. [Google Scholar] [CrossRef] [PubMed]
- Gaudriault, P.; Fassini, D.; Homs-Corbera, A. Chapter 8—Heart-on-a-chip. In Organ-on-a-Chip; Hoeng, J., Bovard, D., Peitsch, M.C., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 255–293. ISBN 978-0-12-817202-5. [Google Scholar] [CrossRef]
- Cui, H.; Miao, S.; Esworthy, T.; Zhou, X.; Lee, S.; Liu, C.; Yu, Z.; Fisher, J.P.; Mohiuddin, M.; Zhang, L.G. 3D bioprinting for cardiovascular regeneration and pharmacology. Adv. Drug Deliv. Rev. 2018, 132, 252–269. [Google Scholar] [CrossRef] [PubMed]
- Jafari, A.; Ajji, Z.; Mousavi, A.; Naghieh, S.; Bencherif, S.A.; Savoji, H. Latest Advances in 3D Bioprinting of Cardiac Tissues. Adv. Mater. Technol. 2022, 7, 2101636. [Google Scholar] [CrossRef] [PubMed]
- Akter, M.Z.; Tufail, F.; Ahmad, A.; Oh, Y.W.; Kim, J.M.; Kim, S.; Hasan, M.M.; Li, L.; Lee, D.-W.; Kim, Y.S.; et al. Harnessing native blueprints for designing bioinks to bioprint functional cardiac tissue. iScience 2025, 28, 111882. [Google Scholar] [CrossRef]
- Kim, M.; Hwang, D.G.; Jang, J. Bioprinting approaches in cardiac tissue engineering to reproduce blood-pumping heart function. iScience 2025, 28, 111664. [Google Scholar] [CrossRef]
- Arai, K.; Iwanaga, S.; Toda, H.; Genci, C.; Nishiyama, Y.; Nakamura, M. Three-dimensional inkjet biofabrication based on designed images. Biofabrication 2011, 3, 034113. [Google Scholar] [CrossRef]
- Lee, W.; Lee, V.; Polio, S.; Keegan, P.; Lee, J.-H.; Fischer, K.; Park, J.-K.; Yoo, S.-S. On-demand three-dimensional freeform fabrication of multi-layered hydrogel scaffold with fluidic channels. Biotechnol. Bioeng. 2010, 105, 1178–1186. [Google Scholar] [CrossRef]
- Moon, S.; Hasan, S.K.; Song, Y.S.; Xu, F.; Keles, H.O.; Manzur, F.; Mikkilineni, S.; Hong, J.W.; Nagatomi, J.; Haeggstrom, E.; et al. Layer by Layer Three-dimensional Tissue Epitaxy by Cell-Laden Hydrogel Droplets. Tissue Eng. Part C Methods 2010, 16, 157–166. [Google Scholar] [CrossRef]
- Snyder, J.E.; Hamid, Q.; Wang, C.; Chang, R.; Emami, K.; Wu, H.; Sun, W. Bioprinting cell-laden matrigel for radioprotection study of liver by pro-drug conversion in a dual-tissue microfluidic chip. Biofabrication 2011, 3, 034112. [Google Scholar] [CrossRef]
- Cui, X.; Boland, T. Human microvasculature fabrication using thermal inkjet printing technology. Biomaterials 2009, 30, 6221–6227. [Google Scholar] [CrossRef]
- Song, S.-J.; Choi, J.; Park, Y.-D.; Lee, J.-J.; Hong, S.Y.; Sun, K. A Three-Dimensional Bioprinting System for Use With a Hydrogel-Based Biomaterial and Printing Parameter Characterization. Artif. Organs 2010, 34, 1044–1048. [Google Scholar] [CrossRef] [PubMed]
- Jakab, K.; Norotte, C.; Marga, F.; Murphy, K.; Vunjak-Novakovic, G.; Forgacs, G. Tissue engineering by self-assembly and bio-printing of living cells. Biofabrication 2010, 2, 022001. [Google Scholar] [CrossRef] [PubMed]
- Almeida, C.R.; Serra, T.; Oliveira, M.I.; Planell, J.A.; Barbosa, M.A.; Navarro, M. Impact of 3-D printed PLA- and chitosan-based scaffolds on human monocyte/macrophage responses: Unraveling the effect of 3-D structures on inflammation. Acta Biomater. 2014, 10, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Hockaday, L.A.; Kang, K.H.; Colangelo, N.W.; Cheung, P.Y.C.; Duan, B.; Malone, E.; Wu, J.; Girardi, L.N.; Bonassar, L.J.; Lipson, H.; et al. Rapid 3D printing of anatomically accurate and mechanically heterogeneous aortic valve hydrogel scaffolds. Biofabrication 2012, 4, 035005. [Google Scholar] [CrossRef]
- Rimann, M.; Bono, E.; Annaheim, H.; Bleisch, M.; Graf-Hausner, U. Standardized 3D Bioprinting of Soft Tissue Models with Human Primary Cells. J. Lab. Autom. 2016, 21, 496–509. [Google Scholar] [CrossRef]
- Zhao, Y.; Rafatian, N.; Feric, N.T.; Cox, B.J.; Aschar-Sobbi, R.; Wang, E.Y.; Aggarwal, P.; Zhang, B.; Conant, G.; Ronaldson-Bouchard, K.; et al. A Platform for Generation of Chamber-Specific Cardiac Tissues and Disease Modeling. Cell 2019, 176, 913–927.e18. [Google Scholar] [CrossRef]
- Wolfe, J.T.; He, W.; Kim, M.-S.; Liang, H.-L.; Shradhanjali, A.; Jurkiewicz, H.; Freudinger, B.P.; Greene, A.S.; LaDisa, J.F.; Tayebi, L.; et al. 3D-bioprinting of patient-derived cardiac tissue models for studying congenital heart disease. Front. Cardiovasc. Med. 2023, 10, 1162731. [Google Scholar] [CrossRef]
- Lee, A.; Hudson, A.R.; Shiwarski, D.J.; Tashman, J.W.; Hinton, T.J.; Yerneni, S.; Bliley, J.M.; Campbell, P.G.; Feinberg, A.W. 3D bioprinting of collagen to rebuild components of the human heart. Science 2019, 365, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Arneri, A.; Bersini, S.; Shin, S.-R.; Zhu, K.; Goli-Malekabadi, Z.; Aleman, J.; Colosi, C.; Busignani, F.; Dell’Erba, V.; et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 2016, 110, 45–59, Correction in Biomaterials 2016, 322, 123363. [Google Scholar] [CrossRef]
- Masson-Meyers, D.S.; Tayebi, L. Vascularization strategies in tissue engineering approaches for soft tissue repair. J. Tissue Eng. Regen. Med. 2021, 15, 747–762. [Google Scholar] [CrossRef]
- Chandra, P.; Atala, A. Engineering blood vessels and vascularized tissues: Technology trends and potential clinical applications. Clin. Sci. 2019, 133, 1115–1135. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Yahya, R.; Hassan, A.; Yar, M.; Abd Halim, A.A.; Rageh Al-Maleki, A.; Shahzadi, L.; Zubairi, W. Novel chitosan derivative based composite scaffolds with enhanced angiogenesis; potential candidates for healing chronic non-healing wounds. J. Mater. Sci. Mater. Med. 2019, 30, 72. [Google Scholar] [CrossRef]
- Ong, C.S.; Nam, L.; Ong, K.; Krishnan, A.; Huang, C.Y.; Fukunishi, T.; Hibino, N. 3D and 4D Bioprinting of the Myocardium: Current Approaches, Challenges, and Future Prospects. BioMed Res. Int. 2018, 2018, 6497242. [Google Scholar] [CrossRef]
- Jain, R.K.; Au, P.; Tam, J.; Duda, D.G.; Fukumura, D. Engineering vascularized tissue. Nat. Biotechnol. 2005, 23, 821–823. [Google Scholar] [CrossRef]
- Cho, S.; Discher, D.E.; Leong, K.W.; Vunjak-Novakovic, G. Challenges and opportunities for the next generation of cardiovascular tissue engineering. Nat. Methods 2022, 19, 1064–1071. [Google Scholar] [CrossRef] [PubMed]
- Hooper, R.; Cummings, C.; Beck, A.; Vazquez-Armendariz, J.; Rodriguez, C.; Dean, D. Sheet-based extrusion bioprinting: A new multi-material paradigm providing mid-extrusion micropatterning control for microvascular applications. Biofabrication 2024, 16, 025032. [Google Scholar] [CrossRef] [PubMed]
- Davoodi, E.; Sarikhani, E.; Montazerian, H.; Ahadian, S.; Costantini, M.; Swieszkowski, W.; Willerth, S.; Walus, K.; Mofidfar, M.; Toyserkani, E.; et al. Extrusion and Microfluidic-based Bioprinting to Fabricate Biomimetic Tissues and Organs. Adv. Mater. Technol. 2020, 5, 1901044. [Google Scholar] [CrossRef]
- Kjar, A.; McFarland, B.; Mecham, K.; Harward, N.; Huang, Y. Engineering of tissue constructs using coaxial bioprinting. Bioact. Mater. 2021, 6, 460–471. [Google Scholar] [CrossRef]
- Bhusal, A.; Dogan, E.; Nguyen, H.-A.; Labutina, O.; Nieto, D.; Khademhosseini, A.; Miri, A.K. Multi-Material Digital Light Processing Bioprinting of Hydrogel-Based Microfluidic Chips. Biofabrication 2021, 14, 014103. [Google Scholar] [CrossRef]
- Colosi, C.; Costantini, M.; Barbetta, A.; Dentini, M. Microfluidic Bioprinting of Heterogeneous 3D Tissue Constructs. In 3D Cell Culture: Methods and Protocols; Koledova, Z., Ed.; Springer: New York, NY, USA, 2017; pp. 369–380. ISBN 978-1-4939-7021-6. [Google Scholar] [CrossRef]
- Blaeser, A.; Duarte Campos, D.F.; Puster, U.; Richtering, W.; Stevens, M.M.; Fischer, H. Controlling Shear Stress in 3D Bioprinting is a Key Factor to Balance Printing Resolution and Stem Cell Integrity. Adv. Healthc. Mater. 2016, 5, 326–333. [Google Scholar] [CrossRef]
- Shiwarski, D.J.; Hudson, A.R.; Tashman, J.W.; Feinberg, A.W. Emergence of FRESH 3D printing as a platform for advanced tissue biofabrication. APL Bioeng. 2021, 5, 010904. [Google Scholar] [CrossRef] [PubMed]
- Hinton, T.J.; Jallerat, Q.; Palchesko, R.N.; Park, J.H.; Grodzicki, M.S.; Shue, H.-J.; Ramadan, M.H.; Hudson, A.R.; Feinberg, A.W. Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci. Adv. 2015, 1, e1500758. [Google Scholar] [CrossRef]
- Skylar-Scott, M.A.; Uzel, S.G.M.; Nam, L.L.; Ahrens, J.H.; Truby, R.L.; Damaraju, S.; Lewis, J.A. Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Sci. Adv. 2019, 5, eaaw2459. [Google Scholar] [CrossRef]
- Ze, Y.; Li, Y.; Huang, L.; Shi, Y.; Li, P.; Gong, P.; Lin, J.; Yao, Y. Biodegradable Inks in Indirect Three-Dimensional Bioprinting for Tissue Vascularization. Front. Bioeng. Biotechnol. 2022, 10, 856398. [Google Scholar] [CrossRef]
- Tsui, J.H.; Leonard, A.; Camp, N.D.; Long, J.T.; Nawas, Z.Y.; Chavanachat, R.; Smith, A.S.T.; Choi, J.S.; Dong, Z.; Ahn, E.H.; et al. Tunable electroconductive decellularized extracellular matrix hydrogels for engineering human cardiac microphysiological systems. Biomaterials 2021, 272, 120764. [Google Scholar] [CrossRef]
- Asaro, G.A.; Solazzo, M.; Suku, M.; Spurling, D.; Genoud, K.; Gonzalez, J.G.; Brien, F.J.O.; Nicolosi, V.; Monaghan, M.G. MXene functionalized collagen biomaterials for cardiac tissue engineering driving iPSC-derived cardiomyocyte maturation. npj 2D Mater. Appl. 2023, 7, 44. [Google Scholar] [CrossRef]
- Navaei, A.; Saini, H.; Christenson, W.; Sullivan, R.T.; Ros, R.; Nikkhah, M. Gold nanorod-incorporated gelatin-based conductive hydrogels for engineering cardiac tissue constructs. Acta Biomater. 2016, 41, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Roshanbinfar, K.; Schiffer, M.; Carls, E.; Angeloni, M.; Koleśnik-Gray, M.; Schruefer, S.; Schubert, D.W.; Ferrazzi, F.; Krstić, V.; Fleischmann, B.K.; et al. Electrically Conductive Collagen-PEDOT:PSS Hydrogel Prevents Post-Infarct Cardiac Arrhythmia and Supports hiPSC-Cardiomyocyte Function. Adv. Mater. 2024, 36, 2403642. [Google Scholar] [CrossRef]
- Xu, Q.; Huang, S.; Lv, Y.; Du, L. Investigation of Ni2P/rGO porous composite catalyst for photocatalytic degradation and electrocataly hydrogen evolution reaction performance. Mater. Res. Bull. 2026, 193, 113657. [Google Scholar] [CrossRef]
- Sanjuan-Alberte, P.; Whitehead, C.; Jones, J.N.; Silva, J.C.; Carter, N.; Kellaway, S.; Hague, R.J.M.; Cabral, J.M.S.; Ferreira, F.C.; White, L.J.; et al. Printing biohybrid materials for bioelectronic cardio-3D-cellular constructs. iScience 2022, 25, 104552. [Google Scholar] [CrossRef] [PubMed]
- Roshanbinfar, K.; Vogt, L.; Greber, B.; Diecke, S.; Boccaccini, A.R.; Scheibel, T.; Engel, F.B. Electroconductive Biohybrid Hydrogel for Enhanced Maturation and Beating Properties of Engineered Cardiac Tissues. Adv. Funct. Mater. 2018, 28, 1803951. [Google Scholar] [CrossRef]
- Park, S.E.; Georgescu, A.; Huh, D. Organoids-on-a-chip. Science 2019, 364, 960–965. [Google Scholar] [CrossRef]
- Gabbin, B.; Meraviglia, V.; Angenent, M.L.; Ward-van Oostwaard, D.; Sol, W.; Mummery, C.L.; Rabelink, T.J.; van Meer, B.J.; van den Berg, C.W.; Bellin, M. Heart and kidney organoids maintain organ-specific function in a microfluidic system. Mater. Today Bio 2023, 23, 100818. [Google Scholar] [CrossRef]
- Rahman, S.M.; Martin, E.C.; Melvin, A.T. Co-culture of Two Different Cell Lines in a Two-Layer Microfluidic Device. In Cancer Drug Resistance: Methods and Protocols; Baiocchi, M., Ed.; Springer: New York, NY, USA, 2022; pp. 33–47. ISBN 978-1-0716-2513-2. [Google Scholar] [CrossRef]
- Min, S.; Kim, S.; Sim, W.-S.; Choi, Y.S.; Joo, H.; Park, J.-H.; Lee, S.-J.; Kim, H.; Lee, M.J.; Jeong, I.; et al. Versatile human cardiac tissues engineered with perfusable heart extracellular microenvironment for biomedical applications. Nat. Commun. 2024, 15, 2564. [Google Scholar] [CrossRef]
- Paez-Mayorga, J.; Hernández-Vargas, G.; Ruiz-Esparza, G.U.; Iqbal, H.M.N.; Wang, X.; Zhang, Y.S.; Parra-Saldivar, R.; Khademhosseini, A. Bioreactors for Cardiac Tissue Engineering. Adv. Healthc. Mater. 2019, 8, 1701504. [Google Scholar] [CrossRef]
- Niculescu, A.-G.; Chircov, C.; Bîrcă, A.C.; Grumezescu, A.M. Fabrication and Applications of Microfluidic Devices: A Review. Int. J. Mol. Sci. 2021, 22, 2011. [Google Scholar] [CrossRef]
- Young, K.M.; Shankles, P.G.; Chen, T.; Ahkee, K.; Bules, S.; Sulchek, T. Scaling microfluidic throughput with flow-balanced manifolds to simply control devices with multiple inlets and outlets. Biomicrofluidics 2022, 16, 034104. [Google Scholar] [CrossRef]
- Mitxelena-Iribarren, O.; Zabalo, J.; Arana, S.; Mujika, M. Improved microfluidic platform for simultaneous multiple drug screening towards personalized treatment. Biosens. Bioelectron. 2019, 123, 237–243. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Hashizume, H.; Takiguchi, S.; Ji, J.; Kawano, R.; Koiwai, K.; Yamamoto, H.; Elbadawy, M.; Omatsu, T.; Abugomaa, A.; et al. A microfluidics platform for simultaneous evaluation of sensitivity and side effects of anti-cancer drugs using a three-dimensional culture method. Sci. Rep. 2025, 15, 39. [Google Scholar] [CrossRef] [PubMed]
- Ugolini, G.S.; Visone, R.; Cruz-Moreira, D.; Mainardi, A.; Rasponi, M. Chapter 4—Generation of functional cardiac microtissues in a beating heart-on-a-chip. In Methods in Cell Biology; Doh, J., Fletcher, D., Piel, M., Eds.; Microfluidics in Cell Biology Part A: Microfluidics for Multicellular Systems; Academic Press: Cambridge, MA, USA, 2018; Volume 146, pp. 69–84. [Google Scholar] [CrossRef]
- Ugolini, G.S.; Pavesi, A.; Rasponi, M.; Fiore, G.B.; Kamm, R.; Soncini, M. Human cardiac fibroblasts adaptive responses to controlled combined mechanical strain and oxygen changes in vitro. eLife 2017, 6, e22847. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Goss, J.A.; Cho, A.; McCain, M.L.; Parker, K.K. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 2013, 13, 3599–3608. [Google Scholar] [CrossRef]
- Bliley, J.M.; Stang, M.A.; Behre, A.; Feinberg, A.W. Advances in 3D Bioprinted Cardiac Tissue Using Stem Cell-Derived Cardiomyocytes. Stem Cells Transl. Med. 2024, 13, 425–435. [Google Scholar] [CrossRef]
- Sidorov, V.Y.; Samson, P.C.; Sidorova, T.N.; Davidson, J.M.; Lim, C.C.; Wikswo, J.P. I-Wire Heart-on-a-Chip I: Three-dimensional cardiac tissue constructs for physiology and pharmacology. Acta Biomater. 2017, 48, 68–78. [Google Scholar] [CrossRef]
- Faulkner-Jones, A.; Zamora, V.; Hortigon-Vinagre, M.P.; Wang, W.; Ardron, M.; Smith, G.L.; Shu, W. A Bioprinted Heart-on-a-Chip with Human Pluripotent Stem Cell-Derived Cardiomyocytes for Drug Evaluation. Bioengineering 2022, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Colosi, C.; Shin, S.R.; Manoharan, V.; Massa, S.; Costantini, M.; Barbetta, A.; Dokmeci, M.R.; Dentini, M.; Khademhosseini, A. Microfluidic Bioprinting of Heterogeneous 3D Tissue Constructs Using Low-Viscosity Bioink. Adv. Mater. 2016, 28, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Di Cio, S.; Marhuenda, E.; Haddrick, M.; Gautrot, J.E. Vascularised cardiac spheroids-on-a-chip for testing the toxicity of therapeutics. Sci. Rep. 2024, 14, 3370. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.; Mendiola, K.; Lightsey, N.K.; Hanjaya-Putra, D. Mimicking blood and lymphatic vasculatures using microfluidic systems. Biomicrofluidics 2024, 18, 031502. [Google Scholar] [CrossRef]
- Thavandiran, N.; Dubois, N.; Mikryukov, A.; Massé, S.; Beca, B.; Simmons, C.A.; Deshpande, V.S.; McGarry, J.P.; Chen, C.S.; Nanthakumar, K.; et al. Design and Formulation of Functional Pluripotent Stem Cell-Derived Cardiac Microtissues. Proc. Natl. Acad. Sci. USA 2013, 110, E4698–E4707. [Google Scholar] [CrossRef]
- Liu, B.; Wang, S.; Ma, H.; Deng, Y.; Du, J.; Zhao, Y.; Chen, Y. Heart-on-a-chip: A revolutionary organ-on-chip platform for cardiovascular disease modeling. J. Transl. Med. 2025, 23, 132. [Google Scholar] [CrossRef]
- Stoppel, W.L.; Kaplan, D.L.; Black, L.D. Electrical and mechanical stimulation of cardiac cells and tissue constructs. Adv. Drug Deliv. Rev. 2016, 96, 135–155. [Google Scholar] [CrossRef]
- Fink, C.; Ergün, S.; Kralisch, D.; Remmers, U.; Weil, J.; Eschenhagen, T. Chronic stretch of engineered heart tissue induces hypertrophy and functional improvement. FASEB J. 2000, 14, 669–679. [Google Scholar] [CrossRef]
- Bhise, N.S.; Manoharan, V.; Massa, S.; Tamayol, A.; Ghaderi, M.; Miscuglio, M.; Lang, Q.; Shrike Zhang, Y.; Shin, S.R.; Calzone, G.; et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 2016, 8, 014101. [Google Scholar] [CrossRef]
- Oleaga, C.; Riu, A.; Rothemund, S.; Lavado, A.; McAleer, C.W.; Long, C.J.; Persaud, K.; Narasimhan, N.S.; Tran, M.; Roles, J.; et al. Investigation of the effect of hepatic metabolism on off-target cardiotoxicity in a multi-organ human-on-a-chip system. Biomaterials 2018, 182, 176–190. [Google Scholar] [CrossRef] [PubMed]
- Charbe, N.B.; Zacconi, F.C.; Amnerkar, N.; Pardhi, D.; Shukla, P.; Mukattash, T.L.; McCarron, P.A.; Tambuwala, M.M. Emergence of Three Dimensional Printed Cardiac Tissue: Opportunities and Challenges in Cardiovascular Diseases. Curr. Cardiol. Rev. 2019, 15, 188–204. [Google Scholar] [CrossRef] [PubMed]
- Tijore, A.; Irvine, S.A.; Sarig, U.; Mhaisalkar, P.; Baisane, V.; Venkatraman, S. Contact guidance for cardiac tissue engineering using 3D bioprinted gelatin patterned hydrogel. Biofabrication 2018, 10, 025003. [Google Scholar] [CrossRef] [PubMed]
- De Spirito, M.; Palmieri, V.; Perini, G.; Papi, M. Bridging the Gap: Integrating 3D Bioprinting and Microfluidics for Advanced Multi-Organ Models in Biomedical Research. Bioengineering 2024, 11, 664. [Google Scholar] [CrossRef]
- Yang, Q.; Xiao, Z.; Lv, X.; Zhang, T.; Liu, H. Fabrication and Biomedical Applications of Heart-on-a-chip. Int. J. Bioprint. 2021, 7, 370. [Google Scholar] [CrossRef]
- Baptista, L.S.; Mironov, V.; Koudan, E.; Amorim, É.A.; Pampolha, T.P.; Kasyanov, V.; Kovalev, A.; Senatov, F.; Granjeiro, J.M. Bioprinting Using Organ Building Blocks: Spheroids, Organoids, and Assembloids. Tissue Eng. Part A 2024, 30, 377–386. [Google Scholar] [CrossRef]
- Jakab, K.; Neagu, A.; Mironov, V.; Markwald, R.R.; Forgacs, G. Engineering biological structures of prescribed shape using self-assembling multicellular systems. Proc. Natl. Acad. Sci. USA 2004, 101, 2864–2869. [Google Scholar] [CrossRef]
- Roche, C.D.; Lin, H.; Huang, Y.; de Bock, C.E.; Beck, D.; Xue, M.; Gentile, C. 3D bioprinted alginate-gelatin hydrogel patches containing cardiac spheroids recover heart function in a mouse model of myocardial infarction. Bioprinting 2023, 30, e00263. [Google Scholar] [CrossRef]
- Ong, C.S.; Fukunishi, T.; Nashed, A.; Blazeski, A.; Zhang, H.; Hardy, S.; DiSilvestre, D.; Vricella, L.; Conte, J.; Tung, L.; et al. Creation of Cardiac Tissue Exhibiting Mechanical Integration of Spheroids Using 3D Bioprinting. J. Vis. Exp. JoVE 2017, 125, e55438. [Google Scholar] [CrossRef]
- Moldovan, N.I.; Hibino, N.; Nakayama, K. Principles of the Kenzan Method for Robotic Cell Spheroid-Based Three-Dimensional Bioprinting. Tissue Eng. Part B Rev. 2017, 23, 237–244. [Google Scholar] [CrossRef]
- Ahrens, J.H.; Uzel, S.G.M.; Skylar-Scott, M.; Mata, M.M.; Lu, A.; Kroll, K.T.; Lewis, J.A. Programming Cellular Alignment in Engineered Cardiac Tissue via Bioprinting Anisotropic Organ Building Blocks. Adv. Mater. 2022, 34, 2200217. [Google Scholar] [CrossRef]
- Heinzelmann, E.; Piraino, F.; Costa, M.; Roch, A.; Norkin, M.; Garnier, V.; Homicsko, K.; Brandenberg, N. iPSC-derived and Patient-Derived Organoids: Applications and challenges in scalability and reproducibility as pre-clinical models. Curr. Res. Toxicol. 2024, 7, 100197. [Google Scholar] [CrossRef]
- Paik, D.T.; Chandy, M.; Wu, J.C. Patient and Disease–Specific Induced Pluripotent Stem Cells for Discovery of Personalized Cardiovascular Drugs and Therapeutics. Pharmacol. Rev. 2020, 72, 320–342. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Song, G.; Wang, H.; Zhang, Q.; Wang, M.; Zhao, M.; Wang, B.; Zhu, H.; Ranzhi, L.; et al. Revolutionizing cardiovascular research: Human organoids as a Beacon of hope for understanding and treating cardiovascular diseases. Mater. Today Bio 2025, 30, 101396. [Google Scholar] [CrossRef]
- Zhao, D.; Lei, W.; Hu, S. Cardiac organoid—A promising perspective of preclinical model. Stem Cell Res. Ther. 2021, 12, 272. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Liu, K.; Feng, Q.; Liao, Y. Cardiac Organoids: A 3D Technology for Modeling Heart Development and Disease. Stem Cell Rev. Rep. 2022, 18, 2593–2605. [Google Scholar] [CrossRef] [PubMed]
- Cetnar, A.D.; Tomov, M.L.; Ning, L.; Jing, B.; Theus, A.S.; Kumar, A.; Wijntjes, A.N.; Bhamidipati, S.R.; Do, K.P.; Mantalaris, A.; et al. Patient-Specific 3D Bioprinted Models of Developing Human Heart. Adv. Healthc. Mater. 2021, 10, e2001169. [Google Scholar] [CrossRef] [PubMed]
- Colter, J.; Murari, K.; Biernaskie, J.; Kallos, M.S. Induced pluripotency in the context of stem cell expansion bioprocess development, optimization, and manufacturing: A roadmap to the clinic. npj Regen. Med. 2021, 6, 72. [Google Scholar] [CrossRef]
- Wolf, K.J.; Weiss, J.D.; Uzel, S.G.M.; Skylar-Scott, M.A.; Lewis, J.A. Biomanufacturing Human Tissues via Organ Building Blocks. Cell Stem Cell 2022, 29, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Hookway, T.A.; Butts, J.C.; Lee, E.; Tang, H.; McDevitt, T.C. Aggregate formation and suspension culture of human pluripotent stem cells and differentiated progeny. Methods 2016, 101, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.C.; Hookway, T.A.; Wu, Q.; Jha, R.; Preininger, M.K.; Chen, X.; Easley, C.A.; Spearman, P.; Deshpande, S.R.; Maher, K.; et al. Microscale Generation of Cardiospheres Promotes Robust Enrichment of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Stem Cell Rep. 2014, 3, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Kengla, C.; Atala, A.; Lee, S.J. Chapter 15—Bioprinting of Organoids. In Essentials of 3D Biofabrication and Translation; Atala, A., Yoo, J.J., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 271–282. ISBN 978-0-12-800972-7. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, Y.; Shao, L.; Xie, M.; Nie, J.; Qiu, J.; Zhao, P.; Ramezani, H.; Fu, J.; Ouyang, H.; et al. Airflow-Assisted 3D Bioprinting of Human Heterogeneous Microspheroidal Organoids with Microfluidic Nozzle. Small 2018, 14, 1802630. [Google Scholar] [CrossRef]
- Quintard, C.; Tubbs, E.; Jonsson, G.; Jiao, J.; Wang, J.; Werschler, N.; Laporte, C.; Pitaval, A.; Bah, T.-S.; Pomeranz, G.; et al. A microfluidic platform integrating functional vascularized organoids-on-chip. Nat. Commun. 2024, 15, 1452. [Google Scholar] [CrossRef]
- Walcott, J.C.; Davis, M.E. Bioprinting organoids for functional cardiac constructs: Progress and unmet challenges. Int. J. Bioprint. 2025, 11, 85–114. [Google Scholar] [CrossRef]
- Schmidt, C.; Deyett, A.; Ilmer, T.; Haendeler, S.; Caballero, A.T.; Novatchkova, M.; Netzer, M.A.; Ginistrelli, L.C.; Juncosa, E.M.; Bhattacharya, T.; et al. Multi-chamber cardioids unravel human heart development and cardiac defects. Cell 2023, 186, 5587–5605.e27. [Google Scholar] [CrossRef]
- Volmert, B.; Kiselev, A.; Juhong, A.; Wang, F.; Riggs, A.; Kostina, A.; O’Hern, C.; Muniyandi, P.; Wasserman, A.; Huang, A.; et al. A patterned human primitive heart organoid model generated by pluripotent stem cell self-organization. Nat. Commun. 2023, 14, 8245. [Google Scholar] [CrossRef]
- Lee, J.; Sutani, A.; Kaneko, R.; Takeuchi, J.; Sasano, T.; Kohda, T.; Ihara, K.; Takahashi, K.; Yamazoe, M.; Morio, T.; et al. In vitro generation of functional murine heart organoids via FGF4 and extracellular matrix. Nat. Commun. 2020, 11, 4283. [Google Scholar] [CrossRef]
- Wang, F.; Zou, X.; Zheng, H.; Kong, T.; Pei, D. Human epicardial organoids from pluripotent stem cells resemble fetal stage with potential cardiomyocyte- transdifferentiation. Cell Biosci. 2025, 15, 4. [Google Scholar] [CrossRef]
- Huang, Z.; Jia, K.; Tan, Y.; Yu, Y.; Xiao, W.; Zhou, X.; Yi, J.; Zhang, C. Advances in cardiac organoid research: Implications for cardiovascular disease treatment. Cardiovasc. Diabetol. 2025, 24, 25. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Mikail, N.; Bengs, S.; Haider, A.; Treyer, V.; Buechel, R.R.; Wegener, S.; Rauen, K.; Tawakol, A.; Bairey Merz, C.N.; et al. Heart–brain interactions in cardiac and brain diseases: Why sex matters. Eur. Heart J. 2022, 43, 3971–3980. [Google Scholar] [CrossRef]
- Matsuura, T.R.; Puchalska, P.; Crawford, P.A.; Kelly, D.P. Ketones and the Heart: Metabolic Principles and Therapeutic Implications. Circ. Res. 2023, 132, 882–898. [Google Scholar] [CrossRef]
- Buliga-Finis, O.N.; Ouatu, A.; Badescu, M.C.; Dima, N.; Tanase, D.M.; Richter, P.; Rezus, C. Beyond the Cardiorenal Syndrome: Pathophysiological Approaches and Biomarkers for Renal and Cardiac Crosstalk. Diagnostics 2022, 12, 773. [Google Scholar] [CrossRef]
- Kanton, S.; Paşca, S.P. Human assembloids. Development 2022, 149, dev201120. [Google Scholar] [CrossRef] [PubMed]
- Kostina, A.; Kiselev, A.; Huang, A.; Lankerd, H.; Caywood, S.; Jurado-Fernandez, A.; Volmert, B.; O’Hern, C.; Juhong, A.; Liu, Y.; et al. Self-organizing human heart assembloids with autologous and developmentally relevant cardiac neural crest-derived tissues. bioRxiv 2024. [Google Scholar] [CrossRef]
- Daly, A.C.; Davidson, M.D.; Burdick, J.A. 3D bioprinting of high cell-density heterogeneous tissue models through spheroid fusion within self-healing hydrogels. Nat. Commun. 2021, 12, 753. [Google Scholar] [CrossRef]
- Song, S.-S.; Park, H.-J.; Kim, Y.K.; Kang, S.-W. Revolutionizing biomedical research: The imperative need for heart–kidney-connected organoids. APL Bioeng. 2024, 8, 010902. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.H.; Varghese, B.; Jia, H.; Ren, X. Alliance of Heart and Endoderm: Multilineage Organoids to Model Co-development. Circ. Res. 2023, 132, 511–518. [Google Scholar] [CrossRef]
- Jaconi, M.E.; Puceat, M. Cardiac Organoids and Gastruloids to Study Physio-Pathological Heart Development. J. Cardiovasc. Dev. Dis. 2021, 8, 178. [Google Scholar] [CrossRef]
- Gu, M.; Zorn, A.M. Follow your heart and trust your gut: Co-development of heart and gut tissue in organoids. Cell Stem Cell 2021, 28, 2037–2038. [Google Scholar] [CrossRef] [PubMed]
- Drakhlis, L.; Biswanath, S.; Farr, C.M.; Lupanow, V.; Teske, J.; Ritzenhoff, K.; Franke, A.; Manstein, F.; Bolesani, E.; Kempf, H.; et al. Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol. 2021, 39, 737–746. [Google Scholar] [CrossRef]
- Branco, M.A.; Dias, T.P.; Cabral, J.M.S.; Pinto-do-Ó, P.; Diogo, M.M. Human multilineage pro-epicardium/foregut organoids support the development of an epicardium/myocardium organoid. Nat. Commun. 2022, 13, 6981. [Google Scholar] [CrossRef]
- Fontanelli, L.; Nisini, N.; Pirola, S.; Recchia, F.A. Neuromuscular and cardiac organoids and assembloids: Advanced platforms for drug testing. Pharmacol. Ther. 2025, 272, 108876. [Google Scholar] [CrossRef]
- Coyle, R.C.; Barrs, R.W.; Richards, D.J.; Ladd, E.P.; Menick, D.R.; Mei, Y. Targeting HIF-α for robust prevascularization of human cardiac organoids. J. Tissue Eng. Regen. Med. 2021, 15, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Yousafzai, M.S.; Hammer, J.A. Using Biosensors to Study Organoids, Spheroids and Organs-on-a-Chip: A Mechanobiology Perspective. Biosensors 2023, 13, 905. [Google Scholar] [CrossRef]
- Liu, S.; Kumari, S.; He, H.; Mishra, P.; Singh, B.N.; Singh, D.; Liu, S.; Srivastava, P.; Li, C. Biosensors integrated 3D organoid/organ-on-a-chip system: A real-time biomechanical, biophysical, and biochemical monitoring and characterization. Biosens. Bioelectron. 2023, 231, 115285. [Google Scholar] [CrossRef]
- Yin, J.; Lees, J.G.; Gong, S.; Nguyen, J.T.; Phang, R.J.; Shi, Q.; Huang, Y.; Kong, A.M.; Dyson, J.M.; Lim, S.Y.; et al. Real-time electro-mechanical profiling of dynamically beating human cardiac organoids by coupling resistive skins with microelectrode arrays. Biosens. Bioelectron. 2025, 267, 116752. [Google Scholar] [CrossRef] [PubMed]
- Caluori, G.; Pribyl, J.; Pesl, M.; Jelinkova, S.; Rotrekl, V.; Skladal, P.; Raiteri, R. Non-invasive electromechanical cell-based biosensors for improved investigation of 3D cardiac models. Biosens. Bioelectron. 2019, 124–125, 129–135. [Google Scholar] [CrossRef]
- Ghosheh, M.; Ehrlich, A.; Ioannidis, K.; Ayyash, M.; Goldfracht, I.; Cohen, M.; Fischer, A.; Mintz, Y.; Gepstein, L.; Nahmias, Y. Electro-metabolic coupling in multi-chambered vascularized human cardiac organoids. Nat. Biomed. Eng. 2023, 7, 1493–1513. [Google Scholar] [CrossRef]
- Kim, M.; Hwang, J.C.; Min, S.; Park, Y.-G.; Kim, S.; Kim, E.; Seo, H.; Chung, W.G.; Lee, J.; Cho, S.-W.; et al. Multimodal Characterization of Cardiac Organoids Using Integrations of Pressure-Sensitive Transistor Arrays with Three-Dimensional Liquid Metal Electrodes. Nano Lett. 2022, 22, 7892–7901. [Google Scholar] [CrossRef]
- Lind, J.U.; Yadid, M.; Perkins, I.; O’Connor, B.B.; Eweje, F.; Chantre, C.O.; Hemphill, M.A.; Yuan, H.; Campbell, P.H.; Vlassak, J.J.; et al. Cardiac microphysiological devices with flexible thin-film sensors for higher-throughput drug screening. Lab Chip 2017, 17, 3692–3703. [Google Scholar] [CrossRef]
- Kowalczewski, A.; Sun, S.; Mai, N.Y.; Song, Y.; Hoang, P.; Liu, X.; Yang, H.; Ma, Z. Design optimization of geometrically confined cardiac organoids enabled by machine learning techniques. Cell Rep. Methods 2024, 4, 100798. [Google Scholar] [CrossRef] [PubMed]
- Belzil, A.; Gélinas, R.; Comtois, P. Development of a high-speed imaging system for real time evaluation and monitoring of cardiac engineered tissues. Front. Bioeng. Biotechnol. 2024, 12, 1403044. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.S.; Sundaram, S.; Lou, L.; Agarwal, A.; Chen, C.S.; Bifano, T.G. High throughput screening system for engineered cardiac tissues. Front. Bioeng. Biotechnol. 2023, 11, 1177688. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Ren, C.; Wang, F.; Park, K.; Volmert, B.D.; Aguirre, A.; Zhou, C. Dual-modality imaging system for monitoring human heart organoids beating in vitro. Opt. Lett. 2023, 48, 3929–3932. [Google Scholar] [CrossRef]
- Ming, Y.; Hao, S.; Wang, F.; Lewis-Israeli, Y.R.; Volmert, B.D.; Xu, Z.; Goestenkors, A.; Aguirre, A.; Zhou, C. Longitudinal morphological and functional characterization of human heart organoids using optical coherence tomography. Biosens. Bioelectron. 2022, 207, 114136. [Google Scholar] [CrossRef]
- Seguret, M.; Davidson, P.; Robben, S.; Jouve, C.; Pereira, C.; Lelong, Q.; Deshayes, L.; Cerveau, C.; Le Berre, M.; Ribeiro, R.S.R.; et al. A versatile high-throughput assay based on 3D ring-shaped cardiac tissues generated from human induced pluripotent stem cell-derived cardiomyocytes. eLife 2024, 12, RP87739. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, G.; Ma, T.; Simmons, C.A.; Santerre, J.P. A critical review on advances and challenges of bioprinted cardiac patches. Acta Biomater. 2024, 189, 1–24. [Google Scholar] [CrossRef]
- Roche, C.D.; Brereton, R.J.L.; Ashton, A.W.; Jackson, C.; Gentile, C. Current challenges in three-dimensional bioprinting heart tissues for cardiac surgery. Eur. J. Cardio-Thorac. Surg. 2020, 58, 500–510. [Google Scholar] [CrossRef]
- Tabury, K.; Rehnberg, E.; Baselet, B.; Baatout, S.; Moroni, L. Bioprinting of Cardiac Tissue in Space: Where Are We? Adv. Healthc. Mater. 2023, 12, 2203338, Correction in Adv. Healthc. Mater. 2025, e04518. https://doi.org/10.1002/adhm.202504518.. [Google Scholar] [CrossRef]
- Zuppinger, C. 3D Cardiac Cell Culture: A Critical Review of Current Technologies and Applications. Front. Cardiovasc. Med. 2019, 6, 87. [Google Scholar] [CrossRef]
- Mannhardt, I.; Saleem, U.; Benzin, A.; Schulze, T.; Klampe, B.; Eschenhagen, T.; Hansen, A. Automated Contraction Analysis of Human Engineered Heart Tissue for Cardiac Drug Safety Screening. J. Vis. Exp. JoVE 2017, 122, 55461. [Google Scholar] [CrossRef]
- Mannhardt, I.; Breckwoldt, K.; Letuffe-Brenière, D.; Schaaf, S.; Schulz, H.; Neuber, C.; Benzin, A.; Werner, T.; Eder, A.; Schulze, T.; et al. Human Engineered Heart Tissue: Analysis of Contractile Force. Stem Cell Rep. 2016, 7, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Brenière-Letuffe, D.; Domke-Shibamiya, A.; Hansen, A.; Eschenhagen, T.; Fehse, B.; Riecken, K.; Stenzig, J. Clonal dynamics studied in cultured induced pluripotent stem cells reveal major growth imbalances within a few weeks. Stem Cell Res. Ther. 2018, 9, 165. [Google Scholar] [CrossRef] [PubMed]
- Andrews, M.G.; Kriegstein, A.R. Challenges of Organoid Research. Annu. Rev. Neurosci. 2022, 45, 23–39. [Google Scholar] [CrossRef]
- Mannhardt, I.; Saleem, U.; Mosqueira, D.; Loos, M.F.; Ulmer, B.M.; Lemoine, M.D.; Larsson, C.; Améen, C.; de Korte, T.; Vlaming, M.L.H.; et al. Comparison of 10 Control hPSC Lines for Drug Screening in an Engineered Heart Tissue Format. Stem Cell Rep. 2020, 15, 983–998. [Google Scholar] [CrossRef]
- Ogle, B.M.; Bursac, N.; Domian, I.; Huang, N.F.; Menasché, P.; Murry, C.E.; Pruitt, B.; Radisic, M.; Wu, J.C.; Wu, S.M.; et al. Distilling complexity to advance cardiac tissue engineering. Sci. Transl. Med. 2016, 8, 342ps13. [Google Scholar] [CrossRef]
- Stein, J.M.; Mummery, C.L.; Bellin, M. Engineered models of the human heart: Directions and challenges. Stem Cell Rep. 2021, 16, 2049–2057. [Google Scholar] [CrossRef]
- Sun, X.; Altalhi, W.; Nunes, S.S. Vascularization strategies of engineered tissues and their application in cardiac regeneration. Adv. Drug Deliv. Rev. 2016, 96, 183–194. [Google Scholar] [CrossRef]
- Liew, A.W.L.; Zhang, Y. In vitro pre-vascularization strategies for tissue engineered constructs–Bioprinting and others. Int. J. Bioprint. 2017, 3, 008. [Google Scholar] [CrossRef]
- Li, H.; Shadrin, I.; Helfer, A.; Heman, K.; Rao, L.; Curtis, C.; Palmer, G.M.; Bursac, N. In vitro vascularization improves in vivo functionality of human engineered cardiac tissues. Acta Biomater. 2024; in press. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, M.; Zhang, W.; Liu, H.; Jin, F.; Mao, S.; Han, C.; Wang, X. Mechanical strategies to promote vascularization for tissue engineering and regenerative medicine. Burn. Trauma 2024, 12, tkae039. [Google Scholar] [CrossRef]
- Singh, A.V.; Romeo, A.; Scott, K.; Wagener, S.; Leibrock, L.; Laux, P.; Luch, A.; Kerkar, P.; Balakrishnan, S.; Dakua, S.P.; et al. Emerging Technologies for In Vitro Inhalation Toxicology. Adv. Healthc. Mater. 2021, 10, e2100633. [Google Scholar] [CrossRef]
- Singh, A.V.; Bhardwaj, P.; Laux, P.; Pradeep, P.; Busse, M.; Luch, A.; Hirose, A.; Osgood, C.J.; Stacey, M.W. AI and ML-based risk assessment of chemicals: Predicting carcinogenic risk from chemical-induced genomic instability. Front. Toxicol. 2024, 6, 1461587. [Google Scholar] [CrossRef]
- Chandrasekar, V.; Mohammad, S.; Aboumarzouk, O.; Singh, A.V.; Dakua, S.P. Quantitative prediction of toxicological points of departure using two-stage machine learning models: A new approach methodology (NAM) for chemical risk assessment. J. Hazard. Mater. 2025, 487, 137071, Correction in J. Hazard. Mater. 2025, 497, 139853. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Yin, J.; Lu, X.; Jiang, D.; Chen, R.; Cui, K.; He, W.; Huang, N.; Xu, G. Influence of experimental variables on spheroid attributes. Sci. Rep. 2025, 15, 9751. [Google Scholar] [CrossRef]
- van Neste, C.C.; Wiley, K.A.; Chang, S.W.; Borrello, J.; Turnbull, I.C.; Costa, K.D. Designing a Bioreactor to Improve Data Acquisition and Model Throughput of Engineered Cardiac Tissues. J. Vis. Exp. JoVE 2023, 196, e64368, Erratum in J. Vis. Exp. JoVE 2024, 203, 10.3791/6589.. [Google Scholar] [CrossRef] [PubMed]
- Visone, R.; Talò, G.; Lopa, S.; Rasponi, M.; Moretti, M. Enhancing all-in-one bioreactors by combining interstitial perfusion, electrical stimulation, on-line monitoring and testing within a single chamber for cardiac constructs. Sci. Rep. 2018, 8, 16944. [Google Scholar] [CrossRef]
- Hollweck, T.; Akra, B.; Häussler, S.; Überfuhr, P.; Schmitz, C.; Pfeifer, S.; Eblenkamp, M.; Wintermantel, E.; Eissner, G. A Novel Pulsatile Bioreactor for Mechanical Stimulation of Tissue Engineered Cardiac Constructs. J. Funct. Biomater. 2011, 2, 107–118. [Google Scholar] [CrossRef]
- Lei, W.; Yuan, M.; Long, M.; Zhang, T.; Huang, Y.; Liu, H.; Jiang, W. scDR: Predicting Drug Response at Single-Cell Resolution. Genes 2023, 14, 268. [Google Scholar] [CrossRef] [PubMed]
- Biendarra-Tiegs, S.M.; Li, X.; Ye, D.; Brandt, E.B.; Ackerman, M.J.; Nelson, T.J. Single-Cell RNA-Sequencing and Optical Electrophysiology of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes Reveal Discordance Between Cardiac Subtype-Associated Gene Expression Patterns and Electrophysiological Phenotypes. Stem Cells Dev. 2019, 28, 659–673. [Google Scholar] [CrossRef]
- Su, X.; Wang, L.; Ma, N.; Yang, X.; Liu, C.; Yang, F.; Li, J.; Yi, X.; Xing, Y. Immune heterogeneity in cardiovascular diseases from a single-cell perspective. Front. Cardiovasc. Med. 2023, 10, 1057870. [Google Scholar] [CrossRef]
- Regev, A.; Teichmann, S.A.; Lander, E.S.; Amit, I.; Benoist, C.; Birney, E.; Bodenmiller, B.; Campbell, P.; Carninci, P.; Clatworthy, M.; et al. The Human Cell Atlas. eLife 2017, 6, e27041. [Google Scholar] [CrossRef]
- Asp, M.; Giacomello, S.; Larsson, L.; Wu, C.; Fürth, D.; Qian, X.; Wärdell, E.; Custodio, J.; Reimegård, J.; Salmén, F.; et al. A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart. Cell 2019, 179, 1647–1660.e19. [Google Scholar] [CrossRef]
- Nguyen, Q.; Tung, L.W.; Lin, B.; Sivakumar, R.; Sar, F.; Singhera, G.; Wang, Y.; Parker, J.; Le Bihan, S.; Singh, A.; et al. Spatial Transcriptomics in Human Cardiac Tissue. Int. J. Mol. Sci. 2025, 26, 995. [Google Scholar] [CrossRef]
- Farah, E.N.; Diaz, J.T.; Bloomekatz, J.; Chi, N.C. Charting the cardiac landscape: Advances in spatial transcriptomics for heart biology. Semin. Cell Dev. Biol. 2025, 175, 103648. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.; Frisén, J.; Lundeberg, J. Spatially resolved transcriptomics adds a new dimension to genomics. Nat. Methods 2021, 18, 15–18. [Google Scholar] [CrossRef]
- Litviňuková, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Worth, C.L.; Lindberg, E.L.; Kanda, M.; Polanski, K.; Heinig, M.; Lee, M.; et al. Cells of the adult human heart. Nature 2020, 588, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Lázár, E.; Mauron, R.; Andrusivová, Ž.; Foyer, J.; He, M.; Larsson, L.; Shakari, N.; Salas, S.M.; Avenel, C.; Sariyar, S.; et al. Spatial Dynamics of the Developing Human Heart. bioRxiv 2024. [Google Scholar] [CrossRef]
- Rodriques, S.G.; Stickels, R.R.; Goeva, A.; Martin, C.A.; Murray, E.; Vanderburg, C.R.; Welch, J.; Chen, L.M.; Chen, F.; Macosko, E.Z. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 2019, 363, 1463–1467. [Google Scholar] [CrossRef] [PubMed]
- Vickovic, S.; Eraslan, G.; Salmén, F.; Klughammer, J.; Stenbeck, L.; Schapiro, D.; Äijö, T.; Bonneau, R.; Bergenstråhle, L.; Navarro, J.F.; et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 2019, 16, 987–990. [Google Scholar] [CrossRef] [PubMed]
- Kanemaru, K.; Cranley, J.; Muraro, D.; Miranda, A.M.A.; Ho, S.Y.; Wilbrey-Clark, A.; Patrick Pett, J.; Polanski, K.; Richardson, L.; Litvinukova, M.; et al. Spatially resolved multiomics of human cardiac niches. Nature 2023, 619, 801–810, Correction in Nature 2025, 640, E4.. [Google Scholar] [CrossRef] [PubMed]
- Vyas, V.; Sandhar, B.; Keane, J.M.; Wood, E.G.; Blythe, H.; Jones, A.; Shahaj, E.; Fanti, S.; Williams, J.; Metic, N.; et al. Tissue-resident memory T cells in epicardial adipose tissue comprise transcriptionally distinct subsets that are modulated in atrial fibrillation. Nat. Cardiovasc. Res. 2024, 3, 1067–1082. [Google Scholar] [CrossRef] [PubMed]
- Amrute, J.M.; Luo, X.; Penna, V.; Yang, S.; Yamawaki, T.; Hayat, S.; Bredemeyer, A.; Jung, I.-H.; Kadyrov, F.F.; Heo, G.S.; et al. Targeting immune-fibroblast cell communication in heart failure. Nature 2024, 635, 423–433. [Google Scholar] [CrossRef]
- Kuppe, C.; Ramirez Flores, R.O.; Li, Z.; Hayat, S.; Levinson, R.T.; Liao, X.; Hannani, M.T.; Tanevski, J.; Wünnemann, F.; Nagai, J.S.; et al. Spatial multi-omic map of human myocardial infarction. Nature 2022, 608, 766–777. [Google Scholar] [CrossRef]
- Linna-Kuosmanen, S.; Schmauch, E.; Galani, K.; Ojanen, J.; Boix, C.A.; Örd, T.; Toropainen, A.; Singha, P.K.; Moreau, P.R.; Harju, K.; et al. Transcriptomic and spatial dissection of human ex vivo right atrial tissue reveals proinflammatory microvascular changes in ischemic heart disease. Cell Rep. Med. 2024, 5, 101556. [Google Scholar] [CrossRef]
- Gastanadui, M.G.; Margaroli, C.; Litovsky, S.; Richter, R.P.; Wang, D.; Xing, D.; Wells, J.M.; Gaggar, A.; Nanda, V.; Patel, R.P.; et al. Spatial Transcriptomic Approach to Understanding Coronary Atherosclerotic Plaque Stability. Arterioscler. Thromb. Vasc. Biol. 2024, 44, e264–e276. [Google Scholar] [CrossRef]
- Ninh, V.K.; Calcagno, D.M.; Yu, J.D.; Zhang, B.; Taghdiri, N.; Sehgal, R.; Mesfin, J.M.; Chen, C.J.; Kalhor, K.; Toomu, A.; et al. Spatially clustered type I interferon responses at injury borderzones. Nature 2024, 633, 174–181. [Google Scholar] [CrossRef]
| Technique | Resolution | Range of Compatible Material | Scalability | Advantages | Limitations |
|---|---|---|---|---|---|
| Extrusion-Based Bioprinting | 200–500 µm filament precision [32] | Broad (viscous, cell-dense bioinks) [33] | Moderate (can be adjusted with automation) | Suitable for facilitating anisotropic filament alignment [34], supports high cell density and scalability [35], automation capability [33], ease of operation [35], affordable [33] | Shear stress can cause damage to cells, difficult to get lower resolution, needle clogging [35,36] |
| Laser-Assisted Bioprinting | 20–100 µm [37,38] | Low viscosity bioinks [39] | Low; suitable for patterning but limited in bulk tissue [40] | Relatively fast deposition, high cell viability, heterocellular patterning, multi-material printing [38], nozzle-free technique that prevents clumps [41] | Weak in mechanical stability, can cause thermal damage to cells [39] |
| Stereolithography/Digital Light Processing | 10 [42]–50 µm [43] (DLP) | Photo-cross linkable bioinks | High; may slow down with larger constructs | High speed and fidelity, suitable for complex microfeatures [43], large volume of initial bioink required [44] | Curing by UV can be detrimental to cells, light penetration is limited to the thickness of the construct, larger and softer structures are prone to deformation [43] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, S.; Laksman, Z. Integrating Bioprinting and Increased Throughput: Next-Generation Models for Cardiac Research. Int. J. Mol. Sci. 2025, 26, 11589. https://doi.org/10.3390/ijms262311589
Nguyen S, Laksman Z. Integrating Bioprinting and Increased Throughput: Next-Generation Models for Cardiac Research. International Journal of Molecular Sciences. 2025; 26(23):11589. https://doi.org/10.3390/ijms262311589
Chicago/Turabian StyleNguyen, Stephanie, and Zachary Laksman. 2025. "Integrating Bioprinting and Increased Throughput: Next-Generation Models for Cardiac Research" International Journal of Molecular Sciences 26, no. 23: 11589. https://doi.org/10.3390/ijms262311589
APA StyleNguyen, S., & Laksman, Z. (2025). Integrating Bioprinting and Increased Throughput: Next-Generation Models for Cardiac Research. International Journal of Molecular Sciences, 26(23), 11589. https://doi.org/10.3390/ijms262311589






