Multimodal Integration of Genomic Data Reveals Regulatory Mechanisms at the Polycystic Ovary Syndrome (PCOS)-Associated 12q13.2 Locus
Abstract
1. Introduction
2. Results
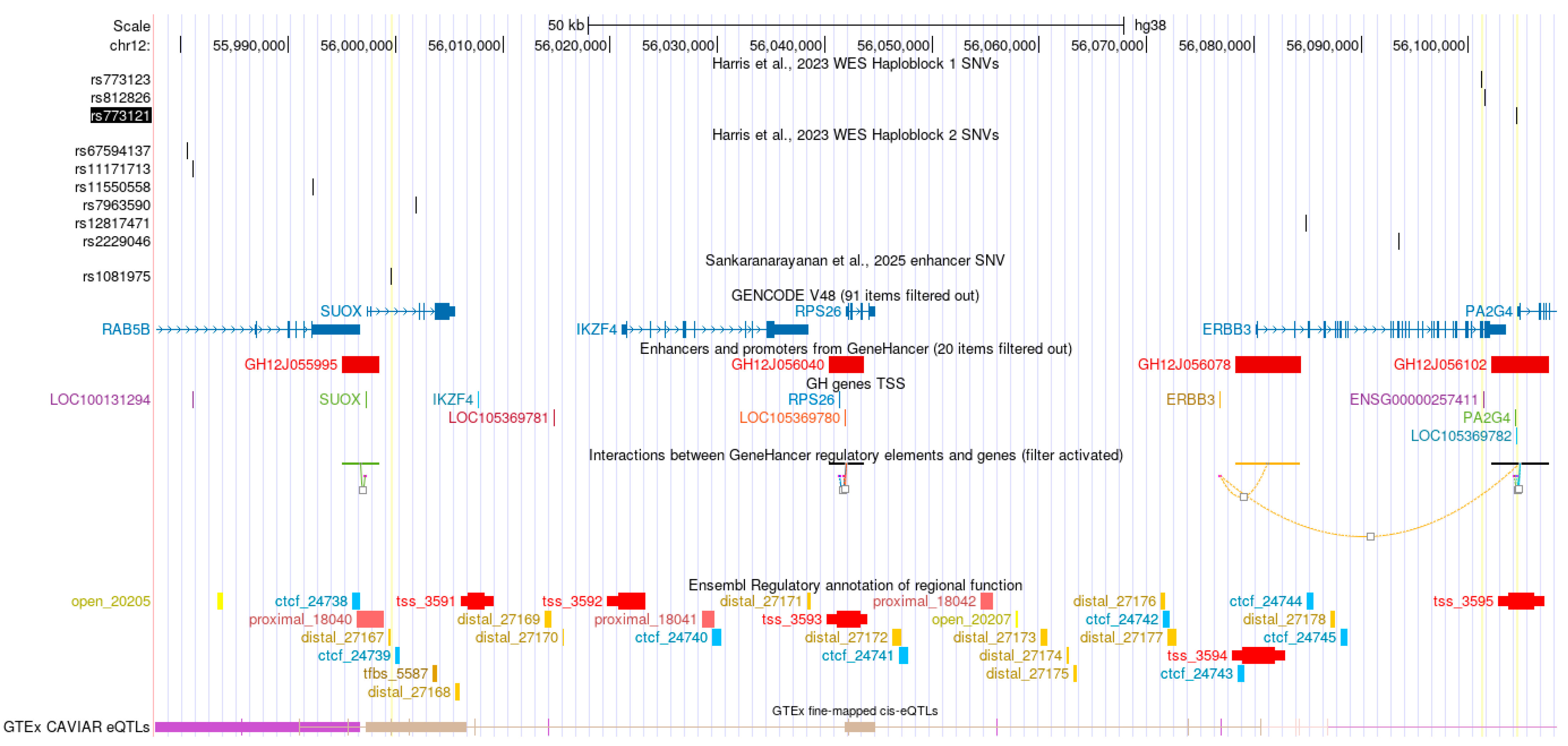
2.1. Genetic Associations at 12q13.2
2.2. Enhancer Activity and eQTL Colocalization
2.3. Single-Cell Expression Patterns
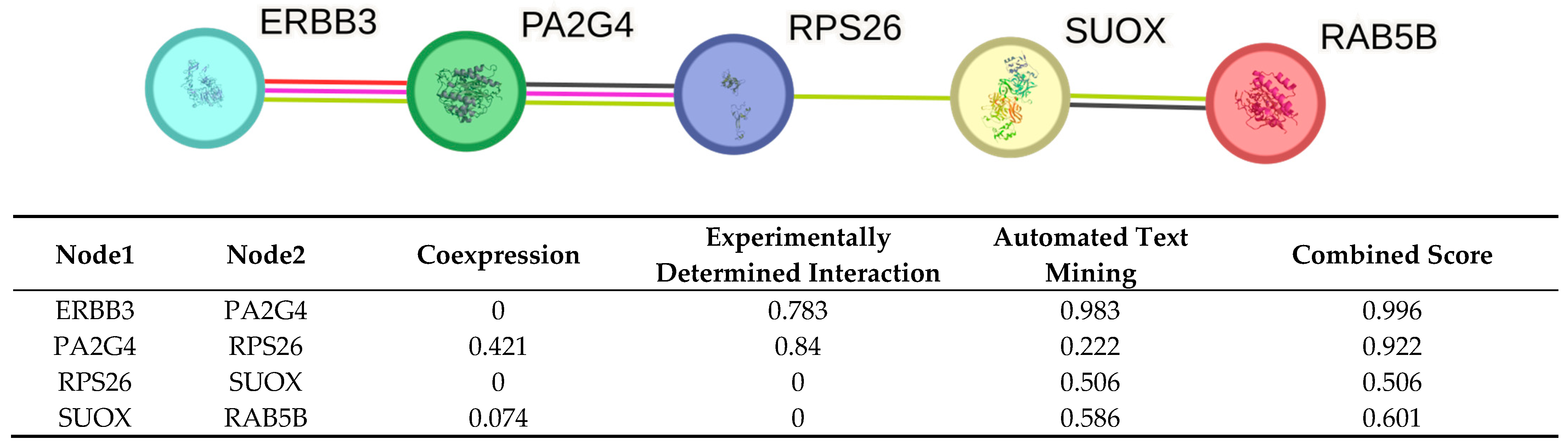
2.4. STRING Analysis
- ERBB3–PA2G4 (0.996): Despite no coexpression evidence, the very high experimental and text mining scores yield a near maximal combined score. This suggests a well validated association between ERBB3 and PA2G4, heavily supported by curated experiments and literature.
- PA2G4–RPS26 (0.922): Moderate coexpression and strong experimental support results in a high combined score. Text mining contributes less, indicating fewer co-mentions.
- RPS26–SUOX (0.506): With no coexpression or experimental support, this association relies solely on text mining at a borderline combined score. This suggests a putative link from literature.
- SUOX–RAB5B (0.601): This pair has low coexpression and no direct experimental evidence. The interaction is inferred primarily through literature co-mentions (0.586), yielding a moderate combined score.
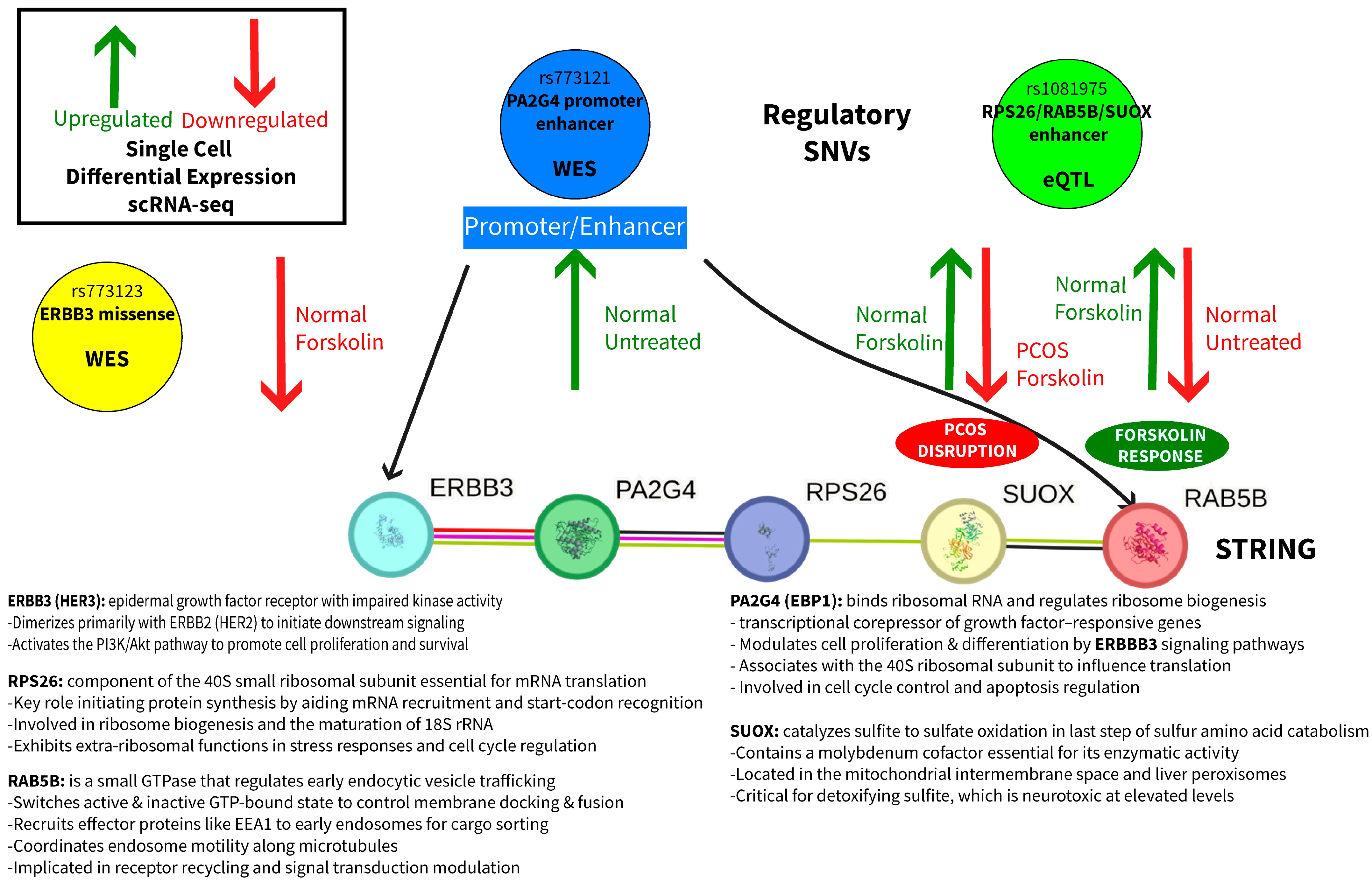
2.5. Integrated Mechanistic Model
- Genetic: SNVs in a promoter/enhancer (rs773121) and enhancer (rs1081975) affect baseline gene expression and forskolin treatment responsiveness.
- Regulatory disruption: Promoter/enhancer SNVs alter the normal coordination between RAB5B vesicular trafficking, PA2G4 androgen receptor regulation, and SUOX metabolic functions.
- Cellular dysfunction: Loss of coordinated responses to hormonal stimulation results in impaired androgen regulation and cellular homeostasis in PCOS theca cells.
3. Discussion
4. Materials and Methods
- WES-identified SNVs from PCOS and normal theca cells with association testing for forskolin-stimulated androgen production (Harris, et al., 2023 [6])
- STARR-seq enhancer activity data with eQTL colocalization analysis (Sankaranarayanan, et al., 2025 [11])
- scRNA-seq expression data comparing forskolin-stimulated PCOS and normal theca cells (Harris, et al., 2023 [10])
Harris, et al., 2023 [6]—Loci on chromosome 12q13.2 encompassing ERBB3, PA2G4, and RAB5B are associated with polycystic ovary syndrome.
4.1. Theca Cell Preparations and Culture
4.2. Whole Exome Sequence Analysis of Normal and PCOS Theca Cell DNA
4.3. Linkage Disequilibrium
4.4. Statistical Analysis
4.5. SNV Analyses of a PCOS Cohort
4.6. Functional Annotations
Sankaranarayanan, et al. 2025 [11]—Gene regulatory activity associated with polycystic ovary syndrome revealed DENND1A-dependent testosterone production.
4.7. Selection of GWAS Regions for Targeted STARR-Seq Assays
4.8. STARR-Seq Reporter Plasmid Construction
4.9. STARR-Seq Assay Library Sequencing
4.10. Alignments and STARR-Seq Data Analysis
4.11. PCOS Case-Control Variant Association Testing Within STARR-Seq Regions
4.12. Colocalization Testing
Harris, et al., 2023 [10]—Single-Cell RNA-Seq Identifies Pathways and Genes Contributing to the Hyperandrogenemia Associated with Polycystic Ovary Syndrome.
4.13. Theca Cell Preparations and Culture
4.14. Single-Cell RNA Sequencing (scRNA-Seq)
4.15. Cell Ranger Data Processing
4.16. Seurat Pre-Processing and Quality Control
4.17. STACAS Integration
4.18. Identification of Differentially Expressed Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dapas, M.; Dunaif, A. Deconstructing a Syndrome: Genomic Insights into PCOS Causal Mechanisms and Classification. Endocr. Rev. 2022, 43, 927–965. [Google Scholar] [CrossRef]
- Vink, J.M.; Sadrzadeh, S.; Lambalk, C.B.; Boomsma, D.I. Heritability of Polycystic Ovary Syndrome in a Dutch Twin-Family Study. J. Clin. Endocrinol. Metab. 2006, 91, 2100–2104. [Google Scholar] [CrossRef]
- Ewens, K.G.; Stewart, D.R.; Ankener, W.; Urbanek, M.; McAllister, J.M.; Baig, K.M.; Parker, S.C.J.; Margulies, E.H.; Legro, R.S.; Dunaif, A.; et al. Family-Based Analysis of Candidate Genes for Polycystic Ovary Syndrome. J. Clin. Endocrinol. Metab. 2010, 95, 2306–2315. [Google Scholar] [CrossRef]
- Chen, Z.J.; Zhao, H.; He, L.; Shi, Y.; Qin, Y.; Shi, Y.; Li, Z.; You, L.; Zhao, J.; Liu, J.; et al. Genome-Wide Association Study Identifies Susceptibility Loci for Polycystic Ovary Syndrome on Chromosome 2p16.3, 2p21 and 9q33.3. Nat. Genet. 2011, 43, 55–59. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, H.; Shi, Y.; Cao, Y.; Yang, D.; Li, Z.; Zhang, B.; Liang, X.; Li, T.; Chen, J.; et al. Genome-Wide Association Study Identifies Eight New Risk Loci for Polycystic Ovary Syndrome. Nat. Genet. 2012, 44, 1020–1025. [Google Scholar] [CrossRef]
- Harris, R.A.; Archer, K.J.; Goodarzi, M.O.; York, T.P.; Rogers, J.; Dunaif, A.; McAllister, J.M.; Strauss, J.F. Loci on Chromosome 12q13.2 Encompassing ERBB3, PA2G4 and RAB5B Are Associated with Polycystic Ovary Syndrome. Gene 2023, 852, 147062. [Google Scholar] [CrossRef] [PubMed]
- Nelson, V.L.; Legro, R.S.; Strauss, J.F., 3rd; McAllister, J.M. Augmented Androgen Production Is a Stable Steroidogenic Phenotype of Propagated Theca Cells from Polycystic Ovaries. Mol. Endocrinol. 1999, 13, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.R.; Nelson, V.L.; Ho, C.; Jansen, E.; Wang, C.Y.; Urbanek, M.; McAllister, J.M.; Mosselman, S.; Strauss, J.F. The Molecular Phenotype of Polycystic Ovary Syndrome (PCOS) Theca Cells and New Candidate PCOS Genes Defined by Microarray Analysis. J. Biol. Chem. 2003, 278, 26380–26390. [Google Scholar] [CrossRef] [PubMed]
- McAllister, J.M.; Modi, B.; Miller, B.A.; Biegler, J.; Bruggeman, R.; Legro, R.S.; Strauss, J.F. Overexpression of a DENND1A Isoform Produces a Polycystic Ovary Syndrome Theca Phenotype. Proc. Natl. Acad. Sci. USA 2014, 111, E1519–E1527. [Google Scholar] [CrossRef]
- Harris, R.A.; McAllister, J.M.; Strauss, J.F. Single-Cell RNA-Seq Identifies Pathways and Genes Contributing to the Hyperandrogenemia Associated with Polycystic Ovary Syndrome. Int. J. Mol. Sci. 2023, 24, 10611. [Google Scholar] [CrossRef]
- Sankaranarayanan, L.; Brewer, K.J.; Morrow, S.; Johnson, G.D.; Barrera, A.; Venukuttan, R.; Sisk, R.; Dunaif, A.; Reddy, T.E. Gene Regulatory Activity Associated with Polycystic Ovary Syndrome Revealed DENND1A-Dependent Testosterone Production. Nat. Commun. 2025, 16, 7697. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, M.; Della Corte, L.; Colacurci, D.; Ascione, M.; D’Angelo, G.; Baldini, G.M.; Giampaolino, P.; Bifulco, G. PCOS and the Genome: Is the Genetic Puzzle Still Worth Solving? Biomedicines 2025, 13, 1912. [Google Scholar] [CrossRef]
- Perez, G.; Barber, G.P.; Benet-Pages, A.; Casper, J.; Clawson, H.; Diekhans, M.; Fischer, C.; Gonzalez, J.N.; Hinrichs, A.S.; Lee, C.M.; et al. The UCSC Genome Browser Database: 2025 Update. Nucleic Acids Res. 2025, 53, D1243–D1249. [Google Scholar] [CrossRef]
- Dapas, M.; Sisk, R.; Legro, R.S.; Urbanek, M.; Dunaif, A.; Hayes, M.G. Family-Based Quantitative Trait Meta-Analysis Implicates Rare Noncoding Variants in DENND1A in Polycystic Ovary Syndrome. J. Clin. Endocrinol. Metab. 2019, 104, 3835–3850. [Google Scholar] [CrossRef]
- Horvath, S.; Xu, X.; Laird, N.M. The Family Based Association Test Method: Strategies for Studying General Genotype--Phenotype Associations. Eur. J. Hum. Genet. 2001, 9, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Fishilevich, S.; Nudel, R.; Rappaport, N.; Hadar, R.; Plaschkes, I.; Stein, T.I.; Rosen, N.; Kohn, A.; Twik, M.; Safran, M.; et al. GeneHancer: Genome-Wide Integration of Enhancers and Target Genes in GeneCards. Database 2017, 2017, bax028. [Google Scholar] [CrossRef] [PubMed]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the Deleteriousness of Variants throughout the Human Genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Johnson, N.; Juetteman, T.; Sheppard, D.; Wilder, S.P.; Lavidas, I.; Nuhn, M.; Perry, E.; Raffaillac-Desfosses, Q.; Sobral, D.; et al. Ensembl Regulation Resources. Database 2016, 2016, bav119. [Google Scholar] [CrossRef]
- Aguet, F.; Barbeira, A.N.; Bonazzola, R.; Brown, A.; Castel, S.E.; Jo, B.; Kasela, S.; Kim-Hellmuth, S.; Liang, Y.; Oliva, M.; et al. The GTEx Consortium Atlas of Genetic Regulatory Effects across Human Tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Kulkarni, R.; Teves, M.E.; Han, A.X.; McAllister, J.M.; Strauss, J.F., 3rd. Colocalization of Polycystic Ovary Syndrome Candidate Gene Products in Theca Cells Suggests Novel Signaling Pathways. J. Endocr. Soc. 2019, 3, 2204–2223. [Google Scholar] [CrossRef]
- Townsend, H.A.; Rosenberger, K.J.; Vanderlinden, L.A.; Inamo, J.; Zhang, F. Evaluating Methods for Integrating Single-Cell Data and Genetics to Understand Inflammatory Disease Complexity. Front. Immunol. 2024, 15, 1454263. [Google Scholar] [CrossRef] [PubMed]
- Alamin, M.; Humaira Sultana, M.; Babarinde, I.A.; Azad, A.K.M.; Moni, M.A.; Xu, H. Single-Cell RNA-Seq Data Analysis Reveals Functionally Relevant Biomarkers of Early Brain Development and Their Regulatory Footprints in Human Embryonic Stem Cells (HESCs). Brief Bioinform. 2024, 25, bbae230. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein-Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Shi, J.; Gao, Q.; Cao, Y.; Fu, J. Dennd1a, a Susceptibility Gene for Polycystic Ovary Syndrome, Is Essential for Mouse Embryogenesis. Dev. Dyn. 2019, 248, 351–362. [Google Scholar] [CrossRef]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated Analysis of Multimodal Single-Cell Data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef] [PubMed]
- Andreatta, M.; Carmona, S.J. STACAS: Sub-Type Anchor Correction for Alignment in Seurat to Integrate Single-Cell RNA-Seq Data. Bioinformatics 2021, 37, 882–884. [Google Scholar] [CrossRef] [PubMed]
| Gene | Condition | Average log2 Fold Change | Adjusted p Value | Regulation vs. All Other Conditions |
|---|---|---|---|---|
| RAB5B | Normal Untreated | −0.084 | 3.51 × 10−48 | Downregulated |
| RAB5B | Normal Forskolin | 0.093 | 5.70 × 10−6 | Upregulated |
| SUOX | Normal Forskolin | 0.021 | 2.72 × 10−16 | Upregulated |
| SUOX | PCOS Forskolin | −0.018 | 8.28 × 10−167 | Downregulated |
| PA2G4 | Normal Untreated | 0.038 | 0.0047 | Upregulated |
| ERBB3 | Normal Forskolin | −0.011 | 1.07 × 10−252 | Downregulated |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harris, R.A.; McAllister, J.M.; Strauss, J.F., III. Multimodal Integration of Genomic Data Reveals Regulatory Mechanisms at the Polycystic Ovary Syndrome (PCOS)-Associated 12q13.2 Locus. Int. J. Mol. Sci. 2025, 26, 11184. https://doi.org/10.3390/ijms262211184
Harris RA, McAllister JM, Strauss JF III. Multimodal Integration of Genomic Data Reveals Regulatory Mechanisms at the Polycystic Ovary Syndrome (PCOS)-Associated 12q13.2 Locus. International Journal of Molecular Sciences. 2025; 26(22):11184. https://doi.org/10.3390/ijms262211184
Chicago/Turabian StyleHarris, R. Alan, Jan M. McAllister, and Jerome F. Strauss, III. 2025. "Multimodal Integration of Genomic Data Reveals Regulatory Mechanisms at the Polycystic Ovary Syndrome (PCOS)-Associated 12q13.2 Locus" International Journal of Molecular Sciences 26, no. 22: 11184. https://doi.org/10.3390/ijms262211184
APA StyleHarris, R. A., McAllister, J. M., & Strauss, J. F., III. (2025). Multimodal Integration of Genomic Data Reveals Regulatory Mechanisms at the Polycystic Ovary Syndrome (PCOS)-Associated 12q13.2 Locus. International Journal of Molecular Sciences, 26(22), 11184. https://doi.org/10.3390/ijms262211184






