Development and Optimization of an RNA-Isolating Protocol for Mammalian Spermatozoa
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
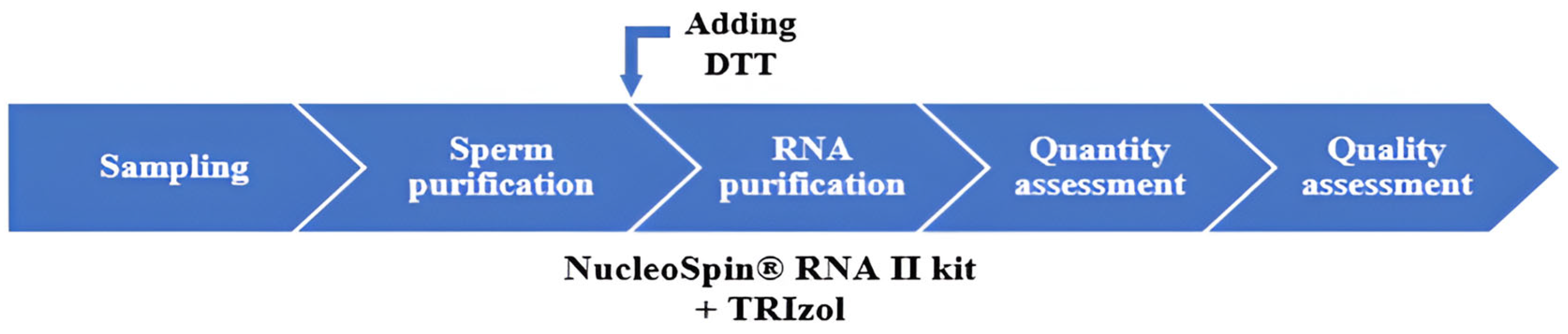
4.1. Sampling
4.2. Sperm Cell Purification via Density Gradient
4.3. Sperm RNA Purification
- 1.
- DTT (100 mM final concentration) was added to the RA1 lysis buffer to reduce protamine disulfide bonds. This promoted nuclear decompaction and facilitated the release of RNAs.
- 2.
- Prior to silica column purification, the samples were incubated with the TRIzol™ reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) to enhance membrane lysis, inhibit RNases, and stabilize the RNAs during extraction.
4.4. RNA Purification Protocol
- -
- Pre-lysis of Sperm Cells: Each sample was centrifuged at 13,000 rpm for 5 min, and then 350 µL of RA1 buffer from the NucleoSpin RNA II kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany) and 70 µL of DTT (100 mM) were added to the pellet. The mixture was incubated and vortexed at room temperature for 4 min.
- -
- TRIzol/Chloroform Treatment: A total of 400 µL of the TRIzol reagent was added to each biological sample; the mixture was then shaken with a vortex for 10 min at room temperature. Next, 300 µL of chloroform (Sigma-Aldrich, St. Louis, MO, USA) was added and the sample was vortexed for 15 to 30 s. The sample was then centrifuged at 13,000 rpm for 4 min to separate the aqueous and organic phases.
- -
- Nucleic Acid Precipitation: The aqueous phase was transferred to a sterile 1.5 mL tube, and 350 µL of 70% ethanol was added to precipitate nucleic acids, followed by mixing briefly with a vortex.
- -
- Column Binding: A total of 600 µL of the precipitated sample was loaded onto a purification column (blue) placed in a collection tube from the NucleoSpin RNA II kit. This was centrifuged at 8000 rpm for 30 s to allow for nucleic acid adsorption onto the column membrane. The lysate was then discarded, and this step was repeated for the remaining sample volume.
- -
- Membrane Desalting: A total of 350 µL of MDB desalting solution from the NucleoSpin RNA II kit was added to the column. Then, the mixture was centrifuged at 11,000 rpm for 1 min to remove saline impurities.
- -
- DNase Treatment: The DNase mixture was prepared by adding 10 µL of reconstituted DNase I to 90 µL of DNase buffer. Next, 95 µL of this DNase mixture was added to the purification column to degrade the residual genomic DNA, and the mixture was incubated for 15 min at room temperature.
- -
- Washing Steps: A total of 200 µL of RA2 wash solution from the NucleoSpin RNA II kit was added to the sample, and the mixture was centrifuged at 8000 rpm for 1 min. The filtrate was then discarded and the column was replaced in a new collection tube. Next, 600 µL of RA3 wash solution from the kit was added, the sample was centrifuged at 8000 rpm for 1 min, the filtrate was discarded, and the column was replaced in the collection tube. The washing step was repeated by adding 500 µL of RA3 and centrifuging at 8000 rpm for 1 min. The filtrate was then discarded and the column was replaced in a new collection tube.
- -
- Drying step: The tube was centrifuged for 2 min at 13,000 rpm to completely dry the column membrane.
- -
- Elution: During the drying step, the DNase/RNase-free water of the NucleoSpin RNA II kit was heated in a water bath at 55 °C for approximately 5 min to enhance the RNA elution efficiency. Next, 40 to 60 µL of DNase/RNase-free water was added to the center of the column for RNA elution. The sample was incubated at room temperature for 2 min and then centrifuged for 1 min at 11,000 rpm to increase the RNA yield. The RNA solution was stored at −80 °C until use.
4.5. Evaluation of Sperm RNA Quantity and Quality
Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Larose, H.; Shami, A.N.; Abbott, H.; Manske, G.; Lei, L.; Hammoud, S.S. Gametogenesis: A journey from inception to conception. Curr. Top. Dev. Biol. 2019, 132, 257–310. [Google Scholar] [CrossRef] [PubMed]
- Cannarella, R.; Crafa, A.; Curto, R.; Mongioì, L.M.; Garofalo, V.; Cannarella, V.; Condorelli, R.A.; La Vignera, S.; Calogero, A.E. Human sperm RNA in male infertility. Nat. Rev. Urol. 2025, 22, 92–115. [Google Scholar] [CrossRef] [PubMed]
- Braun, R.E. Post-transcriptional control of gene expression during spermatogenesis. Semin. Cell Dev. Biol. 1998, 9, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Eddy, E.M. Male germ cell gene expression. Recent Prog. Horm. Res. 2002, 57, 103–128. [Google Scholar] [CrossRef]
- Jodar, M.; Kalko, S.; Castillo, J.; Ballescà, J.L.; Oliva, R. Differential RNAs in the sperm cells of asthenozoospermic patients. Hum. Reprod. 2012, 27, 1431–1438. [Google Scholar] [CrossRef]
- Johnson, G.D.; Lalancette, C.; Linnemann, A.K.; Leduc, F.; Boissonneault, G.; Krawetz, S.A. The sperm nucleus: Chromatin, RNA, and the nuclear matrix. Reproduction 2011, 141, 21–36. [Google Scholar] [CrossRef]
- Flegel, C.; Vogel, F.; Hofreuter, A.; Schreiner, B.S.P.; Osthold, S.; Veitinger, S.; Becker, C.; Brockmeyer, N.H.; Muschol, M.; Wennemuth, G.; et al. Characterization of the olfactory receptors expressed in human spermatozoa. Front. Mol. Biosci. 2016, 2, 73. [Google Scholar] [CrossRef]
- Montjean, D.; Neyroud, A.S.; Yefimova, M.G.; Benkhalifa, M.; Cabry, R.; Ravel, C. Impact of endocrine disruptors upon non-genetic inheritance. Int. J. Mol. Sci. 2022, 23, 3350. [Google Scholar] [CrossRef]
- Joshi, M.; Sethi, S.; Mehta, P.; Kumari, A.; Singh, R. Small RNAs, spermatogenesis, and male infertility: A decade of retrospect. Reprod. Biol. Endocrinol. 2023, 21, 103. [Google Scholar] [CrossRef]
- Yan, Q.; Wang, Q. Exploring the Characters of Non-Coding RNAs in Spermatogenesis and Male Infertility. Int. J. Mol. Sci. 2025, 26, 1128. [Google Scholar] [CrossRef]
- Krawetz, S.A.; Kruger, A.; Lalancette, C.; Tagett, R.; Anton, E.; Draghici, S.; Diamond, M.P. A survey of small RNAs in human sperm. Hum. Reprod. 2011, 26, 3401–3412. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Schuster, A.; Tang, C.; Yu, T.; Ortogero, N.; Bao, J.; Zheng, H.; Yan, W. Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development 2016, 143, 635–647. [Google Scholar] [CrossRef]
- Dadoune, J.P.; Pawlak, A.; Alfonsi, M.F.; Siffroi, J.P. Identification of transcripts by macroarrays, RT-PCR and in situ hybridization in human ejaculate spermatozoa. Mol. Hum. Reprod. 2005, 11, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Patel, D.; Naz, R.K. C-MYC mRNA is present in human sperm cells. Cell. Mol. Biol. Res. 1993, 39, 111–117. [Google Scholar] [PubMed]
- Sendler, E.; Johnson, G.D.; Mao, S.; Goodrich, R.J.; Diamond, M.P.; Hauser, R.; Krawetz, S.A. Stability, delivery and functions of human sperm RNAs at fertilization. Nucleic Acids Res. 2013, 41, 4104–4117. [Google Scholar] [CrossRef]
- Rassoulzadegan, M.; Grandjean, V.; Gounon, P.; Vincent, S.; Gillot, I.; Cuzin, F. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature 2006, 441, 469–474. [Google Scholar] [CrossRef]
- Hamilton, M.; Russell, S.; Swanson, G.M.; Krawetz, S.A.; Menezes, K.; Moskovtsev, S.I.; Librach, C. A comprehensive analysis of spermatozoal RNA elements in idiopathic infertile males undergoing fertility treatment. Sci. Rep. 2024, 14, 10316. [Google Scholar] [CrossRef]
- Mehta, P.; Singh, R. The Composition of Human Sperm sncRNAome: A Cross-Country Small RNA Profiling. Reprod. Biol. Endocrinol. 2025, 23, 36. [Google Scholar] [CrossRef]
- Jodar, M.; Selvaraju, S.; Sendler, E.; Diamond, M.P.; Krawetz, S.A. The presence, role and clinical use of spermatozoal RNAs. Hum. Reprod. Update 2013, 19, 604–624. [Google Scholar] [CrossRef]
- Pessot, C.A.; Brito, M.; Figueroa, J.; Concha, I.I.; Yáñez, A.; Burzio, L.O. Presence of RNA in the sperm nucleus. Biochem. Biophys. Res. Commun. 1989, 158, 272–278. [Google Scholar] [CrossRef]
- Goodrich, R.J.; Anton, E.; Krawetz, S.A. Isolating mRNA and small noncoding RNAs from human sperm. Methods Mol. Biol. 2013, 927, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, Q.; Yao, C.; Wang, Z.; Zhou, Y.; Wang, Y.; Liu, L.; Wang, Y.; Wang, L.; Qiao, Z. Characterization and quantification of mRNA transcripts in ejaculated spermatozoa of fertile men by serial analysis of gene expression. Hum. Reprod. 2006, 21, 1583–1590. [Google Scholar] [CrossRef]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F.; et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016, 351, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Dewry, R.K.; Mohanty, T.K.; Nath, S.; Bhakat, M.; Yadav, H.P.; Baithalu, R.K. Comparative RNA isolation methods from fresh ejaculated spermatozoa in Sahiwal cattle (Bos indicus) and Murrah buffalo (Bubalus bubalis) bulls for high quality and enhanced RNA yield. Anim. Biotechnol. 2023, 34, 5180–5191. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, R.; Johnson, G.; Krawetz, S.A. The preparation of human spermatozoal RNA for clinical analysis. Arch. Androl. 2007, 53, 161–167. [Google Scholar] [CrossRef]
- Selvaraju, S.; Parthipan, S.; Somashekar, L.; Kolte, A.P.; Binsila, B.K.; Arangasamy, A.; Ravindra, J.P. Occurrence and functional significance of the transcriptome in bovine (Bos taurus) spermatozoa. Sci. Rep. 2017, 7, 42392. [Google Scholar] [CrossRef]
- Das, P.J.; Paria, N.; Gustafson-Seabury, A.; Vishnoi, M.; Chaki, S.P.; Love, C.C.; Varner, D.D.; Chowdhary, B.P.; Raudsepp, T. Total RNA isolation from stallion sperm and testis biopsies. Theriogenology 2010, 74, 1099–1106. [Google Scholar] [CrossRef]
- Bianchi, E.; Stermer, A.; Boekelheide, K.; Sigman, M.; Sall, S.J.; Reyes, G.; Dere, E.; Hwang, K. High-quality human and rat spermatozoal RNA isolation for functional genomic studies. Andrology 2018, 6, 374–383. [Google Scholar] [CrossRef]
- Gòdia, M.; Mayer, F.Q.; Nafissi, J.; Castelló, A.; Rodríguez-Gil, J.E.; Sánchez, A.; Clop, A. A technical assessment of the porcine ejaculated spermatozoa for a sperm-specific RNA-seq analysis. Syst. Biol. Reprod. Med. 2018, 64, 291–303. [Google Scholar] [CrossRef]
- Sahoo, B.; Guttula, P.K.; Gupta, M.K. Comparison of spermatozoal RNA extraction methods in goats. Anal. Biochem. 2020, 614, 114059. [Google Scholar] [CrossRef]
- Umar, S.I.U.; Prasad, S.; Naskar, S.; Chowdhury, P.; Rana, A.; Das, P.J. Development and Optimization of an Efficient RNA Isolation Protocol from Bovine (Bos indicus) Spermatozoa. Biochem. Biophys. Rep. 2024, 40, 101862. [Google Scholar] [CrossRef] [PubMed]
- Galibert, F.; Azzouzi, N. Are the olfactory receptors present at the sperm membrane involved in reproduction? Int. J. Mol. Sci. 2023, 24, 11277. [Google Scholar] [CrossRef] [PubMed]
- Georgiadis, A.P.; Kishore, A.; Zorrilla, M.; Jaffe, T.M.; Sanfilippo, J.S.; Volk, E.; Rajkovic, A.; Yatsenko, A.N. High-quality RNA in semen and sperm: Isolation, analysis, and potential application in clinical testing. J. Urol. 2015, 193, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Goodrich, R.J.; Hauser, R.; Schrader, S.M.; Chen, Z.; Krawetz, S.A. Evaluation of the effectiveness of semen storage and sperm purification methods for spermatozoa transcript profiling. Syst. Biol. Reprod. Med. 2013, 59, 287–295. [Google Scholar] [CrossRef]
- Ostermeier, G.C.; Goodrich, R.J.; Moldenhauer, J.S.; Diamond, M.P.; Krawetz, S.A. A suite of novel human spermatozoal RNAs. J. Androl. 2005, 26, 70–74. [Google Scholar] [CrossRef]
- El Fekih, S.; Nguyen, M.-H.; Perrin, A.; Beauvillard, D.; Morel, F.; Saad, A.; Ben Ali, H.; De Braekeleer, M. Sperm RNA preparation for transcriptomic analysis: Review of the techniques and personal experience. Andrologia 2017, 49, e12767. [Google Scholar] [CrossRef]
- Hegedus, R.M.; Donovan, C.E.; Menino, A.R.; Kutzler, M.A. Comparison of canine spermatozoal RNA concentrations and purity using two density gradient centrifugation solutions. Clin. Theriogenol. 2014, 6, 383. [Google Scholar]
- Navarrete-López, P.; Asselstine, V.; Maroto, M.; Lombó, M.; Cánovas, Á.; Gutiérrez-Adán, A. RNA sequencing of sperm from healthy cattle and horses reveals the presence of a large bacterial population. Curr. Issues Mol. Biol. 2024, 46, 10430–10443. [Google Scholar] [CrossRef]
- Gilbert, I.; Bissonnette, N.; Boissonneault, G.; Vallée, M.; Robert, C. A molecular analysis of the population of mRNA in bovine spermatozoa. Reproduction 2007, 133, 1073–1086. [Google Scholar] [CrossRef]
- Shafeeque, C.M.; Singh, R.P.; Sharma, S.K.; Mohan, J.; Sastry, K.V.H.; Kolluri, G.; Saxena, V.K.; Tyagi, J.S.; Kataria, J.M.; Azeez, P.A. Development of a new method for sperm RNA purification in the chicken. Anim. Reprod. Sci. 2014, 149, 259–265. [Google Scholar] [CrossRef]
- Roszkowski, M.; Mansuy, I.M. High efficiency RNA extraction from sperm cells using guanidinium thiocyanate supplemented with Tris (2-Carboxyethyl) Phosphine. Front. Cell Dev. Biol. 2021, 9, 648274. [Google Scholar] [CrossRef]
- Tiwari, S.; Shahat, A.; Kastelic, J.; Thakor, N.; Thundathil, J. Optimized total RNA isolation from bovine sperm with enhanced sperm head lysis. Biochem. Cell Biol. 2023, in press. [CrossRef]
- Chang, J.Y.; Li, L. The unfolding mechanism and the disulfide structures of denatured lysozyme. FEBS Lett. 2002, 511, 73–78. [Google Scholar] [CrossRef]
- Spehr, M.; Gisselmann, G.; Poplawski, A.; Riffell, J.A.; Wetzel, C.H.; Zimmer, R.K.; Hatt, H. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science 2003, 299, 2054–2058. [Google Scholar] [CrossRef] [PubMed]
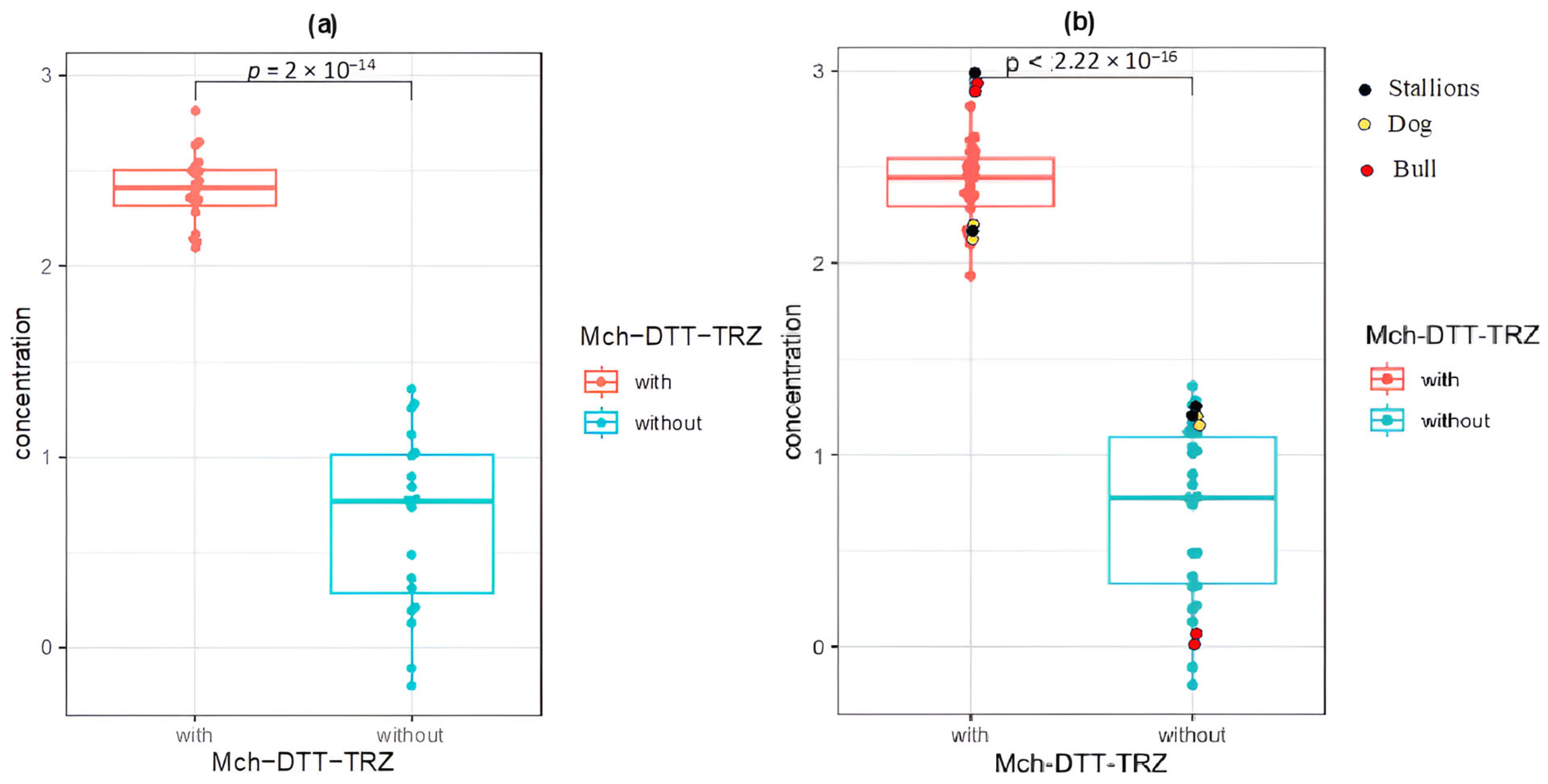
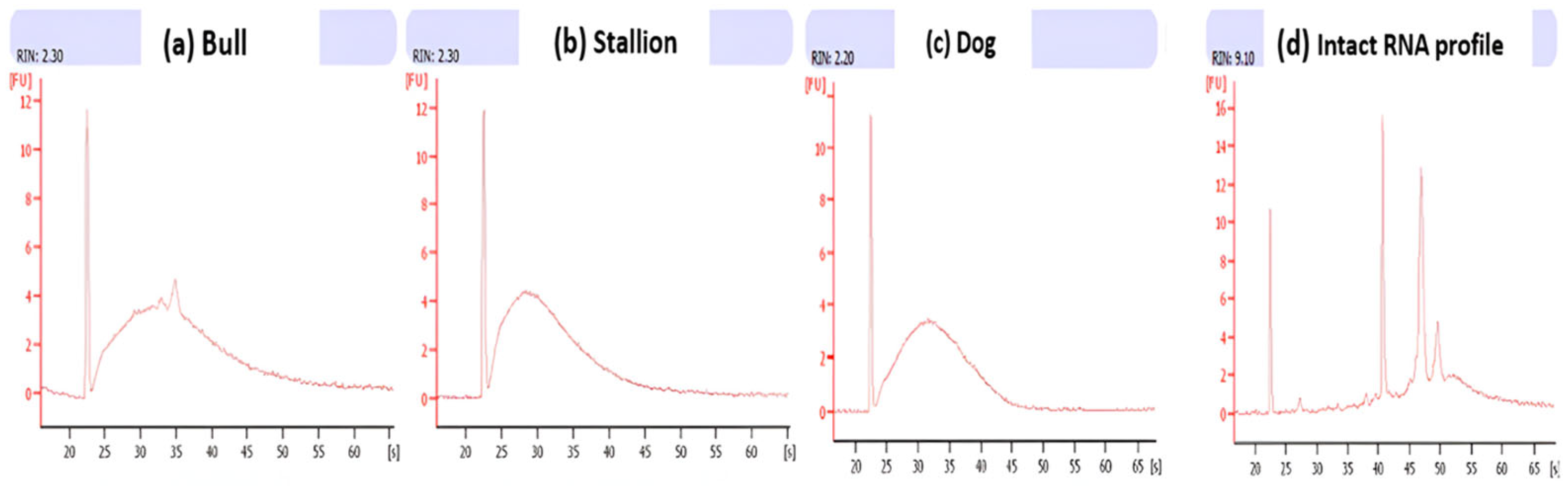
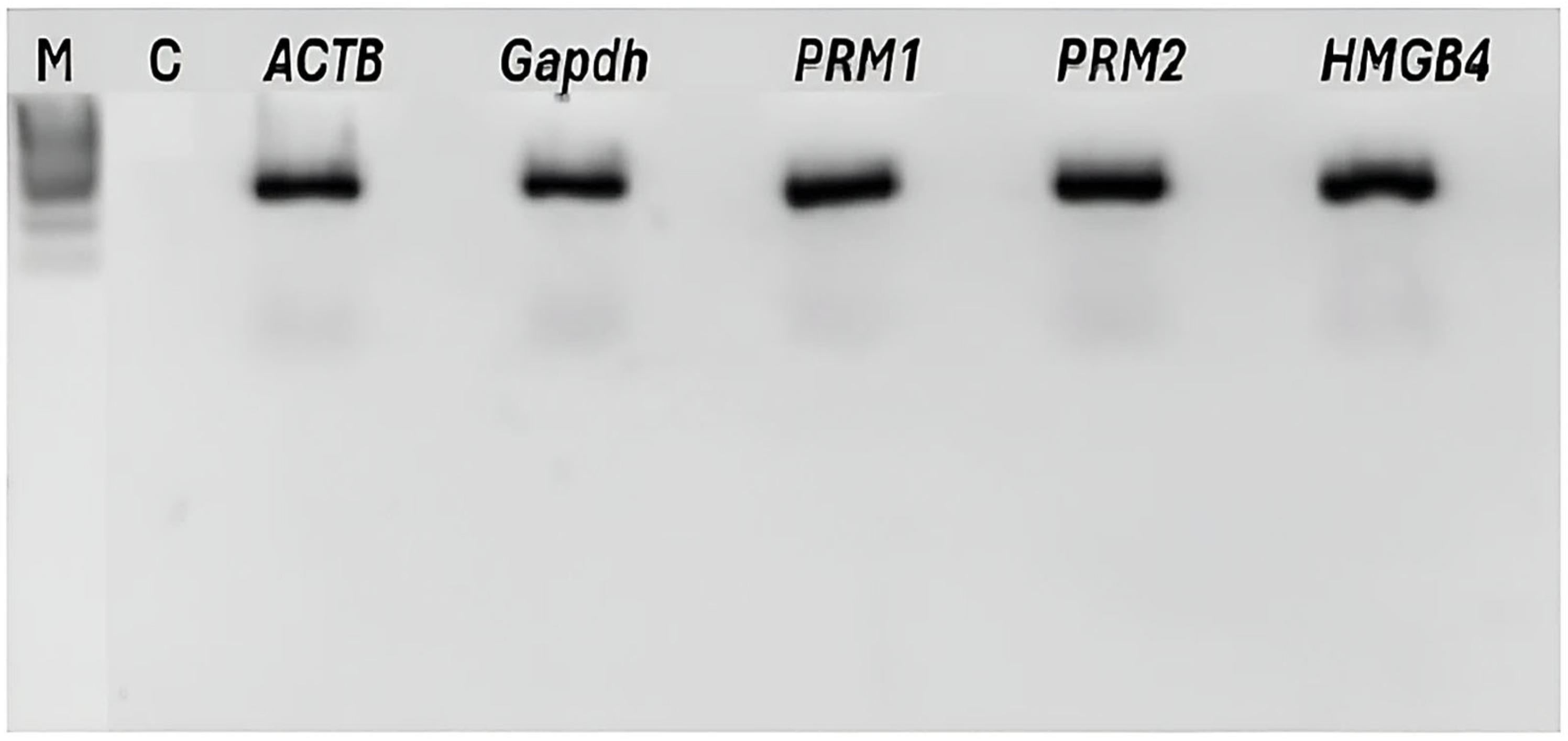

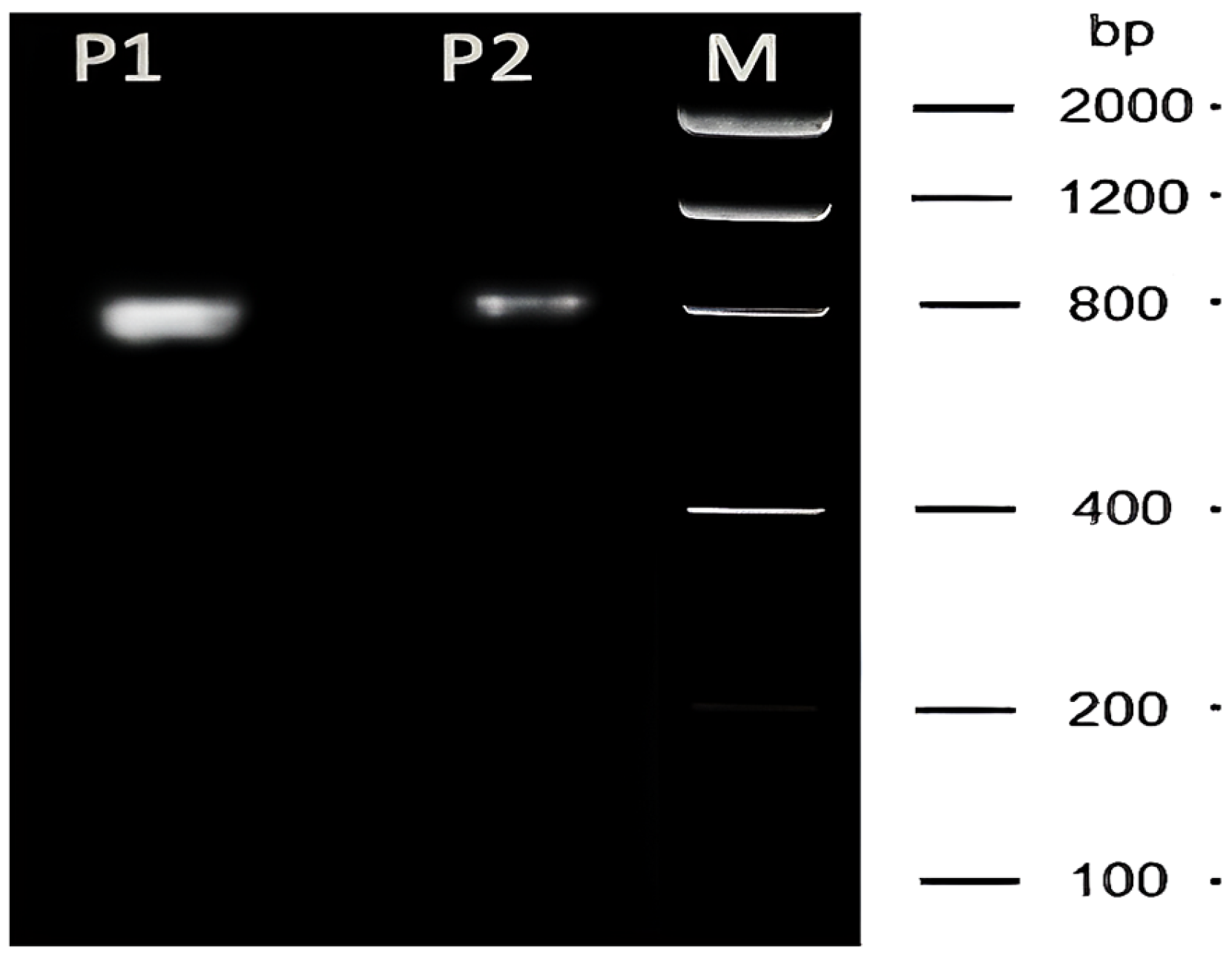
| Species | NucleoSpin RNA II Kit | NucleoSpin RNA II Kit -DTT-TRIzol | ||||
|---|---|---|---|---|---|---|
| RNA Concentration (ng/μL) | OD 260/280 | OD 260/230 | RNA Concentration (ng/μL) | OD 260/280 | OD 260/230 | |
| Human n = 20 | 5.47 | 1.36 | 1.23 | 213.38 | 1.87 | 2.40 |
| 19.10 | 1.53 | 1.68 | 224.76 | 1.86 | 2.37 | |
| 7.00 | 1.49 | 1.40 | 192.88 | 1.85 | 2.38 | |
| 22.83 | 1.43 | 1.04 | 134.05 | 1.83 | 2.37 | |
| 10.21 | 1.54 | 1.22 | 229.29 | 1.85 | 2.43 | |
| 7.91 | 1.47 | 1.53 | 315.66 | 1.87 | 2.41 | |
| 18.13 | 1.41 | 1.43 | 336.17 | 1.87 | 2.40 | |
| 10.50 | 1.61 | 1.56 | 272.00 | 1.86 | 2.42 | |
| 13.16 | 1.40 | 1.34 | 655.25 | 1.91 | 2.35 | |
| 0.63 | 0.76 | 1.22 | 434.55 | 1.89 | 2.38 | |
| 0.78 | 1.05 | 0.00 | 318.57 | 1.85 | 2.42 | |
| 1.64 | 1.00 | 0.27 | 249.45 | 1.84 | 2.43 | |
| 1.35 | 1.13 | 0.75 | 450.53 | 1.87 | 2.40 | |
| 6.02 | 1.53 | 1.14 | 125.66 | 1.78 | 2.37 | |
| 5.86 | 1.46 | 1.80 | 148.06 | 1.79 | 2.29 | |
| 3.09 | 1.27 | 0.28 | 281.85 | 1.84 | 2.34 | |
| 2.06 | 7.32 | 0.35 | 352.44 | 1.88 | 2.38 | |
| 5.95 | 3.13 | 0.60 | 305.42 | 1.84 | 2.31 | |
| 1.57 | 2.35 | 0.13 | 230.17 | 1.87 | 2.28 | |
| 2.32 | 0.00 | 0.28 | 139.35 | 1.77 | 2.21 | |
| Dog n = 2 | 14.63 | 2.01 | 2.02 | 146.90 | 2.10 | 2.04 |
| 12.87 | 1.74 | 1.43 | 85.71 | 1.84 | 2.69 | |
| Stallion n = 2 | 13.01 | 1.18 | 0.67 | 285.50 | 1.92 | 2.17 |
| 11.01 | 0.33 | 0.28 | 380.50 | 1.91 | 2.24 | |
| Bull n = 2 | 3.09 | 4.05 | 0.01 | 402.70 | 1.90 | 2.26 |
| 2.08 | 4.05 | 0.01 | 376.57 | 1.90 | 2.15 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mounia, E.O.; Naoual, A.; Celia, R.; Hind, H.I.; Mouna, H.; Francis, F.; Francis, G.; Omar, A. Development and Optimization of an RNA-Isolating Protocol for Mammalian Spermatozoa. Int. J. Mol. Sci. 2025, 26, 11171. https://doi.org/10.3390/ijms262211171
Mounia EO, Naoual A, Celia R, Hind HI, Mouna H, Francis F, Francis G, Omar A. Development and Optimization of an RNA-Isolating Protocol for Mammalian Spermatozoa. International Journal of Molecular Sciences. 2025; 26(22):11171. https://doi.org/10.3390/ijms262211171
Chicago/Turabian StyleMounia, El Oulidi, Azzouzi Naoual, Ravel Celia, Hassani Idrissi Hind, Habbane Mouna, Fieni Francis, Galibert Francis, and Akhouayri Omar. 2025. "Development and Optimization of an RNA-Isolating Protocol for Mammalian Spermatozoa" International Journal of Molecular Sciences 26, no. 22: 11171. https://doi.org/10.3390/ijms262211171
APA StyleMounia, E. O., Naoual, A., Celia, R., Hind, H. I., Mouna, H., Francis, F., Francis, G., & Omar, A. (2025). Development and Optimization of an RNA-Isolating Protocol for Mammalian Spermatozoa. International Journal of Molecular Sciences, 26(22), 11171. https://doi.org/10.3390/ijms262211171







