Does Platelet Transcriptome Dysregulation Across the Lewy Body Continuum Mirror Neuronal Dysfunction?
Abstract
1. Introduction
2. Results
2.1. Demographic and Clinical Data
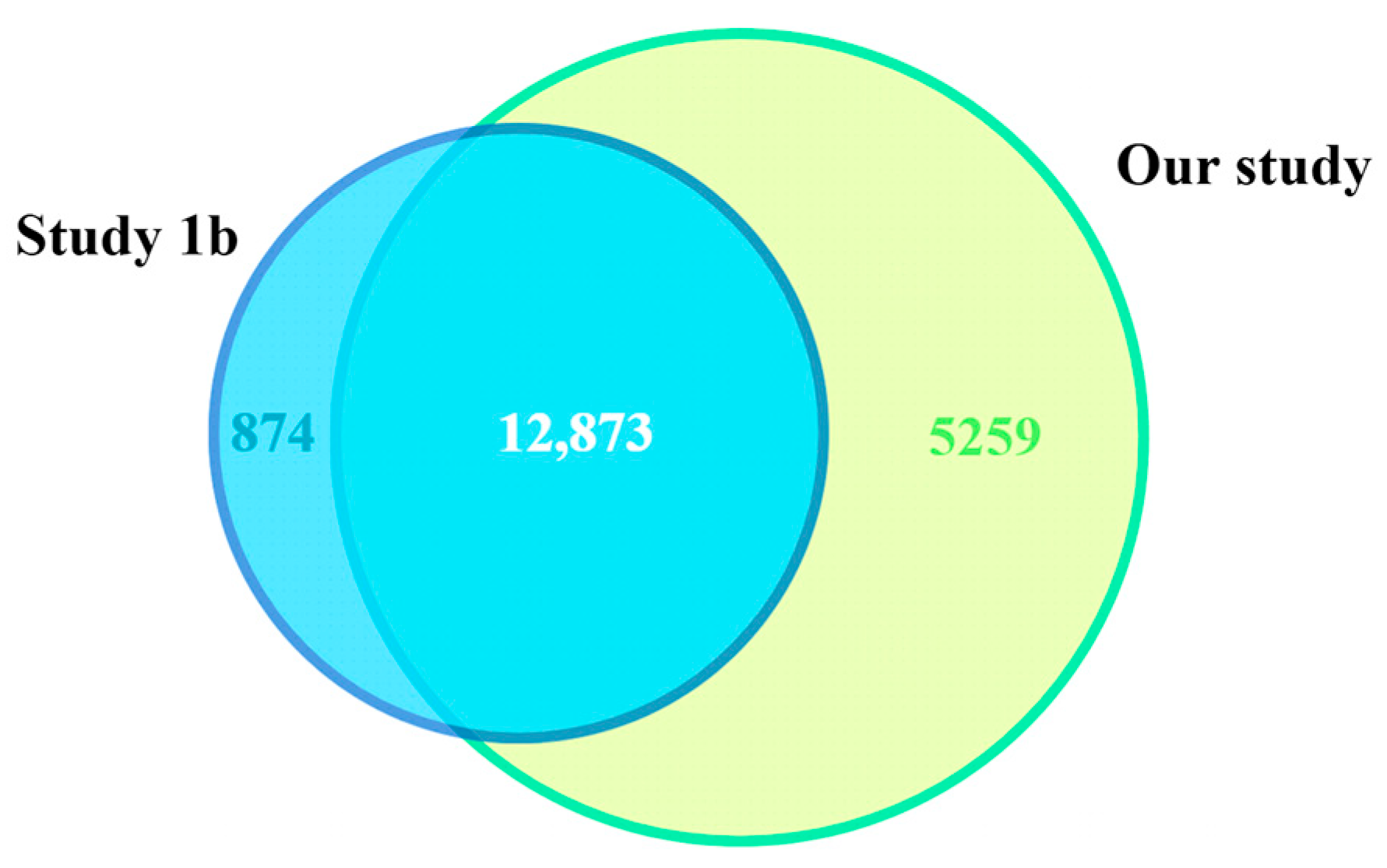
2.2. Comparison of Gene Expression Profiles Across RNA-Seq Studies
2.2.1. CTRLs
2.2.2. PD
2.3. Classification of Transcripts Expressed in PLTs
2.3.1. Long-Non-Coding RNA (lncRNA)
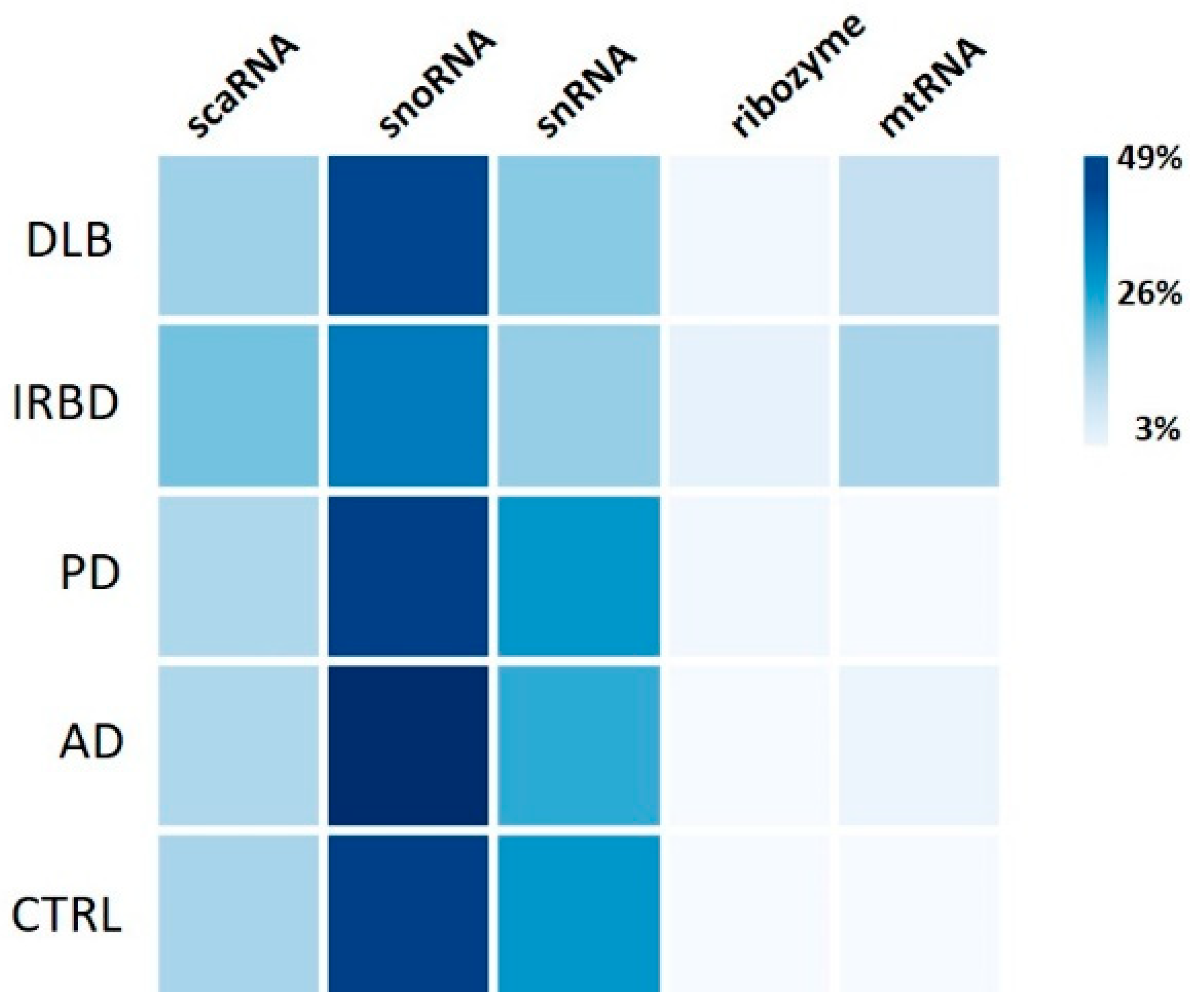
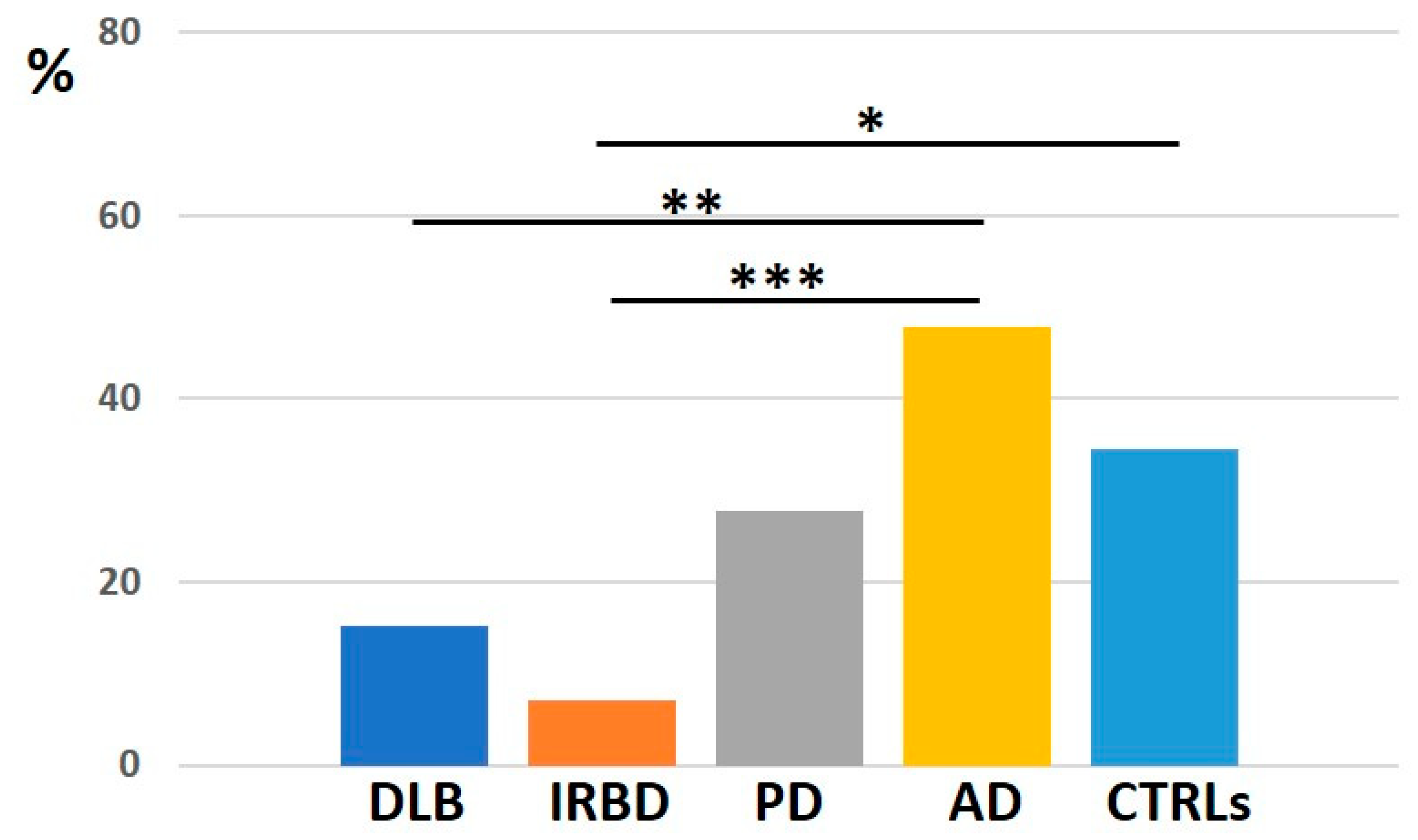
2.3.2. Minor RNAs
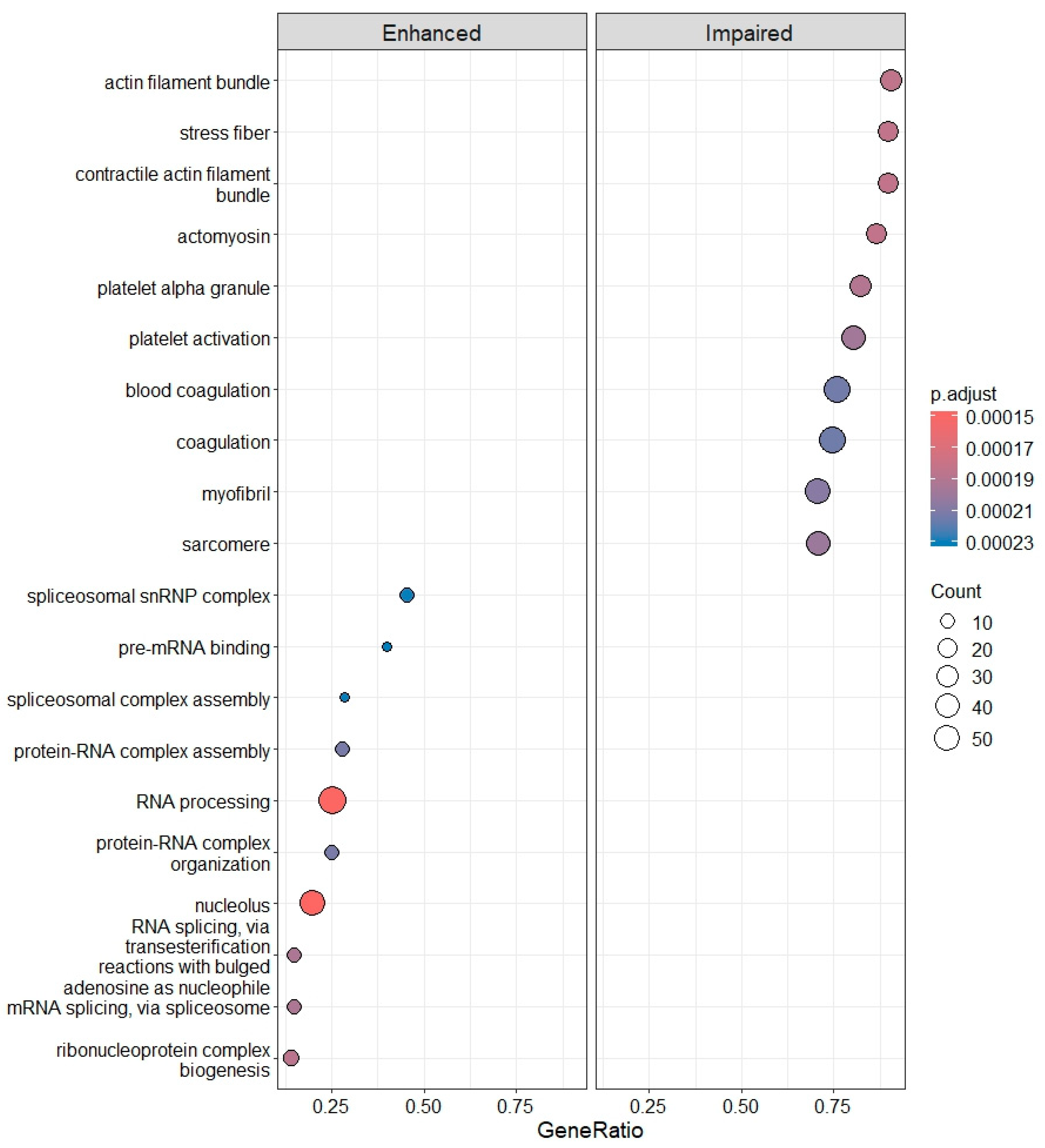
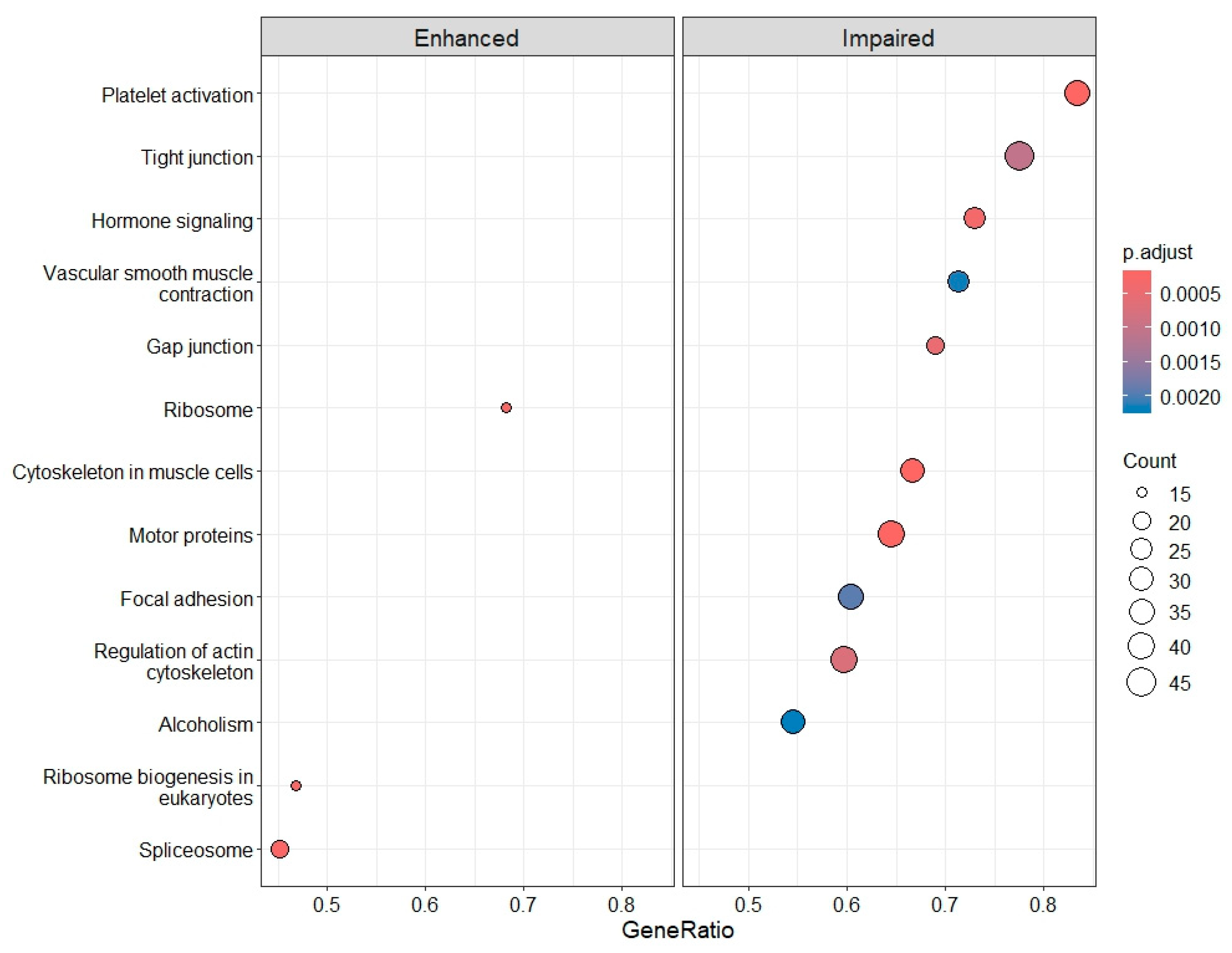
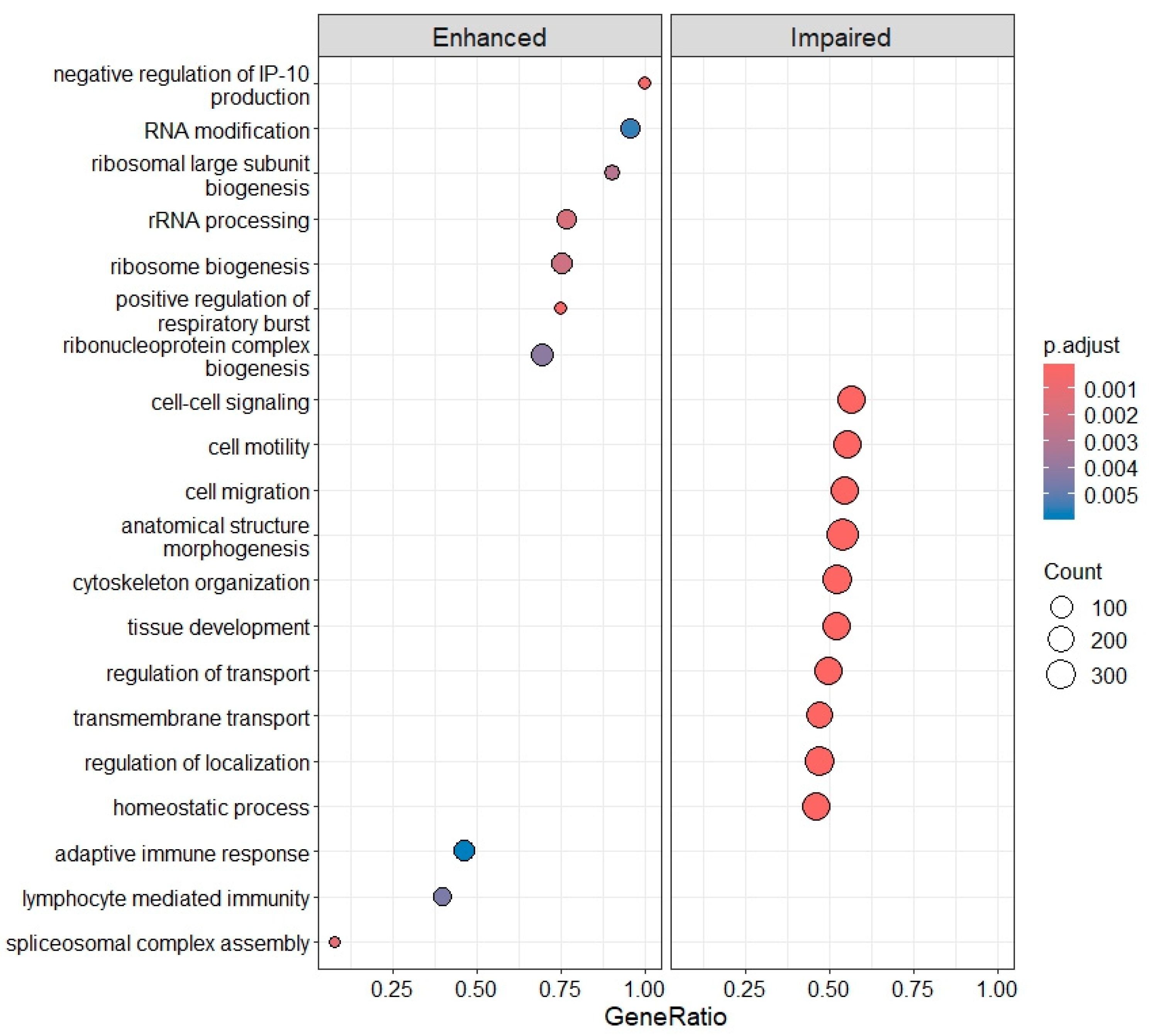
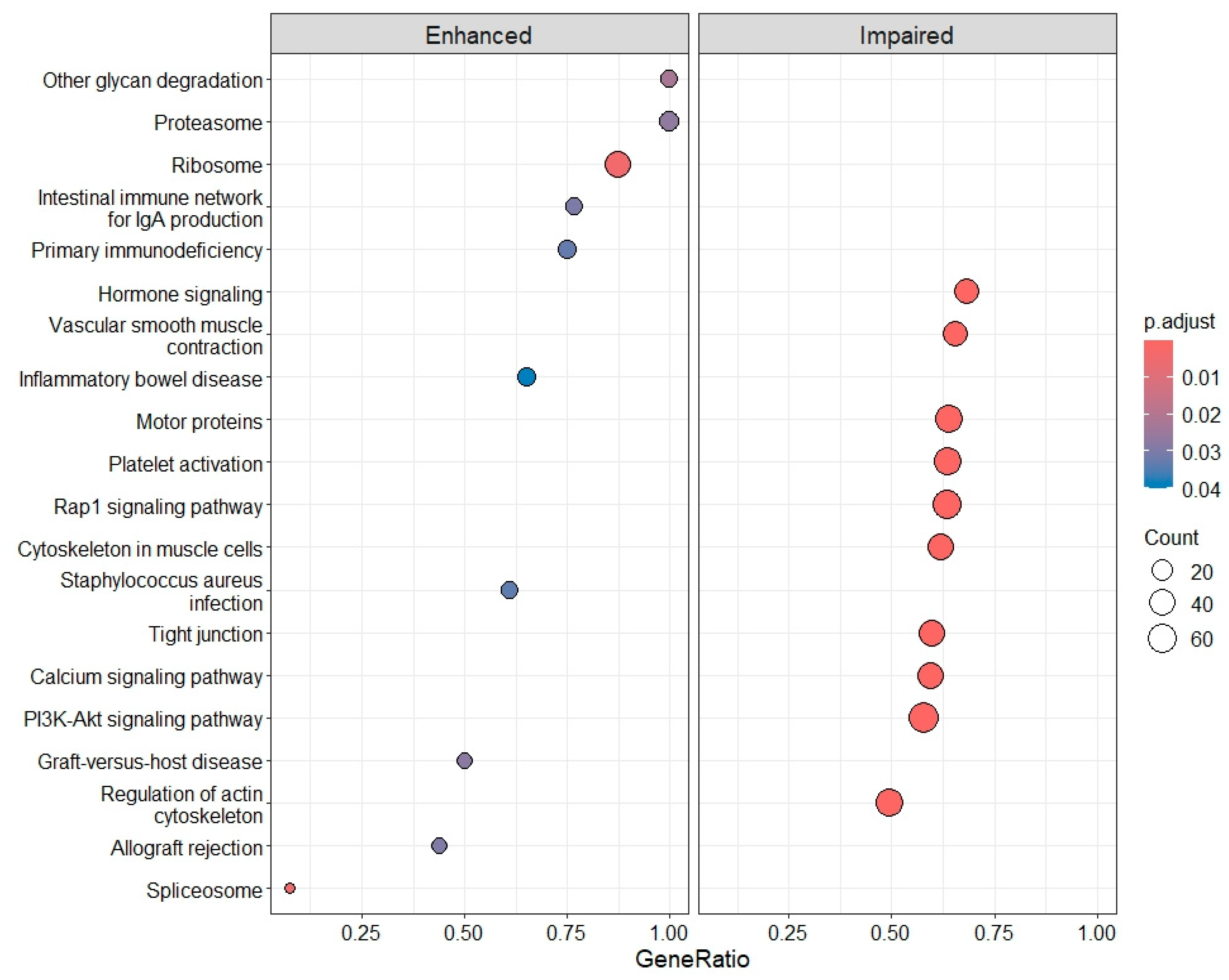
2.4. Differential Gene Expression, Gene Ontology (GO) Enrichment and KEGG Pathway Analysis
3. Discussion
3.1. Comparison of PLT RNA-Seq Studies
3.2. The Composition of the PLT Transcriptome
3.3. Deregulation of Gene Expression in PLTs
3.3.1. IRBD and DLB: Progressive Molecular Impairment and Shared Pathway Dysregulation
3.3.2. PD and AD: Disease Heterogeneity
3.4. Future Biomarker Development
4. Materials and Methods
4.1. Source of PLT Samples
4.2. PLT Obtaining and RNA Purification
4.3. Total RNA Discovery by Next-Generation Sequencing (NGS)
4.4. Sequencing Data Analysis
4.5. Comparison of RNA Expression Between RNA-Seq Studies
4.6. LncRNA and Minor RNA Distribution Analysis
4.7. Gene Ontology (GO) Enrichment and KEGG Pathway Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| APP | amyloid precursor protein |
| Aβ | β-amyloid |
| CSF | cerebrospinal fluid |
| CTRLs | cognitively unaffected controls |
| DAT | dopamine transporter |
| DEA | differential expression analysis |
| DEGs | differentially expressed genes |
| DLB | Dementia with Lewy bodies |
| GDS | Global Deterioration Scale |
| GSEA | Gene Set Enrichment Analysis |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| IRBD | idiopathic REM sleep behavior disorder |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LBD | Lewy body disorders |
| lncRNAs | long non-coding RNAs |
| MMSE | Mini-Mental State Examination |
| mtRNAs | mitochondrial RNAs |
| NGS | Next-Generation Sequencing |
| PD | Parkinson’s disease |
| PLT | Platelets |
| scaRNAs | small Cajal body-specific RNAs |
| snoRNAs | small nucleolar RNAs |
| snRNAS | small nuclear RNAs |
| SRA | Sequence Read Archive |
| UPDRS-III | Unified Parkinson’s Disease Rating Scale Part III |
Appendix A
Appendix A.1
| n | Ensembl Gene ID | External Gene Name | Chromosome | Start Position | End Position | Strand | |
|---|---|---|---|---|---|---|---|
| DLB | 1 | ENSG00000237188 | chr1 | 147172701 | 147295734 | 1 | |
| 2 | ENSG00000289019 | chr4 | 70853613 | 70903213 | –1 | ||
| 3 | ENSG00000226281 | chr6 | 6692737 | 6801186 | –1 | ||
| 4 | ENSG00000287584 | chr7 | 39621253 | 39623201 | –1 | ||
| 5 | ENSG00000272293 | chr8 | 450714 | 451343 | –1 | ||
| 6 | ENSG00000284116 | chr9 | 39931614 | 40106680 | –1 | ||
| 7 | ENSG00000290769 | chr9 | 133079900 | 133087355 | 1 | ||
| 8 | ENSG00000228886 | chr13 | 45350323 | 45351350 | 1 | ||
| 9 | ENSG00000258803 | chr14 | 56514331 | 56551309 | 1 | ||
| 10 | ENSG00000231439 | WASIR2 | chr16 | 22845 | 25191 | 1 | |
| 11 | ENSG00000272884 | chr17 | 7439506 | 7445966 | 1 | ||
| 12 | ENSG00000280800 | chr21 | 8210384 | 8211306 | –1 | ||
| 13 | ENSG00000284391 | chrX | 70427346 | 70435378 | –1 | ||
| AD | 1 | ENSG00000289474 | chr2 | 148881726 | 148881841 | –1 | |
| 2 | ENSG00000242516 | LINC00960 | chr3 | 75672232 | 75742089 | 1 | |
| 3 | ENSG00000290602 | chr7 | 143810373 | 143818699 | 1 | ||
| 4 | ENSG00000289031 | chr9 | 93566714 | 93568075 | 1 | ||
| 5 | ENSG00000288542 | chr13 | 40469955 | 40611127 | –1 | ||
| 6 | ENSG00000288855 | chr14 | 50736300 | 50737517 | –1 | ||
| 7 | ENSG00000259692 | LINC01418 | chr15 | 81610828 | 82013579 | –1 | |
| 8 | ENSG00000290383 | chr16 | 18394972 | 18401925 | –1 | ||
| 9 | ENSG00000185168 | LINC00482 | chr17 | 81303771 | 81311237 | –1 | |
| 10 | ENSG00000288235 | FAM106C | chr17 | 16788879 | 16790501 | 1 | |
| 11 | ENSG00000289172 | chr20 | 45179818 | 45191491 | 1 | ||
| 12 | ENSG00000281181 | chr21 | 8437629 | 8438551 | –1 | ||
| CTRLs | 1 | ENSG00000254154 | CRYZL2P-SEC16B | chr1 | 177928788 | 178038007 | –1 |
| 2 | ENSG00000273382 | TMEM167B-DT | chr1 | 109087971 | 109090858 | –1 | |
| 3 | ENSG00000274769 | chr2 | 61115787 | 61164825 | 1 | ||
| 4 | ENSG00000290614 | PRSS40A | chr2 | 130570829 | 130584161 | 1 | |
| 5 | ENSG00000289929 | chr3 | 195635062 | 195652295 | 1 | ||
| 6 | ENSG00000288473 | chr6 | 30908242 | 30926459 | 1 | ||
| 7 | ENSG00000290972 | chr9 | 64369394 | 64412691 | 1 | ||
| 8 | ENSG00000289381 | chr13 | 31796368 | 31814730 | –1 | ||
| 9 | ENSG00000289049 | chr14 | 101760727 | 101761485 | –1 | ||
| 10 | ENSG00000291023 | chr15 | 32406178 | 32434992 | –1 | ||
| 11 | ENSG00000248101 | chr19 | 36008638 | 36014235 | –1 | ||
| 12 | ENSG00000268744 | chr19 | 12379189 | 12401274 | –1 | ||
| 13 | ENSG00000289298 | chr19 | 41530216 | 41531859 | –1 | ||
| 14 | ENSG00000288861 | chr22 | 22757217 | 22759496 | –1 | ||
| IRBD | 1 | ENSG00000289062 | chr1 | 152897765 | 152913138 | –1 | |
| 2 | ENSG00000289367 | chr1 | 247937142 | 247937864 | 1 | ||
| 3 | ENSG00000291157 | chr1 | 41302911 | 41306148 | –1 | ||
| 4 | ENSG00000228363 | CHMP3-AS1 | chr2 | 86562070 | 86618766 | 1 | |
| 5 | ENSG00000235070 | chr2 | 226086623 | 226185651 | –1 | ||
| 6 | ENSG00000226519 | LINC00390 | chr13 | 44094822 | 44161490 | –1 | |
| 7 | ENSG00000258694 | LINC02319 | chr14 | 52101631 | 52129852 | –1 | |
| 8 | ENSG00000290387 | SORD2P | chr15 | 44825744 | 44884694 | –1 | |
| 9 | ENSG00000290674 | chr16 | 21901552 | 21953031 | –1 | ||
| 10 | ENSG00000261033 | SPECC1-DT | chr17 | 20008051 | 20009234 | –1 | |
| 11 | ENSG00000176728 | TTTY14 | chrY | 18772706 | 19077555 | –1 | |
| 12 | ENSG00000212856 | TTTY2B | chrY | 6406059 | 6462091 | –1 | |
| 13 | ENSG00000229308 | chrY | 4036335 | 4100619 | 1 | ||
| 14 | ENSG00000231535 | LINC00278 | chrY | 3002887 | 3200509 | 1 | |
| 15 | ENSG00000260197 | chrY | 19691941 | 19694606 | –1 | ||
| 16 | ENSG00000288049 | chrY | 19744756 | 19759978 | 1 | ||
| 17 | ENSG00000289707 | chrY | 21138633 | 21257832 | 1 | ||
| 18 | ENSG00000290853 | chrY | 13703902 | 13916244 | 1 | ||
| 19 | ENSG00000291031 | BCORP1 | chrY | 19455431 | 19567280 | –1 | |
| 20 | ENSG00000291033 | TXLNGY | chrY | 19567313 | 19606274 | 1 | |
| PD | 1 | ENSG00000225964 | NRIR | chr2 | 6819463 | 6840464 | –1 |
| 2 | ENSG00000189229 | chr3 | 6490460 | 6736750 | 1 | ||
| 3 | ENSG00000251230 | MIR3945HG | chr4 | 184843296 | 184855751 | –1 | |
| 4 | ENSG00000286274 | chr5 | 129150677 | 129394114 | 1 | ||
| 5 | ENSG00000285492 | chr6 | 159051674 | 159121510 | –1 | ||
| 6 | ENSG00000173862 | chr7 | 33725820 | 33729217 | 1 | ||
| 7 | ENSG00000289725 | chr9 | 64411638 | 64469260 | 1 | ||
| 8 | ENSG00000290717 | ZNF658B | chr9 | 39443589 | 39552802 | –1 | |
| 9 | ENSG00000286715 | chr10 | 75592644 | 75628120 | 1 | ||
| 10 | ENSG00000290690 | chr15 | 84398316 | 84422500 | –1 | ||
| 11 | ENSG00000260280 | SLX1B-SULT1A4 | chr16 | 29455105 | 29464963 | 1 | |
| 12 | ENSG00000290692 | chr16 | 30204316 | 30209071 | –1 | ||
| 13 | ENSG00000286288 | chr20 | 1694082 | 1896406 | –1 | ||
| 14 | ENSG00000291052 | ABCC13 | chr21 | 14236206 | 14338017 | 1 |
Appendix A.2
| n | scaRNA | snoRNA | snRNA | rRNA | mtRNA | |
|---|---|---|---|---|---|---|
| DLB | 55 | 10 (18%) | 25 (45%) | 11 (20%) | 2 (4%) | 7 (13%) |
| IRBD | 36 | 8 (22%) | 13 (36%) | 7 (19%) | 2 (6%) | 6 (17%) |
| PD | 58 | 9 (16%) | 27 (46%) | 18 (31%) | 2 (4%) | 2 (3%) |
| AD | 75 | 12 (16%) | 37 (49%) | 20 (27%) | 2 (3%) | 4 (5%) |
| CTRLs | 65 | 11 (17%) | 30 (46%) | 20 (31%) | 2 (3%) | 2 (3%) |
Appendix A.3
| DLB | IRBD | PD | AD | CTRLs | ||
|---|---|---|---|---|---|---|
| mtRNA | n | 7 | 6 | 2 | 4 | 2 |
| 1 | MT-TL1 | MT-TL1 | ||||
| 2 | MT-TV | MT-TV | MT-TV | |||
| 3 | MT-RNR2 | MT-RNR2 | MT-RNR2 | MT-RNR2 | MT-RNR2 | |
| 4 | MT-TM | MT-TM | ||||
| 5 | MT-TH | MT-TH | ||||
| 6 | MT-TE | MT-TE | ||||
| 7 | MT-RNR1 | MT-RNR1 | MT-RNR1 | MT-RNR1 | MT-RNR1 | |
| Ribozyme | n | 2 | 2 | 2 | 2 | 2 |
| 1 | RMRP | RMRP | RMRP | RMRP | RMRP | |
| 2 | RPPH1 | RPPH1 | RPPH1 | RPPH1 | RPPH1 | |
| scaRNA | n | 10 | 8 | 9 | 12 | 11 |
| 1 | SCARNA7 | SCARNA7 | SCARNA7 | SCARNA7 | SCARNA7 | |
| 2 | SCARNA8 | |||||
| 3 | SCARNA6 | SCARNA6 | SCARNA6 | SCARNA6 | SCARNA6 | |
| 4 | SCARNA5 | SCARNA5 | SCARNA5 | SCARNA5 | SCARNA5 | |
| 5 | SCARNA10 | SCARNA10 | SCARNA10 | SCARNA10 | SCARNA10 | |
| 6 | SCARNA12 | SCARNA12 | SCARNA12 | SCARNA12 | SCARNA12 | |
| 7 | SCARNA13 | SCARNA13 | SCARNA13 | SCARNA13 | SCARNA13 | |
| 8 | SCARNA21 | SCARNA21 | SCARNA21 | |||
| 9 | SCARNA3 | |||||
| 10 | SCARNA1 | SCARNA1 | SCARNA1 | |||
| 11 | SCARNA16 | SCARNA16 | SCARNA16 | SCARNA16 | SCARNA16 | |
| 12 | SCARNA2 | SCARNA2 | SCARNA2 | SCARNA2 | SCARNA2 | |
| 13 | SCARNA4 | SCARNA4 | ||||
| snoRNA | n | 25 | 13 | 27 | 37 | 30 |
| 1 | SNORA10 | |||||
| 2 | SNORA11 | SNORA11 | ||||
| 3 | SNORA12 | SNORA12 | SNORA12 | SNORA12 | ||
| 4 | SNORA20 | SNORA20 | ||||
| 5 | SNORA23 | SNORA23 | ||||
| 6 | SNORA2C | SNORA2C | SNORA2C | SNORA2C | ||
| 7 | SNORA33 | |||||
| 8 | SNORA37 | SNORA37 | SNORA37 | |||
| 9 | SNORA38B | |||||
| 10 | SNORA48 | SNORA48 | ||||
| 11 | SNORA53 | SNORA53 | SNORA53 | SNORA53 | SNORA53 | |
| 12 | SNORA54 | SNORA54 | SNORA54 | SNORA54 | ||
| 13 | SNORA57 | |||||
| 14 | SNORA59B | SNORA59B | SNORA59B | SNORA59B | ||
| 15 | SNORA5C | SNORA5C | ||||
| 16 | SNORA62 | |||||
| 17 | SNORA63 | SNORA63 | SNORA63 | SNORA63 | SNORA63 | |
| 18 | SNORA66 | |||||
| 19 | SNORA73A | SNORA73A | SNORA73A | SNORA73A | SNORA73A | |
| 20 | SNORA73B | SNORA73B | SNORA73B | SNORA73B | SNORA73B | |
| 21 | SNORA74A | SNORA74A | SNORA74A | SNORA74A | ||
| 22 | SNORA74B | SNORA74B | SNORA74B | SNORA74B | ||
| 23 | SNORA79B | |||||
| 24 | SNORA7A | SNORA7A | ||||
| 25 | SNORA7B | SNORA7B | SNORA7B | SNORA7B | ||
| 26 | SNORA8 | SNORA8 | ||||
| 27 | SNORA81 | SNORA81 | SNORA81 | SNORA81 | SNORA81 | |
| 28 | SNORD10 | SNORD10 | SNORD10 | SNORD10 | ||
| 29 | SNORD13 | SNORD13 | SNORD13 | SNORD13 | ||
| 30 | SNORD15B | SNORD15B | SNORD15B | SNORD15B | SNORD15B | |
| 31 | SNORD17 | SNORD17 | SNORD17 | SNORD17 | SNORD17 | |
| 32 | SNORD22 | SNORD22 | SNORD22 | |||
| 33 | SNORD33 | |||||
| 34 | SNORD3A | SNORD3A | SNORD3A | SNORD3A | SNORD3A | |
| 35 | SNORD3B-1 | SNORD3B-1 | SNORD3B-1 | SNORD3B-1 | SNORD3B-1 | |
| 36 | SNORD3B-2 | SNORD3B-2 | SNORD3B-2 | SNORD3B-2 | ||
| 37 | SNORD3C | SNORD3C | SNORD3C | SNORD3C | ||
| 38 | SNORD89 | SNORD89 | SNORD89 | SNORD89 | SNORD89 | |
| 39 | SNORD94 | SNORD94 | SNORD94 | |||
| 40 | SNORD97 | SNORD97 | SNORD97 | SNORD97 | ||
| 41 | U3 | U3 | U3 | |||
| snRNA | n | 11 | 7 | 18 | 20 | 20 |
| 1 | RNU5A-1 | RNU5A-1 | RNU5A-1 | RNU5A-1 | RNU5A-1 | |
| 2 | RNU5B-1 | RNU5B-1 | RNU5B-1 | |||
| 3 | RNU4-1 | RNU4-1 | RNU4-1 | RNU4-1 | RNU4-1 | |
| 4 | RNU4-2 | RNU4-2 | RNU4-2 | RNU4-2 | RNU4-2 | |
| 5 | RNVU1-7 | |||||
| 6 | RNU1-28P | RNU1-28P | RNU1-28P | |||
| 7 | RNU1-27P | RNU1-27P | RNU1-27P | |||
| 8 | RNU1-1 | RNU1-1 | RNU1-1 | RNU1-1 | ||
| 9 | RNVU1-18 | RNVU1-18 | RNVU1-18 | |||
| 10 | RNU1-2 | RNU1-2 | RNU1-2 | |||
| 11 | RNU1-4 | RNU1-4 | RNU1-4 | |||
| 12 | RNVU1-14 | |||||
| 13 | RNU1-3 | RNU1-3 | RNU1-3 | |||
| 14 | RNU6ATAC | |||||
| 15 | RNU2-2P | RNU2-2P | RNU2-2P | RNU2-2P | ||
| 16 | RNU6ATAC | |||||
| 17 | RNVU1-2 | RNVU1-2 | ||||
| 18 | RNU2-2P | |||||
| 19 | RNVU1-31 | RNVU1-31 | RNVU1-31 | RNVU1-31 | ||
| 20 | RNVU1-29 | RNVU1-29 | RNVU1-29 | |||
| 21 | RNVU1-27 | RNVU1-27 | RNVU1-27 | RNVU1-27 | RNVU1-27 | |
| 22 | RNU12 | RNU12 | RNU12 | RNU12 | ||
| 23 | RNVU1-28 | RNVU1-28 | RNVU1-28 | RNVU1-28 | ||
| 24 | RN7SK | RN7SK | RN7SK | RN7SK | RN7SK | |
Appendix B
Appendix B.1
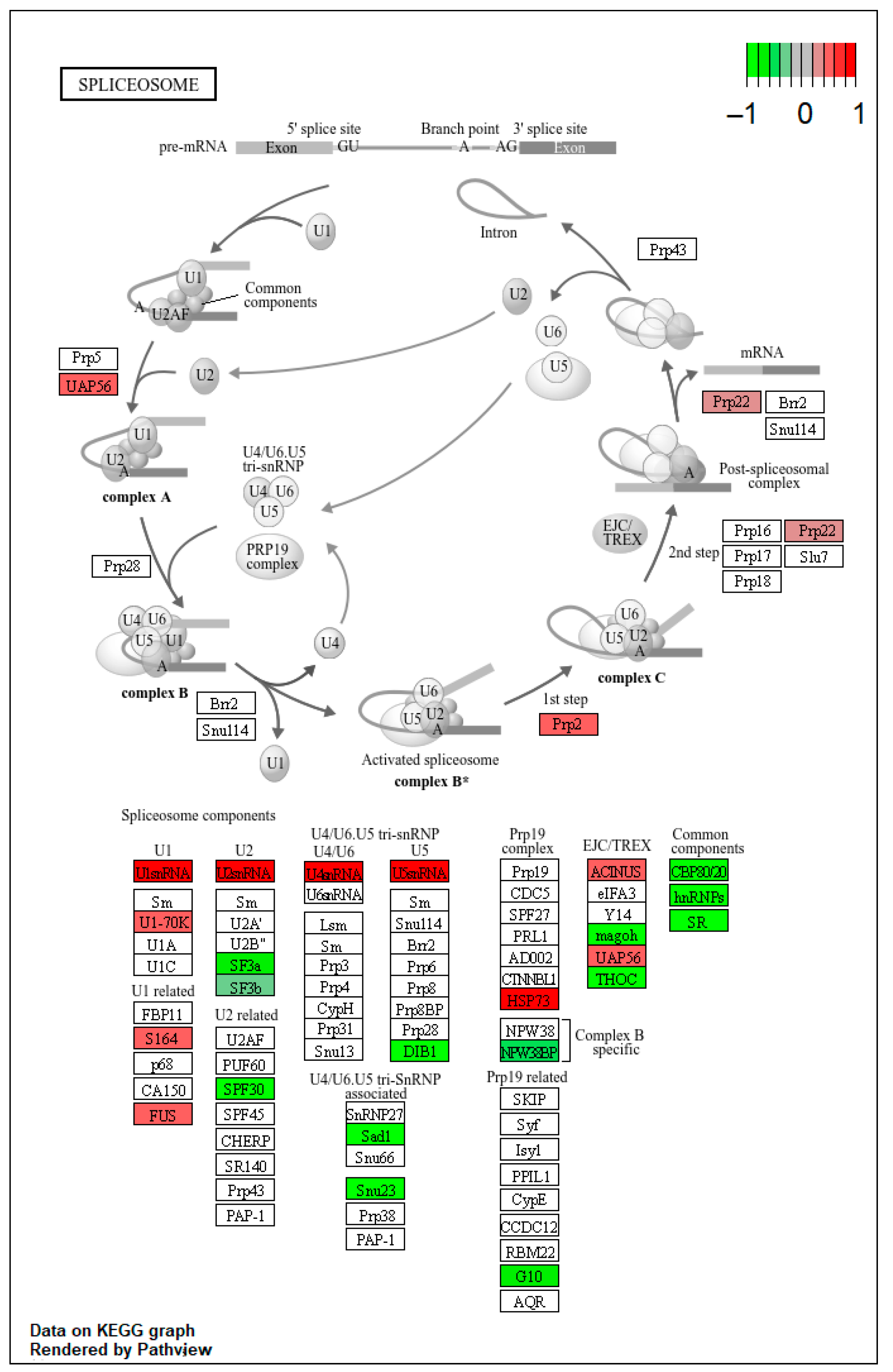
Appendix B.2

Appendix B.3
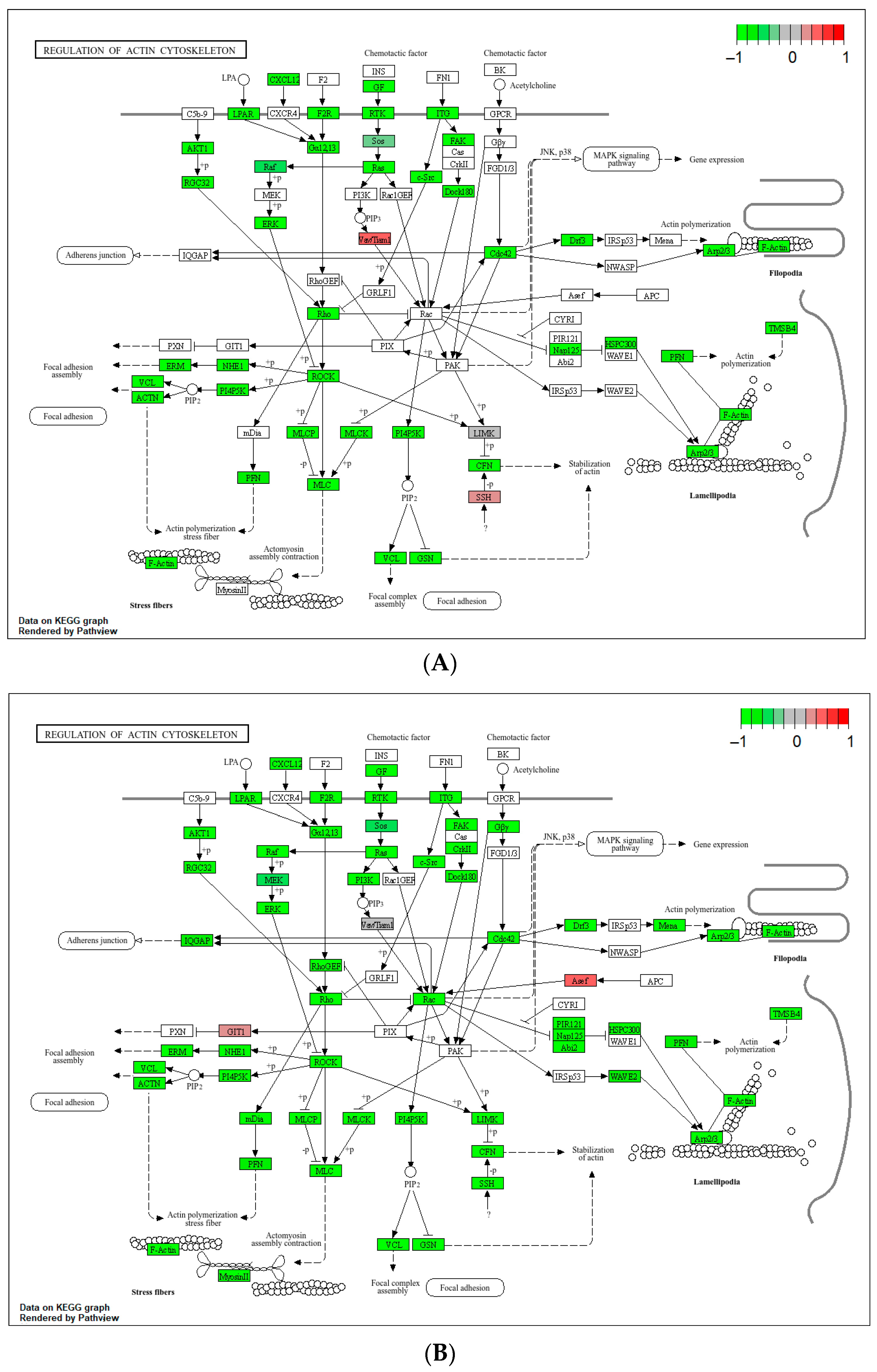
Appendix B.4
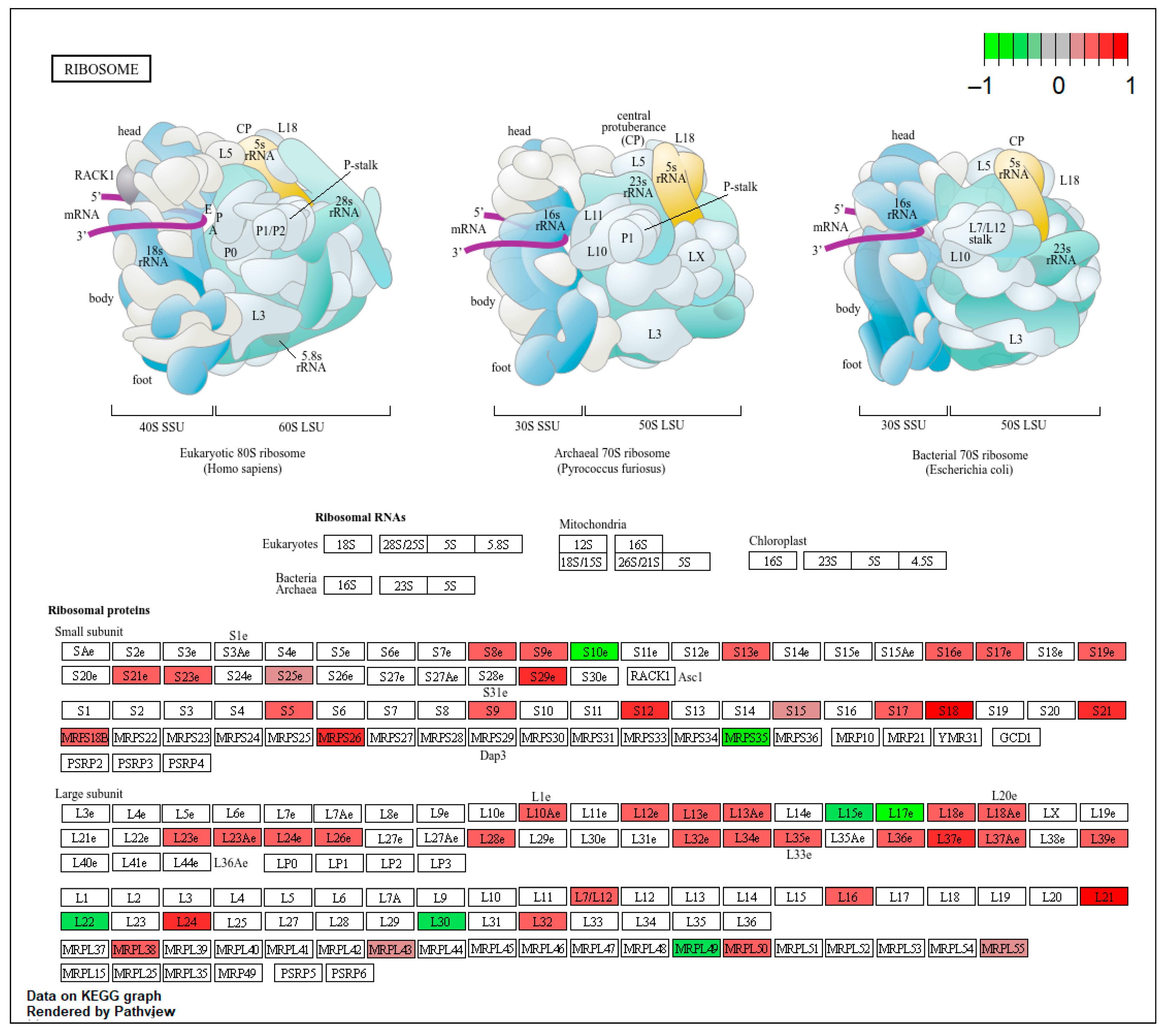
References
- Nichols, E.; Steinmetz, J.D.; Vollset, S.E.; Fukutaki, K.; Chalek, J.; Abd-Allah, F.; Abdoli, A.; Abualhasan, A.; Abu-Gharbieh, E.; Akram, T.T.; et al. GBD 2019 Dementia Forecasting Collaborators. Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: An analysis for the Global Burden of Disease Study 2019. Lancet Public Health 2022, 7, e105–e125. [Google Scholar] [CrossRef]
- DeTure, M.A.; Dickson, D.W. The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 32. [Google Scholar] [CrossRef]
- McKeith, I.G.; Boeve, B.F.; Dickson, D.W.; Halliday, G.; Taylor, J.P.; Weintraub, D.; Aarsland, D.; Galvin, J.; Attems, J.; Ballard, C.G.; et al. Diagnosis and management of dementia with Lewy bodies: Fourth consensus report of the DLB Consortium. Neurology 2017, 89, 88–100. [Google Scholar] [CrossRef]
- Jellinger, K.A. Dementia with Lewy bodies and Parkinson’s disease-dementia: Current concepts and controversies. J. Neural. Transm. 2018, 125, 615–650. [Google Scholar] [CrossRef] [PubMed]
- Outeiro, T.F.; Koss, D.J.; Erskine, D.; Walker, L.; Kurzawa-Akanbi, M.; Burn, D.; Donaghy, P.; Morris, C.; Taylor, J.-P.; Thomas, A. Dementia with Lewy bodies: An update and outlook. Mol. Neurodegener. 2019, 14, 5. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Crowther, R.A.; Jakes, R.; Hasegawa, M.; Godert, M. alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with lewy bodies. Proc. Natl. Acad. Sci. USA 1998, 95, 6469–6473. [Google Scholar] [CrossRef] [PubMed]
- Erskine, D.; Koss, D.; Korolchuk, V.I.; Outeiro, T.F.; Attems, J.; McKeith, I. Lipids, lysosomes and mitochondria: Insights into Lewy body formation from rare monogenic disorders. Acta Neuropathol. 2021, 141, 511–526. [Google Scholar] [CrossRef]
- Postuma, R.B.; Gagnon, J.F.; Vendette, M.; Fantini, M.L.; Massicotte-Marquez, J.; Montplaisir, J. Quantifying the risk of neurodegenerative disease in idiopathic REM sleep behavior disorder. Neurology 2009, 72, 1296–1300. [Google Scholar] [CrossRef]
- Iranzo, A.; Fernández-Arcos, A.; Tolosa, E.; Serradell, M.; Molinuevo, J.L.; Valldeoriola, F.; Gelpi, E.; Vilaseca, I.; Sánchez-Valle, R.; Lladó, A.; et al. Neurodegenerative disorder risk in idiopathic REM sleep behavior disorder: Study in 174 patients. PLoS ONE 2014, 9, e89741. [Google Scholar] [CrossRef]
- Iranzo, A.; Tolosa, E.; Gelpi, E.; Molinuevo, J.L.; Valldeoriola, F.; Serradell, M.; Sanchez-Valle, R.; Vilaseca, I.; Lomeña, F.; Vilas, D.; et al. Neurodegenerative disease status and post-mortem pathology in idiopathic rapid-eye-movement sleep behaviour disorder: An observational cohort study. Lancet Neurol. 2013, 12, 443–453. [Google Scholar] [CrossRef]
- Iranzo, A.; Fairfoul, G.; Ayudhaya, A.C.N.; Serradell, M.; Gelpi, E.; Vilaseca, I.; Sanchez-Valle, R.; Gaig, C.; Santamaria, J.; Tolosa, E.; et al. Detection of α-synuclein in CSF by RT-QuIC in patients with isolated rapid-eye-movement sleep behaviour disorder: A longitudinal observational study. Lancet Neurol. 2021, 20, 203–212. [Google Scholar] [CrossRef]
- Nording, H.; Langer, H.F. Complement links platelets to innate immunity. Semin. Immunol. 2018, 37, 43–52. [Google Scholar] [CrossRef]
- Leiter, O.; Walker, T.L. Platelets: The missing link between the blood and brain? Prog. Neurobiol. 2019, 183, 101695. [Google Scholar] [CrossRef]
- Gowert, N.S.; Donner, L.; Chatterjee, M.; Eisele, Y.S.; Towhid, S.T.; Münzer, P.; Walker, B.; Ogorek, I.; Borst, O.; Grandoch, M.; et al. Blood platelets in the progression of Alzheimer’s disease. PLoS ONE 2014, 9, e90523. [Google Scholar] [CrossRef] [PubMed]
- McRedmond, J.P.; Park, S.D.; Reilly, D.F.; Coppinger, J.A.; Maguire, P.B.; Shields, D.C.; Fitzgerald, D.J. Integration of proteomics and genomics in platelets: A profile of platelet proteins and platelet-specific genes. Mol. Cell. Proteomics 2004, 3, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Landry, P.; Plante, I.; Ouellet, D.L.; Perron, M.P.; Rousseau, G.; Provost, P. Existence of a microRNA pathway in anucleate platelets. Nat. Struct. Mol. Biol. 2009, 16, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Gámez-Valero, A.; Campdelacreu, J.; Vilas, D.; Ispierto, L.; Gascón-Bayarri, J.; Reñé, R.; Álvarez, R.; Armengol, M.P.; Borràs, F.E.; Beyer, K. Platelet miRNA Biosignature Discriminates between Dementia with Lewy Bodies and Alzheimer’s Disease. Biomedicines 2021, 9, 1272. [Google Scholar] [CrossRef]
- Arnaldo, L.; Mena, J.; Serradell, M.; Gaig, C.; Adamuz, D.; Vilas, D.; Samaniego, D.; Ispierto, L.; Montini, A.; Mayà, G.; et al. Platelet miRNAs as early biomarkers for progression of idiopathic REM sleep behavior disorder to a synucleinopathy. Sci. Rep. 2025, 15, 12136. [Google Scholar] [CrossRef]
- Thibord, F.; Johnson, A.D. Sources of variability in the human platelet transcriptome. Thromb. Res. 2023, 231, 255–263. [Google Scholar] [CrossRef]
- Zhang, L.; Shao, Y.; Tang, C.; Liu, Z.; Tang, D.; Hu, C.; Liang, X. Identification of Novel Biomarkers in Platelets for Diagnosing Parkinson’s Disease. Eur. Neurol. 2022, 85, 122–131. [Google Scholar] [CrossRef]
- In ‘t Veld, S.G.J.G.; Arkani, M.; Post, E.; Antunes-Ferreira, M.; D’Ambrosi, S.; Vessies, D.C.L.; Vermunt, L.; Vancura, A.; Muller, M.; Niemeijer, A.-L.N.; et al. Detection and localization of early- and late-stage cancers using platelet RNA. Cancer Cell 2022, 40, 999–1009.e6. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhang, R.; Duan, M.; Zhou, Y.; Bao, J.; Lu, H.; Wang, J.; Hu, M.; Hu, Z.; Zhou, F.; et al. A polygenic stacking classifier revealed the complicated platelet transcriptomic landscape of adult immune thrombocytopenia. Mol. Ther. Nucleic Acids 2022, 28, 477–487. [Google Scholar] [CrossRef]
- Xu, L.; Li, X.; Li, X.; Wang, X.; Ma, Q.; She, D.; Lu, X.; Zhang, J.; Yang, Q.; Lei, S.; et al. RNA profiling of blood platelets noninvasively differentiates colorectal cancer from healthy donors and noncancerous intestinal diseases: A retrospective cohort study. Genome Med. 2022, 14, 26. [Google Scholar] [CrossRef]
- Koupenova, M.; Clancy, L.; Corkrey, H.A.; Freedman, J.E. Circulating Platelets as Mediators of Immunity, Inflammation, and Thrombosis. Circ. Res. 2018, 122, 337–351. [Google Scholar] [CrossRef]
- Bo, Y.; Lu, Q.; Li, B.; Sha, R.; Yu, H.; Miao, C. The role of platelets in central hubs of inflammation: A literature review. Medicine 2024, 103, e38115. [Google Scholar] [CrossRef]
- Michelson, A.D.; Frelinger, A.L., III; Haynes, R.L.; Kinney, H.C.; Gremmel, T. Platelet Pathophysiology: Unexpected New Research Directions. Semin. Thromb. Hemost. 2024, 50, 1187–1190. [Google Scholar] [CrossRef] [PubMed]
- Ercan, H.; Reumiller, C.M.; Mühlberger, J.; Hsu, F.; Schmidt, G.J.; Umlauf, E.; Miller, I.; Rappold, E.; Attems, J.; Oehler, R.; et al. Platelets mirror changes in the frontal lobe antioxidant system in Alzheimer’s disease. Alzheimers Dement. 2025, 21, e70117. [Google Scholar] [CrossRef]
- Cimmino, G.; Conte, S.; Palumbo, D.; Sperlongano, S.; Torella, M.; Della Corte, A.; Golino, P. The Novel Role of Noncoding RNAs in Modulating Platelet Function: Implications in Activation and Aggregation. Int. J. Mol. Sci. 2023, 24, 7650. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, T.; Lv, Y.; Li, J.; Jiang, X.; Jiang, J.; Zhang, D.; Bian, W.; Zhang, C. MALAT1 promotes platelet activity and thrombus formation through PI3k/Akt/GSK-3β signalling pathway. Stroke Vasc. Neurol. 2023, 8, 181–192. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Palma, A.; Buonaiuto, G.; Ballarino, M.; Laneve, P. Genome biology of long non-coding RNAs in humans: A virtual karyotype. Comput. Struct. Biotechnol. J. 2025, 27, 575–584. [Google Scholar] [CrossRef]
- Li, X.; Zong, Q.; Liu, L.; Liu, Y.; Shen, Y.; Tang, X.; Wing, Y.K.; Li, S.X.; Zhou, J. Sex differences in rapid eye movement sleep behavior disorder: A systematic review and meta-analysis. Sleep Med. Rev. 2023, 71, 101810. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; McIntosh, C.S.; Mastaglia, F.L.; Wilton, S.D.; Aung-Htut, M.T. Neurodegenerative diseases: A hotbed for splicing defects and the potential therapies. Transl. Neurodegener. 2021, 10, 16, Correction in Transl. Neurodegener. 2021, 10, 41. [Google Scholar] [CrossRef]
- Bray, P.F.; McKenzie, S.E.; Edelstein, L.C.; Nagalla, S.; Delgrosso, K.; Ertel, A.; Kupper, J.; Jing, Y.; Londin, E.; Loher, P.; et al. The complex transcriptional landscape of the anucleate human platelet. BMC Genom. 2013, 14, 1. [Google Scholar] [CrossRef]
- Belur, N.R.; Bustos, B.I.; Lubbe, S.J.; Mazzulli, J.R. Nuclear aggregates of NONO/SFPQ and A-to-I-edited RNA in Parkinson’s disease and dementia with Lewy bodies. Neuron 2024, 112, 2558–2580.e13. [Google Scholar] [CrossRef]
- Stefanini, L.; Bergmeier, W. RAP1-GTPase signaling and platelet function. J. Mol. Med. 2016, 94, 13–19. [Google Scholar] [CrossRef]
- Stefanini, L.; Bergmeier, W. RAP GTPases and platelet integrin signaling. Platelets 2019, 30, 41–47. [Google Scholar] [CrossRef]
- Murphy, D.D.; Rueter, S.M.; Trojanowski, J.Q.; Lee, V.M. Synucleins are developmentally expressed, and α-synuclein regulates the size of the presynaptic vesicular pool in primary hippocampal neurons. J. Neurosci. 2000, 20, 3214–3220. [Google Scholar] [CrossRef]
- Pranke, I.M.; Morello, V.; Bigay, J.; Gibson, K.; Verbavatz, J.M.; Antonny, B.; Jackson, C.L. α-Synuclein and ALPS motifs are membrane curvature sensors whose contrasting chemistry mediates selective vesicle binding. J. Cell. Biol. 2011, 194, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Oliveira da Silva, M.I.; Liz, M.A. Linking Alpha-Synuclein to the Actin Cytoskeleton: Consequences to Neuronal Function. Front. Cell. Dev. Biol. 2020, 8, 787. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Nair, M.G.; Jaroszewski, L.; Godzik, A. Deciphering Abnormal Platelet Subpopulations in COVID-19, Sepsis and Systemic Lupus Erythematosus through Machine Learning and Single-Cell Transcriptomics. Int. J. Mol. Sci. 2024, 25, 5941. [Google Scholar] [CrossRef]
- Höglinger, G.U.; Adler, C.H.; Berg, D.; Klein, C.; Outeiro, T.F.; Poewe, W.; Postuma, R.; Stoessl, A.J.; Lang, A.E. A biological classification of Parkinson’s disease: The SynNeurGe research diagnostic criteria. Lancet Neurol. 2024, 23, 191–204, Erratum in Lancet Neurol. 2024, 23, e7. [Google Scholar] [CrossRef]
- Tijms, B.M.; Vromen, E.M.; Mjaavatten, O.; Holstege, H.; Reus, L.M.; van der Lee, S.; Wesenhagen, K.E.J.; Lorenzini, L.; Vermunt, L.; Venkatraghavan, V.; et al. Cerebrospinal fluid proteomics in patients with Alzheimer’s disease reveals five molecular subtypes with distinct genetic risk profiles. Nat. Aging 2024, 4, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, D.; Nordberg, A.; Westman, E. Biological subtypes of Alzheimer disease: A systematic review and meta-analysis. Neurology 2020, 94, 436–448. [Google Scholar] [CrossRef]
- Jones, L.D.; Jackson, J.W.; Maggirwar, S.B. Modeling HIV-1 Induced Neuroinflammation in Mice: Role of Platelets in Mediating Blood-Brain Barrier Dysfunction. PLoS ONE 2016, 11, e0151702. [Google Scholar] [CrossRef]
- Burnouf, T.; Walker, T.L. The multifaceted role of platelets in mediating brain function. Blood 2022, 140, 815–827. [Google Scholar] [CrossRef]
- Hughes, A.J.; Daniel, S.E.; Kilford, L.; Lees, A.J. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: A clinico-pathological study of 100 cases. J. Neurol. Neurosurg. Psychiatry 1992, 55, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Sateia, M.J. International classification of sleep disorders-third edition: Highlights and modifications. Chest 2014, 146, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R., Jr.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.J.; Mayeux, R.; et al. The diagnosis of dementia due to Alzheimer’s disease, recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011, 7, 263–269. [Google Scholar] [CrossRef]
- Lynöe, N.; Sandlund, M.; Dahlqvist, G.; Jacobsson, L. Informed consent, study of quality of information given to participants in a clinical trial. BMJ 1991, 303, 610–613. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 12 June 2023).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Krueger, F.; James, F.; Ewels, P.; Afyounian, E.; Weinstein, M.; Schuster-Boeckler, B.; Hulselmans, G.; Sclamons. FelixKrueger/TrimGalore: v0.6.10—Add Default Decompression Path (0.6.10). Zenodo 2023. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000 Res. 2015, 4, 1521. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
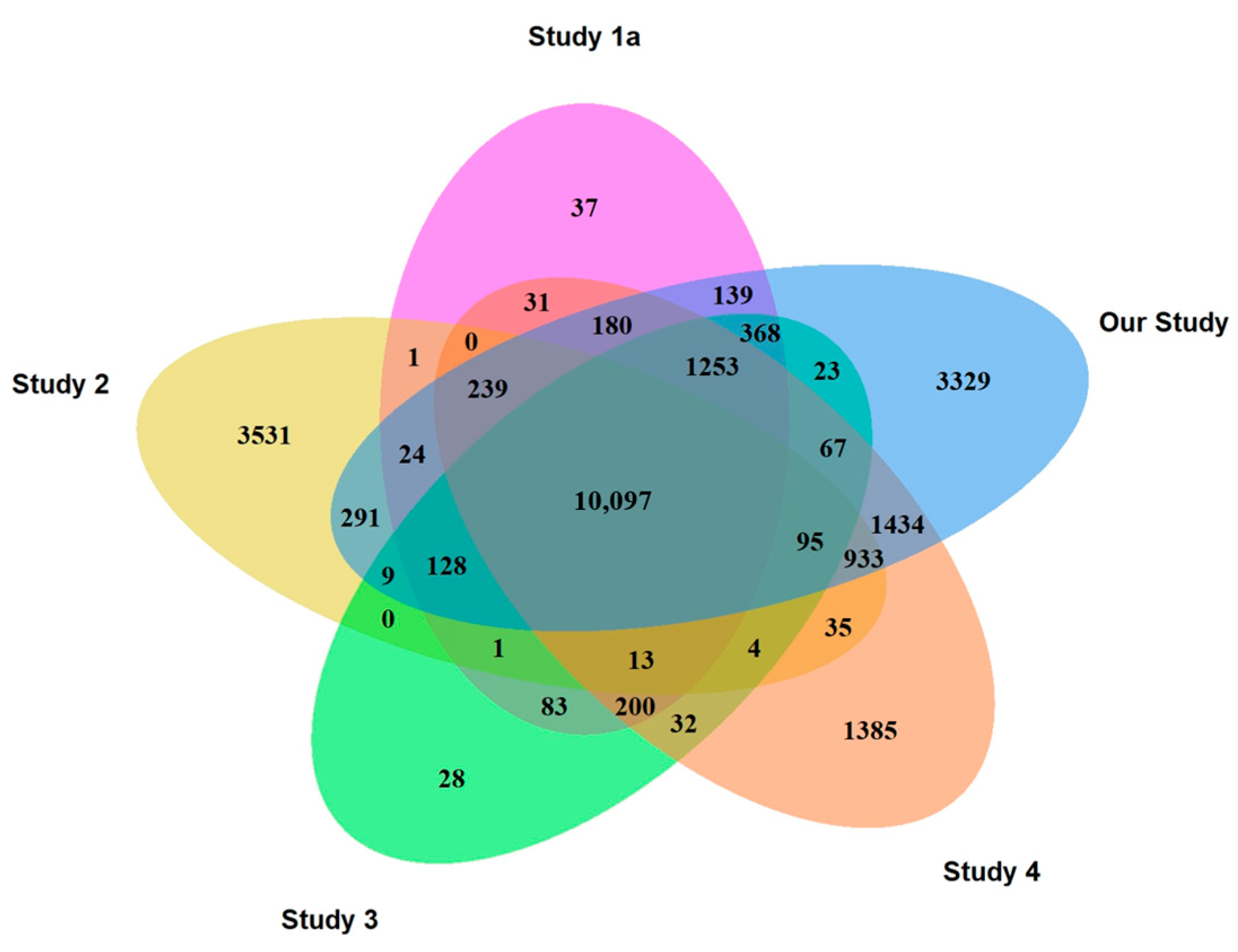


| DLB (n = 12) | PD (n = 12) | IRBD (n = 12) | AD (n = 14) | CTRL (n = 14) | p1 | |
|---|---|---|---|---|---|---|
| Mean age, y 2 (age range, y) | 74.1 (64–85) | 66.9 (44–87) | 74.5 (65–89) | 68.8 (60–80) | 71.3 (61–86) | 0.017 |
| Gender (male/female ratio) | 7/5 | 8/4 | 9/3 | 7/7 | 7/7 | 0.069 |
| Duration 3, years (range) | 5.7 (2.1–10.6) | 15.2 (4.9–23.7) | 8.9 (2.5–18.2) | 5.2 (0.8–8.0) | ||
| MMSE 4, mean (range) | 15.3 (3–27) | n.a. 5 | 19.9 (5–28) | - | 0.189 | |
| UPDRS-III 6, mean (range) | - | 20.9 (5–39) | - | - | - | |
| GDS fast 7, mean (range) | - | - | 4.1 (3–6) | - | - | |
| Parkinsonism, n (%) | 10 (83.3%) | - | - | - | - | |
| Positive DAT imaging, n (%) | 11 (91.6%) | - | - | - | - |
| Study | Year | Accession Number | Samples (n) | Age 1 | Expressed Genes 2 |
|---|---|---|---|---|---|
| 1a | 2021 [20] | PRJNA732990 | 20 CTRLs | 49.2 (21–75) | 12,794 |
| 1b | 2021 [20] | PRJNA732803 | 20 PD | 67.1 (50–86) | 13,747 |
| 2 | 2022 [21] | GSE183635 | 316 CTRLs | 55.4 (18–86) | 15,402 |
| 3 | 2022 [22] | PRJNA668820 | 56 CTRLs | 47.8 (n/a) | 12,401 |
| 4 | 2022 [23] | PRJNA737596 | 190 CTRLs | 54.6 (31–72) | 15,998 |
| 5a | 2025 | Our study | 14 CTRLs | 71.3 (61–86) | 18,609 |
| 5b | 2025 | Our study | 12 PD | 66.9 (44–87) | 18,132 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arnaldo, L.; Mena, J.; Adamuz, D.; Menéndez, A.; Serradell, M.; Samaniego, D.; Gaig, C.; Ispierto, L.; Vilas, D.; Iranzo, A.; et al. Does Platelet Transcriptome Dysregulation Across the Lewy Body Continuum Mirror Neuronal Dysfunction? Int. J. Mol. Sci. 2025, 26, 11169. https://doi.org/10.3390/ijms262211169
Arnaldo L, Mena J, Adamuz D, Menéndez A, Serradell M, Samaniego D, Gaig C, Ispierto L, Vilas D, Iranzo A, et al. Does Platelet Transcriptome Dysregulation Across the Lewy Body Continuum Mirror Neuronal Dysfunction? International Journal of Molecular Sciences. 2025; 26(22):11169. https://doi.org/10.3390/ijms262211169
Chicago/Turabian StyleArnaldo, Laura, Jorge Mena, David Adamuz, Alex Menéndez, Mònica Serradell, Daniela Samaniego, Carles Gaig, Lourdes Ispierto, Dolores Vilas, Alex Iranzo, and et al. 2025. "Does Platelet Transcriptome Dysregulation Across the Lewy Body Continuum Mirror Neuronal Dysfunction?" International Journal of Molecular Sciences 26, no. 22: 11169. https://doi.org/10.3390/ijms262211169
APA StyleArnaldo, L., Mena, J., Adamuz, D., Menéndez, A., Serradell, M., Samaniego, D., Gaig, C., Ispierto, L., Vilas, D., Iranzo, A., Aarsland, D., Pastor, P., & Beyer, K. (2025). Does Platelet Transcriptome Dysregulation Across the Lewy Body Continuum Mirror Neuronal Dysfunction? International Journal of Molecular Sciences, 26(22), 11169. https://doi.org/10.3390/ijms262211169





