Research Progress Regarding the Use of Single-Cell Sequencing Technology in Analyzing Tumor Endothelial Cell Pathophysiology
Abstract
1. Introduction
2. Shared Characteristics: A Comprehensive Single-Cell Analysis of Tumor Endothelial Cells (TECs) Across Various Types of Tumors
2.1. At the Genetic Level
2.2. At the Cellular Functional Level
- (1)
- Arterial ECs, venous ECs, capillary-like ECs, and lymphatic ECs: Pan (2024) [26] constructed the most comprehensive pan-cancer vascular single-cell atlas to date, encompassing approximately 200,000 cells across 31 cancer types. The study revealed that venous endothelial cells serve as the initiation site for tumor angiogenesis. Furthermore, three major vascular endothelial cell types were identified: arterial ECs, venous ECs, and capillary ECs. Capillary-like EC numbers are significantly higher (approximately 1.8-fold) in tumor tissue compared to normal tissue, and these cells constitute a major component of the tumor vascular network [26].
- (2)
- APLN+ tip cells/TipSI ECs represent the most characteristic tumor-specific subset of endothelial cells and can be considered a “hallmark” of tumor angiogenesis. These cells exhibit high migratory activity, respond to gradients of growth factors such as VEGF, and guide the formation of new blood vessel sprouts. Moreover, they express a variety of cytokines and signaling molecules that facilitate communication with other cell types in the tumor microenvironment, including secreted phosphoprotein 1 (SPP1+) macrophages. Usually, tip ECs are expressed during the early phase of blood vessel sprouting, a process that signifies tumor progression and is strongly associated with poor clinical outcomes. The level of its expression may serve as a predictive indicator for the efficacy of anti-angiogenic treatments; an example is bevacizumab, an anti-angiogenic agent that targets VEGF-A. Some recent single-cell studies focusing on tip-like endothelial cells are summarized in Table 2 [27,28,29,30,31].
- (3)
- The endothelial–mesenchymal transition (EndoMT) is a biological process in which endothelial cells acquire mesenchymal cell characteristics, and it has been widely observed in various tumors. Endothelial cells undergoing EndoMT (referred to as EndoMT-like ECs) co-express both endothelial markers (such as PECAM1/CD31 and CDH5/VE-cadherin) and mesenchymal markers (including ACTA2/α-SMA, VIM/Vimentin, FN1/Fibronectin, as well as the transcription factors SNAI1/2 and TWIST1). These cells play a crucial role in promoting tumor cell intravasation into blood vessels [36]. Moreover, they secrete extracellular matrix (ECM) proteins that lead to perivascular matrix sclerosis, thereby impairing drug delivery [37]. This phenomenon is closely associated with poor prognosis in cancer therapy.
- (4)
- There is often a population of endothelial cells exhibiting abnormal function that may contribute to immune regulation. Within the tumor microenvironment, these cells frequently lack key costimulatory molecules, such as CD80 and CD86, which can lead to T cell dysfunction or promote the induction of regulatory T cells (Tregs), thereby facilitating immune tolerance. These endothelial cells highly express MHC class II molecules (e.g., HLA-DRA, HLA-DRB1), as well as costimulatory molecules (e.g., CD74) [38,39].
- (5)
- Immuno-regulatory endothelial cells (ECs) are a subset of endothelial cells that do not directly present antigens but actively modulate immune responses through the expression of various immunosuppressive molecules and adhesion factors. These cells typically exhibit high expression levels of immune checkpoint molecules such as PD-L1 (CD274) [40,41], as well as adhesion molecules including VCAM1, ICAM1, and SELE, which collectively contribute to their immunosuppressive functions. Here, we highlight the pivotal role of immune cells in shaping tumor endothelial heterogeneity. Advances in single-cell technologies have provided deeper insights into the molecular mechanisms underlying immune cell–endothelial cell interactions. Such crosstalk may result in T cells being sequestered at the perivascular regions, limiting their ability to infiltrate the tumor parenchyma effectively. Notably, certain endothelial cell subsets constitutively express high levels of immune checkpoint molecules, including PD-L1, which directly contribute to T cell suppression. Furthermore, cytokines such as IFN-γ, secreted by activated T cells and other immune cells, can induce upregulation of MHC molecules in endothelial cells and enhance their antigen-presenting capacity [34,42]. Collectively, these findings establish immune cells as key regulators driving endothelial heterogeneity within the tumor microenvironment. CXCR4+ tip cells (a predominant angiogenic phenotype) and SELE+ venous endothelial cells (a proinflammatory phenotype) were identified across 19 solid tumor types, revealing significant heterogeneity in endothelial cell composition and functional states among different cancers. Tumor tissues frequently exhibit an increased proportion of CXCR4+ tip cells (promoting angiogenesis) and a reduced presence of SELE+ venous endothelial cells, which may impair immune cell infiltration and subsequently influence treatment response [43]. In-depth investigation of the intricate interactions between immune cells and tumor-associated endothelial cells using single-cell technologies holds promise for identifying novel therapeutic targets and advancing innovative strategies. For instance, research focusing on specific molecules such as CXCR4 is paving the way for new therapeutic approaches.
2.3. Application: Anti-Angiogenic Therapy (AAT)
3. Characteristics of Tumor Endothelial Cells in Different Tumors
3.1. Gene Expression Changes in Tumor Endothelial Cells
3.2. Distinct Cellular Origins of Tumor Endothelial Cells
3.3. Endothelial-Targeted Gene Delivery Therapy with Different Vectors
4. Conclusions and Prospects
5. Literature Search
- Databases: A systematic search was conducted across several authoritative databases, including Web of Science Core Collection, Scopus, PubMed, and China National Knowledge Infrastructure (CNKI).
- Timeframe: The search covered studies published from 2010 to 2025.
- Keywords and Search String: An optimized search string was employed using a combination of subject headings and free-text words, connected by Boolean operators (AND, OR, NOT). For instance, the search string used for Web of Science was (“single cell analysis”) AND (“tumor”) AND (“endothelial cell”).
6. Literature Screening and Eligibility Criteria
- (1)
- Study type: original research articles, reviews.
- (2)
- Subject: involves TECs, focuses on single-cell analysis.
- (3)
- Topic: single-cell studies of TECs.
- (4)
- Language: publications in English and Chinese.
7. Data Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Y.; Liu, Z.; Han, X.; Liang, F.; Zhang, Q.; Huang, X.; Shi, X.; Huo, H.; Han, M.; Liu, X.; et al. Dynamics of Endothelial Cell Generation and Turnover in Arteries During Homeostasis and Diseases. Circulation 2024, 149, 135–154. [Google Scholar] [CrossRef]
- Meyer, A.; Zack, S.R.; Nijim, W.; Burgos, A.; Patel, V.; Zanotti, B.; Volin, M.V.; Amin, M.A.; Lewis, M.J.; Pitzalis, C.; et al. Metabolic reprogramming by Syntenin-1 directs RA FLS and endothelial cell-mediated inflammation and angiogenesis. Cell. Mol. Immunol. 2024, 21, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Bussard, K.M.; Mutkus, L.; Stumpf, K.; Gomez-Manzano, C.; Marini, F.C. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res. 2016, 18, 84. [Google Scholar] [CrossRef]
- Lambrechts, D.; Wauters, E.; Boeckx, B.; Aibar, S.; Nittner, D.; Burton, O.; Bassez, A.; Decaluwe, H.; Pircher, A.; Van den Eynde, K.; et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018, 24, 1277–1289. [Google Scholar] [CrossRef] [PubMed]
- Heidegger, I.; Pircher, A.; Pichler, R. Targeting the Tumor Microenvironment in Renal Cell Cancer Biology and Therapy. Front. Oncol. 2019, 9, 490. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Gao, R.R.; Zhang, X.; Yang, J.X.; Liu, Y.; Ma, J.; Chen, Q. Dissecting endothelial cell heterogeneity with new tools. Cell Regen. 2025, 14, 10. [Google Scholar] [CrossRef]
- Cantelmo, A.R.; Conradi, L.C.; Brajic, A.; Goveia, J.; Kalucka, J.; Pircher, A.; Chaturvedi, P.; Hol, J.; Thienpont, B.; Teuwen, L.A.; et al. Inhibition of the Glycolytic Activator PFKFB3 in Endothelium Induces Tumor Vessel Normalization, Impairs Metastasis, and Improves Chemotherapy. Cancer Cell 2016, 30, 968–985. [Google Scholar] [CrossRef]
- Rohlenova, K.; Veys, K.; Miranda-Santos, I.; De Bock, K.; Carmeliet, P. Endothelial Cell Metabolism in Health and Disease. Trends Cell Biol. 2018, 28, 224–236. [Google Scholar] [CrossRef]
- Heidegger, I.; Fotakis, G.; Offermann, A.; Goveia, J.; Daum, S.; Salcher, S.; Noureen, A.; Timmer-Bosscha, H.; Schafer, G.; Walenkamp, A.; et al. Comprehensive characterization of the prostate tumor microenvironment identifies CXCR4/CXCL12 crosstalk as a novel antiangiogenic therapeutic target in prostate cancer. Mol. Cancer 2022, 21, 132. [Google Scholar] [CrossRef]
- Agnihotri, T.G.; Salave, S.; Shinde, T.; Srikanth, I.; Gyanani, V.; Haley, J.C.; Jain, A. Understanding the role of endothelial cells in brain tumor formation and metastasis: A proposition to be explored for better therapy. J. Natl. Cancer Cent. 2023, 3, 222–235. [Google Scholar] [CrossRef]
- De Sanctis, F.; Ugel, S.; Facciponte, J.; Facciabene, A. The dark side of tumor-associated endothelial cells. Semin. Immunol. 2018, 35, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr. Biol. 2020, 30, R921–R925. [Google Scholar] [CrossRef] [PubMed]
- Hida, K.; Ohga, N.; Akiyama, K.; Maishi, N.; Hida, Y. Heterogeneity of tumor endothelial cells. Cancer Sci. 2013, 104, 1391–1395. [Google Scholar] [CrossRef] [PubMed]
- La Porta, S.; Roth, L.; Singhal, M.; Mogler, C.; Spegg, C.; Schieb, B.; Qu, X.; Adams, R.H.; Baldwin, H.S.; Savant, S.; et al. Endothelial Tie1-mediated angiogenesis and vascular abnormalization promote tumor progression and metastasis. J. Clin. Investig. 2018, 128, 834–845. [Google Scholar] [CrossRef]
- Huijbers, E.J.M.; Khan, K.A.; Kerbel, R.S.; Griffioen, A.W. Tumors resurrect an embryonic vascular program to escape immunity. Sci. Immunol. 2022, 7, eabm6388. [Google Scholar] [CrossRef]
- Young, M.D.; Mitchell, T.J.; Vieira Braga, F.A.; Tran, M.G.B.; Stewart, B.J.; Ferdinand, J.R.; Collord, G.; Botting, R.A.; Popescu, D.M.; Loudon, K.W.; et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 2018, 361, 594–599. [Google Scholar] [CrossRef]
- Dirkx, A.E.; Oude Egbrink, M.G.; Kuijpers, M.J.; van der Niet, S.T.; Heijnen, V.V.; Bouma-ter Steege, J.C.; Wagstaff, J.; Griffioen, A.W. Tumor angiogenesis modulates leukocyte-vessel wall interactions in vivo by reducing endothelial adhesion molecule expression. Cancer Res. 2003, 63, 2322–2329. [Google Scholar]
- Pircher, A.; Hilbe, W.; Heidegger, I.; Drevs, J.; Tichelli, A.; Medinger, M. Biomarkers in tumor angiogenesis and anti-angiogenic therapy. Int. J. Mol. Sci. 2011, 12, 7077–7099. [Google Scholar] [CrossRef]
- Sievert, W.; Tapio, S.; Breuninger, S.; Gaipl, U.; Andratschke, N.; Trott, K.R.; Multhoff, G. Adhesion molecule expression and function of primary endothelial cells in benign and malignant tissues correlates with proliferation. PLoS ONE 2014, 9, e91808. [Google Scholar] [CrossRef]
- Hirsch, H.A.; Iliopoulos, D.; Joshi, A.; Zhang, Y.; Jaeger, S.A.; Bulyk, M.; Tsichlis, P.N.; Shirley Liu, X.; Struhl, K. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell 2010, 17, 348–361. [Google Scholar] [CrossRef]
- Xie, Z.; Niu, L.; Du, K.; Chen, L.; Zheng, G.; Dai, S.; Dan, H.; Duan, L.; Dou, X.; Feng, F.; et al. Endothelial cell heterogeneity in colorectal cancer: Tip cells drive angiogenesis. Cell. Mol. Life Sci. 2024, 81, 365. [Google Scholar] [CrossRef]
- Cervantes-Villagrana, R.D.; Albores-Garcia, D.; Cervantes-Villagrana, A.R.; Garcia-Acevez, S.J. Tumor-induced neurogenesis and immune evasion as targets of innovative anti-cancer therapies. Signal Transduct. Target. Ther. 2020, 5, 99. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, K.; Onoe, T.; Yoshida, T.; Yamashita, Y.; Tanaka, Y.; Ohdan, H. Tumor Endothelial Cell-Mediated Antigen-Specific T-cell Suppression via the PD-1/PD-L1 Pathway. Mol. Cancer Res. 2020, 18, 1427–1440. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.M.; Wiesolek, H.L.; Sumagin, R. ICAM-1: A master regulator of cellular responses in inflammation, injury resolution, and tumorigenesis. J. Leukoc. Biol. 2020, 108, 787–799. [Google Scholar] [CrossRef]
- Biel, N.M.; Siemann, D.W. Targeting the Angiopoietin-2/Tie-2 axis in conjunction with VEGF signal interference. Cancer Lett. 2016, 380, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Li, X.; Dong, L.; Liu, T.; Zhang, M.; Zhang, L.; Zhang, X.; Huang, L.; Shi, W.; Sun, H.; et al. Tumour vasculature at single-cell resolution. Nature 2024, 632, 429–436. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, T.; Lu, S.; Ma, S.; Han, D.; Zhang, K.; Xu, C.; Liu, S.; Gan, L.; Wu, X.; et al. Single-cell analysis of multiple cancer types reveals differences in endothelial cells between tumors and normal tissues. Comput. Struct. Biotechnol. J. 2023, 21, 665–676. [Google Scholar] [CrossRef]
- Lee, S.; Kim, H.; Kim, B.S.; Chae, S.; Jung, S.; Lee, J.S.; Yu, J.; Son, K.; Chung, M.; Kim, J.K.; et al. Angiogenesis-on-a-chip coupled with single-cell RNA sequencing reveals spatially differential activations of autophagy along angiogenic sprouts. Nat. Commun. 2024, 15, 230. [Google Scholar] [CrossRef]
- Funasaki, S.; Miyamura, Y.; Kamei, S.; Rahman, A.; Yamazaki, M.; Usuki, S.; Yasunaga, K.; Satou, Y.; Ohguchi, H.; Minami, T. Protocol for transcriptomic and epigenomic analyses of tip-like endothelial cells using scRNA-seq and ChIP-seq. STAR Protoc. 2025, 6, 103326. [Google Scholar] [CrossRef]
- Tang, H.; Cai, Y.; Yang, M.; Tang, S.; Huang, Q.; Li, H.; Liu, S.; Teng, H.; Xie, T.; He, M.; et al. Single-cell and spatial transcriptomics reveals the key role of MCAM+ tip-like endothelial cells in osteosarcoma metastasis. npj Precis. Oncol. 2025, 9, 104. [Google Scholar] [CrossRef]
- Pang, L.; Sun, Q.; Wang, W.; Song, M.; Wu, Y.; Shi, X.; Shi, X. A novel gene signature for predicting outcome in colorectal cancer patients based on tumor cell-endothelial cell interaction via single-cell sequencing and machine learning. Heliyon 2025, 11, e42237. [Google Scholar] [CrossRef]
- Rohlenova, K.; Goveia, J.; Garcia-Caballero, M.; Subramanian, A.; Kalucka, J.; Treps, L.; Falkenberg, K.D.; de Rooij, L.; Zheng, Y.; Lin, L.; et al. Single-Cell RNA Sequencing Maps Endothelial Metabolic Plasticity in Pathological Angiogenesis. Cell Metab. 2020, 31, 862–877.e14. [Google Scholar] [CrossRef]
- Parte, S.; Kaur, A.B.; Nimmakayala, R.K.; Ogunleye, A.O.; Chirravuri, R.; Vengoji, R.; Leon, F.; Nallasamy, P.; Rauth, S.; Alsafwani, Z.W.; et al. Cancer-Associated Fibroblast Induces Acinar-to-Ductal Cell Transdifferentiation and Pancreatic Cancer Initiation Via LAMA5/ITGA4 Axis. Gastroenterology 2024, 166, 842–858.e5. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Lin, Y.; Liao, Z.; Gao, X.; Lu, C.; Lu, L.; Huang, J.; Huang, X.; Huang, S.; Yu, H.; et al. Single cell-spatial transcriptomics and bulk multi-omics analysis of heterogeneity and ecosystems in hepatocellular carcinoma. npj Precis. Oncol. 2024, 8, 262. [Google Scholar] [CrossRef] [PubMed]
- Yue, N.; Jin, Q.; Li, C.; Zhang, L.; Cao, J.; Wu, C. CD36: A promising therapeutic target in hematologic tumors. Leuk. Lymphoma 2024, 65, 1749–1765. [Google Scholar] [CrossRef]
- Dejana, E.; Lampugnani, M.G. Endothelial cell transitions. Science 2018, 362, 746–747. [Google Scholar] [CrossRef]
- Platel, V.; Faure, S.; Corre, I.; Clere, N. Endothelial-to-Mesenchymal Transition (EndoMT): Roles in Tumorigenesis, Metastatic Extravasation and Therapy Resistance. J. Oncol. 2019, 2019, 8361945. [Google Scholar] [CrossRef]
- Mezyk-Kopec, R.; Potin, L.; Gomez Medellin, J.E.; Salles, C.M.; Swartz, M.A. TGF-beta Signaling Prevents MHC Class II-Expressing Lymphatic Endothelial Cells from Reactivating Human Allogenic Memory CD4+ T Cells. J. Immunol. 2023, 211, 782–790. [Google Scholar] [CrossRef]
- Norder, M.; Gutierrez, M.G.; Zicari, S.; Cervi, E.; Caruso, A.; Guzman, C.A. Lymph node-derived lymphatic endothelial cells express functional costimulatory molecules and impair dendritic cell-induced allogenic T-cell proliferation. FASEB J. 2012, 26, 2835–2846. [Google Scholar] [CrossRef]
- Wu, S.Z.; Al-Eryani, G.; Roden, D.L.; Junankar, S.; Harvey, K.; Andersson, A.; Thennavan, A.; Wang, C.; Torpy, J.R.; Bartonicek, N.; et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 2021, 53, 1334–1347. [Google Scholar] [CrossRef]
- Kurten, C.H.L.; Kulkarni, A.; Cillo, A.R.; Santos, P.M.; Roble, A.K.; Onkar, S.; Reeder, C.; Lang, S.; Chen, X.; Duvvuri, U.; et al. Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing. Nat. Commun. 2021, 12, 7338. [Google Scholar] [CrossRef]
- Chu, X.; Li, X.; Zhang, Y.; Dang, G.; Miao, Y.; Xu, W.; Wang, J.; Zhang, Z.; Cheng, S. Integrative single-cell analysis of human colorectal cancer reveals patient stratification with distinct immune evasion mechanisms. Nat. Cancer 2024, 5, 1409–1426. [Google Scholar] [CrossRef]
- Li, J.; Wang, D.; Tang, F.; Ling, X.; Zhang, W.; Zhang, Z. Pan-cancer integrative analyses dissect the remodeling of endothelial cells in human cancers. Natl. Sci. Rev. 2024, 11, nwae231. [Google Scholar] [CrossRef]
- Perez-Gutierrez, L.; Ferrara, N. Biology and therapeutic targeting of vascular endothelial growth factor A. Nat. Rev. Mol. Cell Biol. 2023, 24, 816–834. [Google Scholar] [CrossRef] [PubMed]
- van der Meel, R.; Symons, M.H.; Kudernatsch, R.; Kok, R.J.; Schiffelers, R.M.; Storm, G.; Gallagher, W.M.; Byrne, A.T. The VEGF/Rho GTPase signalling pathway: A promising target for anti-angiogenic/anti-invasion therapy. Drug Discov. Today 2011, 16, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Coelho, F.; Martins, F.; Pereira, S.A.; Serpa, J. Anti-Angiogenic Therapy: Current Challenges and Future Perspectives. Int. J. Mol. Sci. 2021, 22, 3765. [Google Scholar] [CrossRef] [PubMed]
- Walchli, T.; Ghobrial, M.; Schwab, M.; Takada, S.; Zhong, H.; Suntharalingham, S.; Vetiska, S.; Gonzalez, D.R.; Wu, R.; Rehrauer, H.; et al. Single-cell atlas of the human brain vasculature across development, adulthood and disease. Nature 2024, 632, 603–613. [Google Scholar] [CrossRef]
- Hosaka, K.; Andersson, P.; Wu, J.; He, X.; Du, Q.; Jing, X.; Seki, T.; Gao, J.; Zhang, Y.; Sun, X.; et al. KRAS mutation-driven angiopoietin 2 bestows anti-VEGF resistance in epithelial carcinomas. Proc. Natl. Acad. Sci. USA 2023, 120, e2303740120. [Google Scholar] [CrossRef]
- Efimova, I.; Catanzaro, E.; Van der Meeren, L.; Turubanova, V.D.; Hammad, H.; Mishchenko, T.A.; Vedunova, M.V.; Fimognari, C.; Bachert, C.; Coppieters, F.; et al. Vaccination with early ferroptotic cancer cells induces efficient antitumor immunity. J. Immunother. Cancer 2020, 8, e001369. [Google Scholar] [CrossRef]
- Hicklin, D.J.; Ellis, L.M. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J. Clin. Oncol. 2005, 23, 1011–1027. [Google Scholar] [CrossRef]
- Jayson, G.C.; Kerbel, R.; Ellis, L.M.; Harris, A.L. Antiangiogenic therapy in oncology: Current status and future directions. Lancet 2016, 388, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.; Hurwitz, H.I.; Sandler, A.B.; Miles, D.; Coleman, R.L.; Deurloo, R.; Chinot, O.L. Bevacizumab (Avastin(R)) in cancer treatment: A review of 15 years of clinical experience and future outlook. Cancer Treat. Rev. 2020, 86, 102017. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, Y.; Cao, Z.; Ji, H.; Yang, X.; Iwamoto, H.; Wahlberg, E.; Lanne, T.; Sun, B.; Cao, Y. Anti-VEGF- and anti-VEGF receptor-induced vascular alteration in mouse healthy tissues. Proc. Natl. Acad. Sci. USA 2013, 110, 12018–12023. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y. VEGF-targeted cancer therapeutics-paradoxical effects in endocrine organs. Nat. Rev. Endocrinol. 2014, 10, 530–539. [Google Scholar] [CrossRef]
- Goveia, J.; Rohlenova, K.; Taverna, F.; Treps, L.; Conradi, L.C.; Pircher, A.; Geldhof, V.; de Rooij, L.; Kalucka, J.; Sokol, L.; et al. An Integrated Gene Expression Landscape Profiling Approach to Identify Lung Tumor Endothelial Cell Heterogeneity and Angiogenic Candidates. Cancer Cell 2020, 37, 421. [Google Scholar] [CrossRef]
- Zeng, Q.; Mousa, M.; Nadukkandy, A.S.; Franssens, L.; Alnaqbi, H.; Alshamsi, F.Y.; Safar, H.A.; Carmeliet, P. Understanding tumour endothelial cell heterogeneity and function from single-cell omics. Nat. Rev. Cancer 2023, 23, 544–564. [Google Scholar] [CrossRef]
- Ma, L.; Wang, L.; Khatib, S.A.; Chang, C.W.; Heinrich, S.; Dominguez, D.A.; Forgues, M.; Candia, J.; Hernandez, M.O.; Kelly, M.; et al. Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J. Hepatol. 2021, 75, 1397–1408. [Google Scholar] [CrossRef]
- Liu, Y.; Carson-Walter, E.B.; Cooper, A.; Winans, B.N.; Johnson, M.D.; Walter, K.A. Vascular gene expression patterns are conserved in primary and metastatic brain tumors. J. Neurooncol. 2010, 99, 13–24. [Google Scholar] [CrossRef]
- Denzer, L.; Muranyi, W.; Schroten, H.; Schwerk, C. The role of PLVAP in endothelial cells. Cell Tissue Res. 2023, 392, 393–412. [Google Scholar] [CrossRef]
- Singh, V.; Kaur, R.; Kumari, P.; Pasricha, C.; Singh, R. ICAM-1 and VCAM-1: Gatekeepers in various inflammatory and cardiovascular disorders. Clin. Chim. Acta 2023, 548, 117487. [Google Scholar] [CrossRef]
- Bejarano, L.; Kauzlaric, A.; Lamprou, E.; Lourenco, J.; Fournier, N.; Ballabio, M.; Colotti, R.; Maas, R.; Galland, S.; Massara, M.; et al. Interrogation of endothelial and mural cells in brain metastasis reveals key immune-regulatory mechanisms. Cancer Cell 2024, 42, 378–395.e310. [Google Scholar] [CrossRef]
- Pu, S.; Liu, T.; Gao, Y.; Wu, Z.; Liu, S. The role of tumor-associated endothelial cells in malignant progression and immune evasion of liver cancer. Int. Immunopharmacol. 2025, 161, 115013. [Google Scholar] [CrossRef]
- Sun, X.; Cai, W.; Li, H.; Gao, C.; Ma, X.; Guo, Y.; Fu, D.; Xiao, D.; Zhang, Z.; Wang, Y.; et al. Endothelial-like cancer-associated fibroblasts facilitate pancreatic cancer metastasis via vasculogenic mimicry and paracrine signalling. Gut 2025, 74, 1437–1451. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yang, M.; Deng, C.; Hu, M.; Wu, S.; Lai, K.; Zhang, L.; Chen, Z.; Tang, Q.; Wang, Q.; et al. Single-cell sequencing reveals the role of IL-33+ endothelial subsets in promoting early gastric cancer progression. iMeta 2025, 4, e70050. [Google Scholar] [CrossRef]
- Hou, M.; Chen, J.; Jiang, Y.; Liu, C.; Wang, X.; Liu, E.; Zong, Y.; Gu, M.; Meng, Z.; Wang, S.; et al. Single-cell analysis reveals CD34+CD90+ endothelial cells promote tumor metastasis in gallbladder cancer. npj Precis. Oncol. 2025, 9, 242. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Guo, C.; Wang, C.; Xu, J.; Zheng, S.; Duan, J.; Li, Y.; Bai, H.; Xu, Q.; Ning, F.; et al. Single-cell RNA sequencing reveals tumor heterogeneity, microenvironment, and drug-resistance mechanisms of recurrent glioblastoma. Cancer Sci. 2023, 114, 2609–2621. [Google Scholar] [CrossRef]
- Bosisio, D.; Ronca, R.; Salvi, V.; Presta, M.; Sozzani, S. Dendritic cells in inflammatory angiogenesis and lymphangiogenesis. Curr. Opin. Immunol. 2018, 53, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Havemann, K.; Pujol, B.F.; Adamkiewicz, J. In vitro transformation of monocytes and dendritic cells into endothelial like cells. Adv. Exp. Med. Biol. 2003, 522, 47–57. [Google Scholar]
- Yang, R.; Zhou, Y.; Du, C.; Wu, Y. Bioinformatics analysis of differentially expressed genes in tumor and paracancerous tissues of patients with lung adenocarcinoma. J. Thorac. Dis. 2020, 12, 7355–7364. [Google Scholar] [CrossRef]
- Danoy, M.; Jellali, R.; Tauran, Y.; Bruce, J.; Leduc, M.; Gilard, F.; Gakiere, B.; Scheidecker, B.; Kido, T.; Miyajima, A.; et al. Characterization of the proteome and metabolome of human liver sinusoidal endothelial-like cells derived from induced pluripotent stem cells. Differentiation 2021, 120, 28–35. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhou, J.; Li, L.; Liao, S.; He, J.; Zhou, S.; Zhou, Y. Pericytes in the tumor microenvironment. Cancer Lett. 2023, 556, 216074. [Google Scholar] [CrossRef]
- Ou, Z.; Lin, S.; Qiu, J.; Ding, W.; Ren, P.; Chen, D.; Wang, J.; Tong, Y.; Wu, D.; Chen, A.; et al. Single-Nucleus RNA Sequencing and Spatial Transcriptomics Reveal the Immunological Microenvironment of Cervical Squamous Cell Carcinoma. Adv. Sci. 2022, 9, e2203040. [Google Scholar] [CrossRef]
- Peng, H.; Wu, X.; Liu, S.; He, M.; Xie, C.; Zhong, R.; Liu, J.; Tang, C.; Li, C.; Xiong, S.; et al. Multiplex immunofluorescence and single-cell transcriptomic profiling reveal the spatial cell interaction networks in the non-small cell lung cancer microenvironment. Clin. Transl. Med. 2023, 13, e1155. [Google Scholar] [CrossRef] [PubMed]
- Mugisha, S.; Labhsetwar, S.; Dave, D.; Klemke, R.; Desgrosellier, J.S. A dataset of chronic nicotine-induced genes in breast cancer cells. Data Brief. 2025, 60, 111573. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, X.; Li, S.; Zhao, C.; Sun, Y.; Tian, K.; Wang, J.; Li, W.; Xu, L.; Jing, J.; et al. Resolving the lineage relationship between malignant cells and vascular cells in glioblastomas. Protein Cell 2023, 14, 105–122. [Google Scholar] [CrossRef] [PubMed]
- Zarebkohan, A.; Najafi, F.; Moghimi, H.R.; Hemmati, M.; Deevband, M.R.; Kazemi, B. SRL-Coated PAMAM Dendrimer Nano-Carrier for Targeted Gene Delivery to the Glioma Cells and Competitive Inhibition by Lactoferrin. Iran. J. Pharm. Res. 2016, 15, 629–640. [Google Scholar]
- Wu, G.; Jiang, Q.; Cui, T.; Liu, X.; Gong, D.; Yin, Y.; Wang, C.; Wang, T.; Lu, Y.; Zhu, D.; et al. The glymphatic system delivery enhances the transduction efficiency of AAV1 to brain endothelial cells in adult mice. J. Neurosci. Methods 2019, 328, 108441. [Google Scholar] [CrossRef]
- Barbon, E.; Kawecki, C.; Marmier, S.; Sakkal, A.; Collaud, F.; Charles, S.; Ronzitti, G.; Casari, C.; Christophe, O.D.; Denis, C.V.; et al. Development of a dual hybrid AAV vector for endothelial-targeted expression of von Willebrand factor. Gene Ther. 2023, 30, 245–254. [Google Scholar] [CrossRef]
- Wen, Y.; Ju, C. New insights into liver injury and regeneration from single-cell transcriptomics. eGastroenterology 2025, 3, e100202. [Google Scholar] [CrossRef]
- Qiu, Y.; Wang, Y.; Liu, J.; Liu, B.; Sun, K.; Hou, Q. Single-cell sequencing uncovers a high ESM1-expression endothelial cell subpopulation associated with bladder cancer progression and the immunosuppressive microenvironment. Sci. Rep. 2025, 15, 10946. [Google Scholar] [CrossRef]
- Xu, W.; Liang, T.; Fang, H.; Fu, L.; Deng, D.; Tan, X.; Liu, L.; Tang, D.; Zheng, H.; Ding, Q.; et al. Single-Cell RNA Sequencing Identifies MMP11+ Cancer-Associated Fibroblasts as Drivers of Angiogenesis and Bladder Cancer Progression. Adv. Sci. 2025, 12, e02774. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Ramnarayanan, K.; Sundar, R.; Padmanabhan, N.; Srivastava, S.; Koiwa, M.; Yasuda, T.; Koh, V.; Huang, K.K.; Tay, S.T.; et al. Single-Cell Atlas of Lineage States, Tumor Microenvironment, and Subtype-Specific Expression Programs in Gastric Cancer. Cancer Discov. 2022, 12, 670–691. [Google Scholar] [CrossRef]
- Zhang, P.; Yang, M.; Zhang, Y.; Xiao, S.; Lai, X.; Tan, A.; Du, S.; Li, S. Dissecting the Single-Cell Transcriptome Network Underlying Gastric Premalignant Lesions and Early Gastric Cancer. Cell Rep. 2019, 27, 1934–1947.e1935. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hu, S.; Min, M.; Ni, Y.; Lu, Z.; Sun, X.; Wu, J.; Liu, B.; Ying, X.; Liu, Y. Dissecting transcriptional heterogeneity in primary gastric adenocarcinoma by single cell RNA sequencing. Gut 2021, 70, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, M.; Tang, H.; Xie, T.; Lin, Y.; Liu, S.; Liang, J.; Li, F.; Luo, K.; Yang, M.; et al. Single-cell and spatial transcriptomics reveal metastasis mechanism and microenvironment remodeling of lymph node in osteosarcoma. BMC Med. 2024, 22, 200. [Google Scholar] [CrossRef]
- Marlow, S.A.; Deaville, L.A.; Berrens, R.V. Long-read RNA sequencing of transposable elements from single cells using CELLO-seq. Nat. Protoc. 2025, 20, 3070–3095. [Google Scholar] [CrossRef]
- Fu, Y.; Kim, H.; Adams, J.I.; Grimes, S.M.; Huang, S.; Lau, B.T.; Sathe, A.; Hess, P.; Ji, H.P.; Zhang, N.R. Single cell and spatial alternative splicing analysis with long read sequencing. Res Sq. 2023. [Google Scholar] [CrossRef]
- Ma, W.; Wang, F.; Fan, Y.; Hao, G.; Su, Y.; Wong, K.C.; Li, X. DeepNanoHi-C: Deep learning enables accurate single-cell nanopore long-read data analysis and 3D genome interpretation. Nucleic Acids Res. 2025, 53, gkaf640. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J.; Malcolm, A.A.; Mansfield, W.; Clark, S.J.; Argelaguet, R.; Biggins, L.; Acton, R.J.; Andrews, S.; Reik, W.; et al. Combinatorial profiling of multiple histone modifications and transcriptome in single cells using scMTR-seq. Sci. Adv. 2025, 11, eadu3308. [Google Scholar] [CrossRef]
- Tirosh, I.; Suva, M.L. Cancer cell states: Lessons from ten years of single-cell RNA-sequencing of human tumors. Cancer Cell 2024, 42, 1497–1506. [Google Scholar] [CrossRef]
- Lavie, D.; Ben-Shmuel, A.; Erez, N.; Scherz-Shouval, R. Cancer-associated fibroblasts in the single-cell era. Nat. Cancer 2022, 3, 793–807. [Google Scholar] [CrossRef]
- Cheng, Y.; Chen, X.; Feng, L.; Yang, Z.; Xiao, L.; Xiang, B.; Wang, X.; Liu, D.; Lin, P.; Shi, J.; et al. Stromal architecture and fibroblast subpopulations with opposing effects on outcomes in hepatocellular carcinoma. Cell Discov. 2025, 11, 1. [Google Scholar] [CrossRef]
- Shi, Q.; Chen, Y.; Li, Y.; Qin, S.; Yang, Y.; Gao, Y.; Zhu, L.; Wang, D.; Zhang, Z. Cross-tissue multicellular coordination and its rewiring in cancer. Nature 2025, 643, 529–538. [Google Scholar] [CrossRef]
- Ma, J.; Huang, Y.; Chen, J.; Li, Y.; Yao, R.; Li, X.; Liang, Q.; Chen, X.; Peng, C.; Liu, K.; et al. FAP+ fibroblasts orchestrate tumor microenvironment remodeling in renal cell carcinoma with tumor thrombus. Nat. Commun. 2025, 16, 9387. [Google Scholar] [CrossRef]
- Yang, S.; Deng, C.; Pu, C.; Bai, X.; Tian, C.; Chang, M.; Feng, M. Single-Cell RNA Sequencing and Its Applications in Pituitary Research. Neuroendocrinology 2024, 114, 875–893. [Google Scholar] [CrossRef]
- Han, J.; DePinho, R.A.; Maitra, A. Single-cell RNA sequencing in pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 451–452. [Google Scholar] [CrossRef]
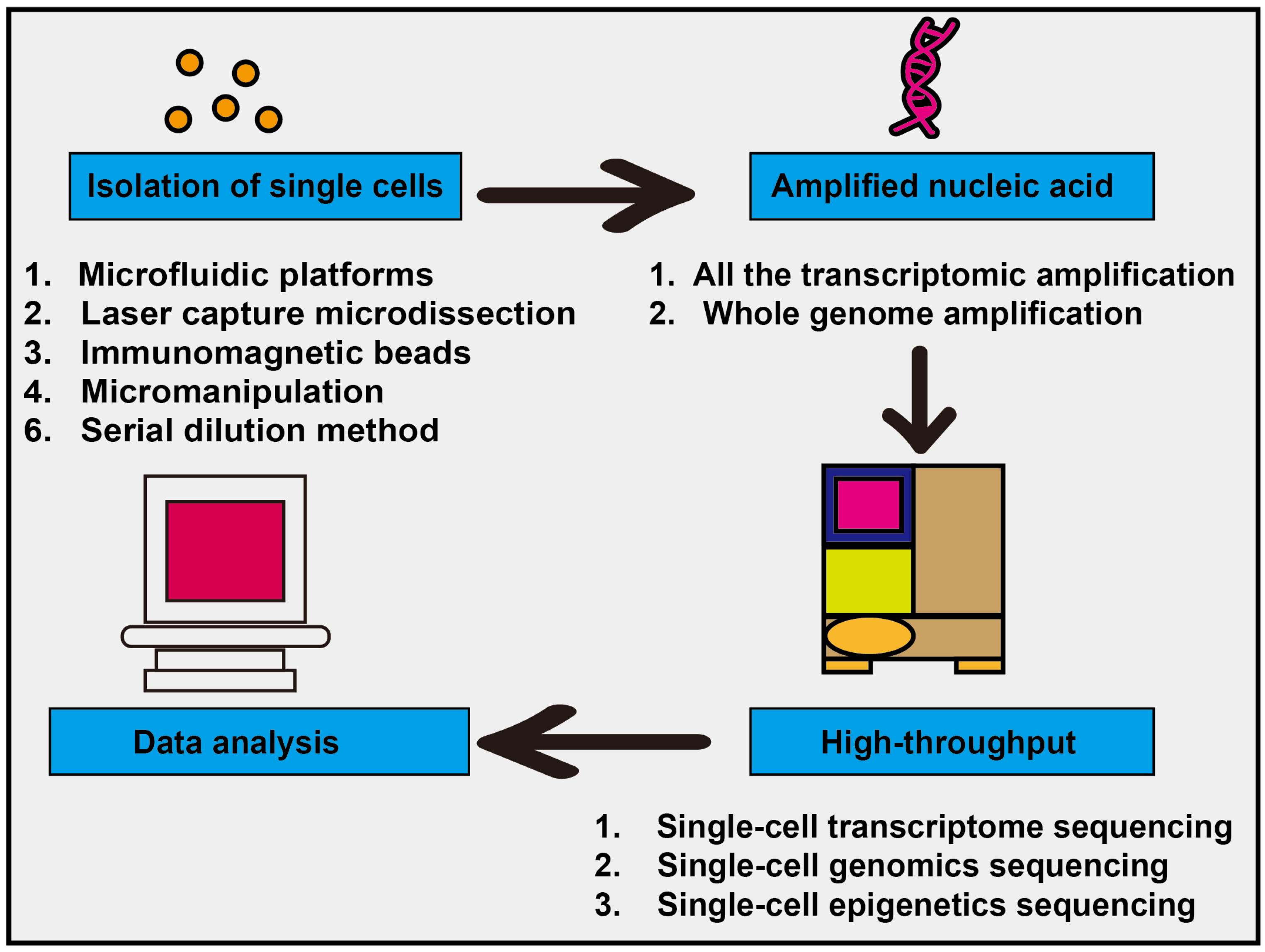
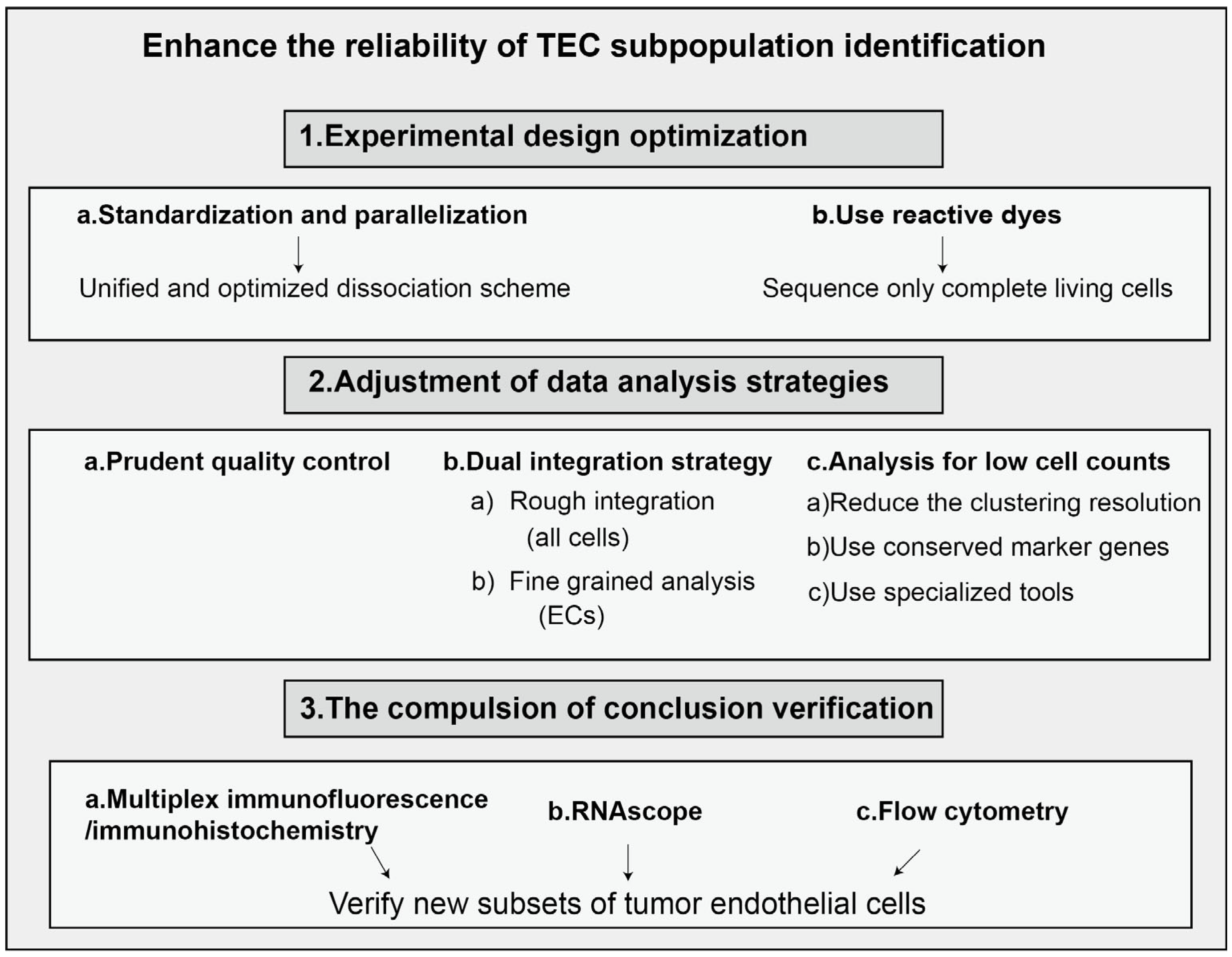
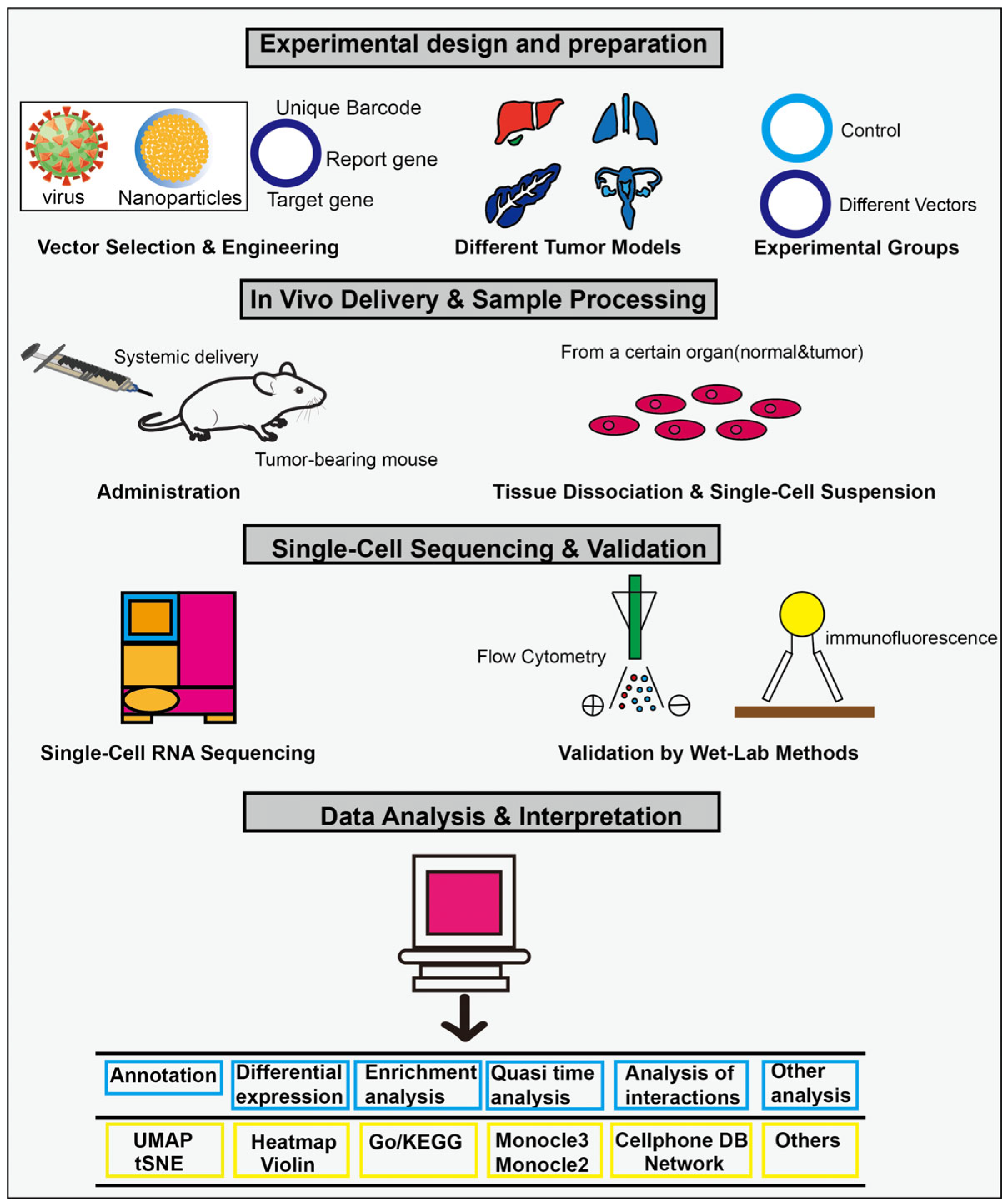
| Gene Symbol | Gene Name | Main Function |
|---|---|---|
| VEGFA | Vascular Endothelial Growth Factor A | Core pro-angiogenic signaling molecules |
| ANGPT2 | Angiopoietin-2 | Regulating vascular stability, promoting sprouting and proliferation |
| SPP1 | Secreted Phosphoprotein 1 | Cell adhesion, migration, and signaling |
| SELE | E-selectin | Mediating leukocyte adhesion |
| ICAM1 | Intercellular Adhesion Molecule-1 | Mediating cell adhesion |
| VCAN | Recombinant Versican | Remodeling of the extracellular matrix |
| POSTN | Periostin | Extracellular matrix proteins |
| COL4A/2 | Type IV Collagen | The main component of the basement membrane |
| Title | Year | Journal | Reference | Datasets | Cell Counts |
|---|---|---|---|---|---|
| Single-cell analysis of multiple cancer types reveals differences in endothelial cells between tumors and normal tissues | 2023 | Computational and Structural Biotechnology Journal | [32] | GSE155698 GSE159115 GSE167297 | 220,075 cells |
| Angiogenesis-on-a-chip coupled with single-cell RNA sequencing reveals spatially differential activations of autophagy along angiogenic sprouts | 2024 | Nature Communications | [33] | GSE155109 PRJNA931762 | 4693 cells |
| Protocol for transcriptomic and epigenomic analyses of tip-like endothelial cells using scRNA-seq and ChIP-seq | 2025 | STAR Protocols | [20] | GSE220509 | 7529 cells |
| Single-cell and spatial transcriptomics reveal the key role of MCAM+ tip-like endothelial cells in osteosarcoma metastasis | 2025 | NPJ Precision Oncology | [34] | GSE162454 GSE152048 GSE21257 HRA007229 | 129,315 cells |
| A novel gene signature for predicting outcome in colorectal cancer patients based on tumor cell–endothelial cell interaction via single-cell sequencing and machine learning | 2025 | Heliyon | [35] | GSE173839 GSE110224 GSE144735 GSE39582 GSE20916 GSE21510 GSE33113 GSE23878 GSE5206 GSE9348 | 27,414 cells |
| Types of Targets | Some Associated Cell Subsets/Molecules | Mechanisms of Function | Treatment Strategies |
|---|---|---|---|
| Specific endothelial cell subsets | APLN+ tip ECs | Guided new angiogenesis; high expression of pro-angiogenic factors; associated with poor prognosis | To develop drugs specifically targeting APLN+ cells, as a biomarker to screen for patients who may respond to AAT |
| Targets related to immune regulation | MHC-II+ ECs | Immunosuppression | Combined with immune checkpoint inhibitors |
| Subpopulation of pericytes | BASP1+ promotes angiogenesis of pericytes (matPCs) | Driven by ER stress, the secretion of VEGF promotes angiogenesis and is associated with poor prognosis | Targeting BASP1+ pericytes or their mediated ER stress pathway |
| Specific signaling pathways | VEGF/VEGFR | The classical angiogenic pathway | Bevacizumab, lenvatinib, and other multi-target TKIs |
| Interactions between cells | Endothelial cell-immune cell interaction (PODXL-SELL, ICAM1-SPN) | Forming an immunosuppressive microenvironment | Combined immunotherapy to destroy the “vaso-immunosuppressive alliance” |
| Specific markers of tumor vessels | Universal TEC markers (ACKR1, PLVAP, IGFBP3) | Prevalent in TECs from most tumor types | A potential target or diagnostic tool |
| Cancer Types | EC Subtypes | Markers |
|---|---|---|
| Hepatic carcinoma (HCC) | CD34+ CLDN5+ TECs [62] | CD34, CLDN5, VEGFR2 |
| CXCL12+ TECs [62] | CXCL12 | |
| PLVAP+ TECs [62] | PLVAP | |
| Pancreatic carcinoma (PDAC) | Endothelioid cancer-associated fibroblasts (endoCAFs) [63] | FAPα, CD144 (VE-cadherin) |
| Gastric carcinoma (EGC) | IL-33+ ECs [64] | IL-33, CD34, PECAM1 |
| Gallbladder carcinoma (GBC) | CD34+ CD90+ (SAEndo2) ECs [65] | CD34, CD90 (THY1), ESM1 |
| Pan-cancer type | APLN+ tip cells (TipSI) [26] | APLN |
| Venous ECs (VenEC) [26] | ACKR1 | |
| Capillary-like ECs (CapEC) [26] | RGCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.; Liu, S.; Shao, W.; Liu, D. Research Progress Regarding the Use of Single-Cell Sequencing Technology in Analyzing Tumor Endothelial Cell Pathophysiology. Int. J. Mol. Sci. 2025, 26, 11128. https://doi.org/10.3390/ijms262211128
Zhao S, Liu S, Shao W, Liu D. Research Progress Regarding the Use of Single-Cell Sequencing Technology in Analyzing Tumor Endothelial Cell Pathophysiology. International Journal of Molecular Sciences. 2025; 26(22):11128. https://doi.org/10.3390/ijms262211128
Chicago/Turabian StyleZhao, Shu, Siyi Liu, Wenxin Shao, and Dong Liu. 2025. "Research Progress Regarding the Use of Single-Cell Sequencing Technology in Analyzing Tumor Endothelial Cell Pathophysiology" International Journal of Molecular Sciences 26, no. 22: 11128. https://doi.org/10.3390/ijms262211128
APA StyleZhao, S., Liu, S., Shao, W., & Liu, D. (2025). Research Progress Regarding the Use of Single-Cell Sequencing Technology in Analyzing Tumor Endothelial Cell Pathophysiology. International Journal of Molecular Sciences, 26(22), 11128. https://doi.org/10.3390/ijms262211128






