The Emerging Role of FAM171A2 in Gynecological Malignancies: Bioinformatic Insights from UCEC and Ovarian Cancer
Abstract
1. Introduction
2. Results
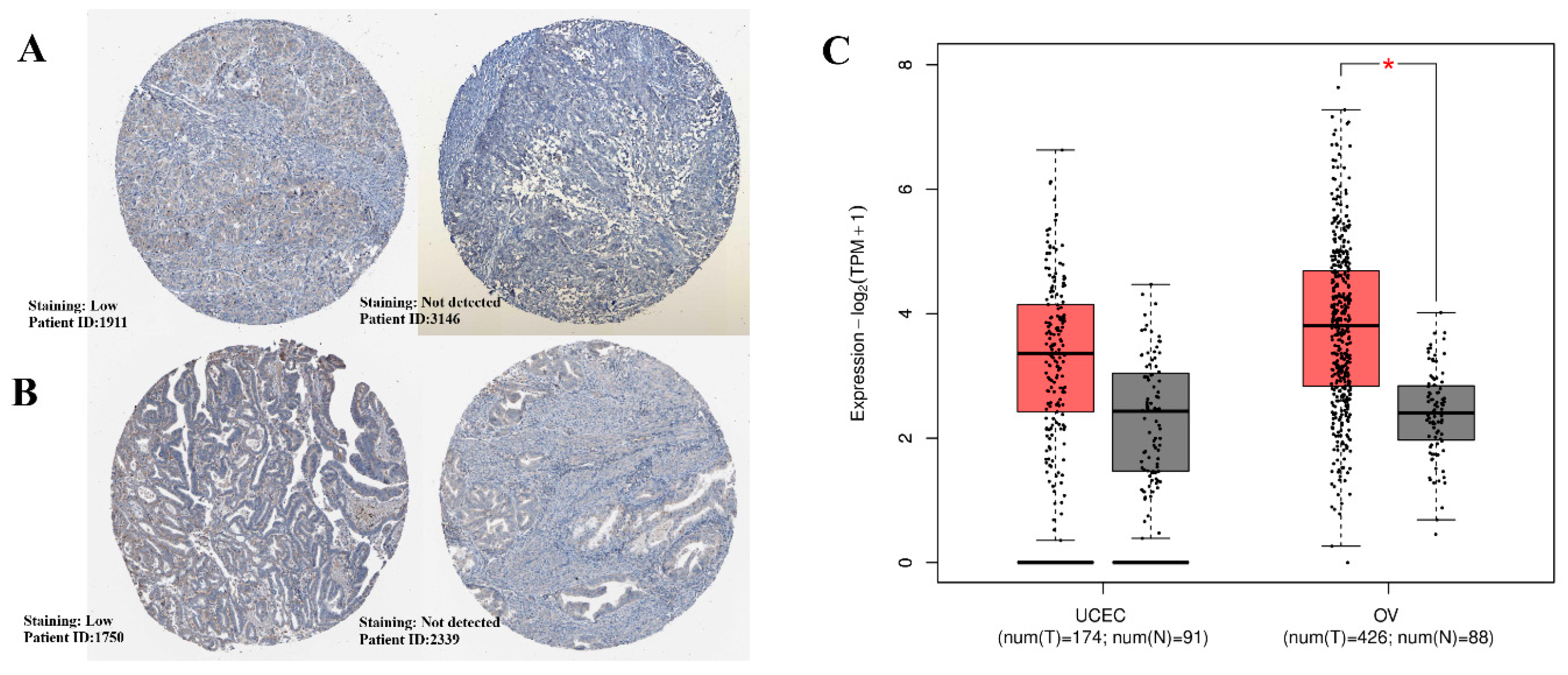
2.1. Expression Patterns of the Gene Across OV and UCEC
2.2. Comparative Expression Analysis Across Cancer Types via TNMplot
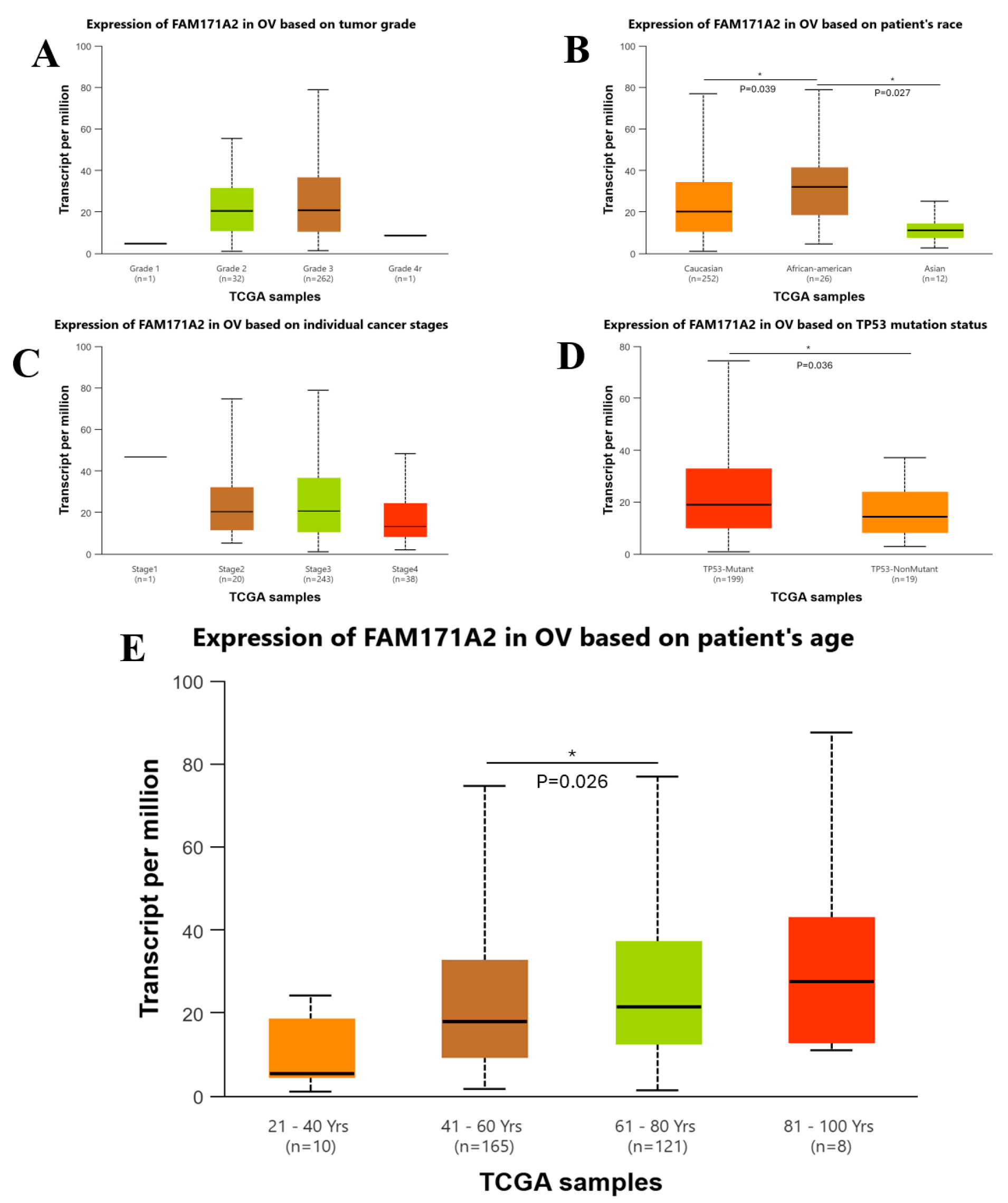
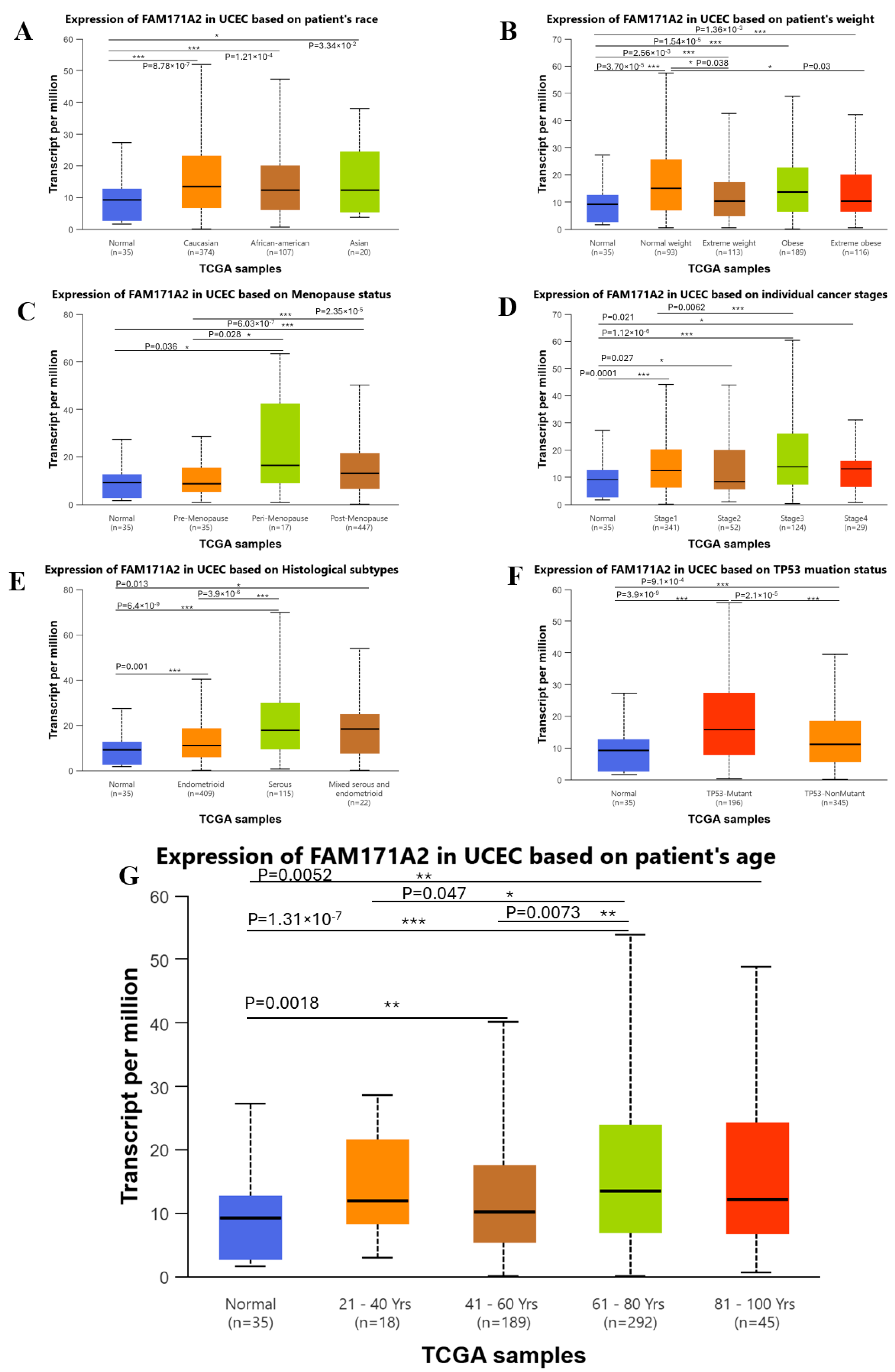
2.3. Comparative Expression Analysis Across Different Clinicopathological Variables via UALCAN
2.3.1. Comparative Expression Analysis of OV
2.3.2. Comparative Expression Analysis of UCEC
2.4. Kaplan–Meier Survival Analysis of FAM171A2 in UCEC and OV
2.5. Network Visualization of FAM171A2 and Its Associated miRNAs
- Green nodes represent miRNAs derived from the 5p arm.
- Blue nodes represent miRNAs derived from the 3p arm.
- Gray nodes correspond to variants or miRNAs without explicit arm annotation.
- Red node highlights the central FAM171A2 gene.
| TargetScan8.0 |
|---|
| hsa-miR-6838-5p, hsa-miR-15b-5p, hsa-miR-497-5p, hsa-miR-16-5p, hsa-miR-424-5p, hsa-miR-195-5p, hsa-miR-15a-5p, hsa-miR-4746-3p, hsa-miR-6816-5p, hsa-miR-3196, hsa-miR-3180-3p, hsa-miR-3180, hsa-miR-423-5p, hsa-miR-3184-5p, hsa-miR-6785-5p, hsa-miR-4728-5p, hsa-miR-6883-5p, hsa-miR-149-3p, hsa-miR-2277-5p, hsa-miR-4767, hsa-miR-4466, hsa-miR-675-5p, hsa-miR-6741-5p, hsa-miR-6776-5p, hsa-miR-6742-3p, hsa-miR-6791-5p, hsa-miR-4292, hsa-miR-504-5p.1, hsa-miR-3620-3p, hsa-miR-3178, hsa-miR-6784-5p, hsa-miR-4532, hsa-miR-5587-3p, hsa-miR-6840-5p, hsa-miR-296-5p, hsa-miR-296-5p, hsa-miR-7160-3p, hsa-miR-3939, hsa-miR-4633-3p, hsa-miR-6500-5p, hsa-miR-6726-5p, hsa-miR-5591-5p, hsa-miR-920, hsa-miR-4300, hsa-miR-6090, hsa-miR-6827-5p, hsa-miR-3192-5p, hsa-miR-4505, hsa-miR-5787, hsa-miR-4492, hsa-miR-5001-5p, hsa-miR-4498, hsa-miR-762, hsa-miR-185-3p, hsa-miR-4489, hsa-miR-4283, hsa-miR-6852-5p, hsa-miR-661, hsa-miR-7107-5p, hsa-miR-1234-3p, hsa-miR-6850-3p, hsa-miR-6892-3p, hsa-miR-4749-3p, hsa-miR-296-5p, hsa-miR-6724-5p, hsa-miR-6773-5p, hsa-miR-939-3p, hsa-miR-4292, hsa-miR-6791-5p, hsa-miR-331-3p, hsa-miR-210-5p, hsa-miR-3922-5p, hsa-miR-5695, hsa-miR-6736-3p, hsa-miR-29c-3p, hsa-miR-29b-3p, hsa-miR-29a-3p, hsa-miR-3065-3p, hsa-miR-551b-3p, hsa-miR-551a, hsa-miR-4696, hsa-miR-6841-3p, hsa-miR-4780, hsa-miR-6780b-3p, hsa-miR-623, hsa-miR-6768-5p, hsa-miR-3166, hsa-miR-6511a-5p, hsa-miR-1910-3p, hsa-miR-1827, hsa-miR-3612, hsa-miR-650, hsa-miR-4443, hsa-miR-6515-5p, hsa-miR-432-5p, hsa-miR-4707-5p, hsa-miR-6763-3p, hsa-miR-5587-3p, hsa-miR-6749-3p, hsa-miR-5193, hsa-miR-4667-3p, hsa-miR-6887-3p, hsa-miR-6859-3p, hsa-miR-711, hsa-miR-4638-3p, hsa-miR-422a, hsa-miR-378f, hsa-miR-378i, hsa-miR-378c, hsa-miR-378e, hsa-miR-378a-3p, hsa-miR-378d, hsa-miR-378h, hsa-miR-378b, hsa-miR-6835-5p, hsa-miR-6803-5p, hsa-miR-6751-5p, hsa-miR-6752-5p, hsa-miR-6842-5p, hsa-miR-7110-5p, hsa-miR-4447, hsa-miR-4472, hsa-miR-1306-5p, hsa-miR-4707-5p, hsa-miR-6763-3p, hsa-miR-5587-3p, hsa-miR-6749-3p, hsa-miR-5193, hsa-miR-4667-3p, hsa-miR-4290, hsa-miR-4687-5p, hsa-miR-361-3p, hsa-miR-3679-3p, hsa-miR-2115-5p, hsa-miR-7162-5p, hsa-miR-516a-3p, hsa-miR-516b-3p, hsa-miR-4269, hsa-miR-6715b-5p, hsa-miR-6768-5p, hsa-miR-4672, hsa-miR-3618, hsa-miR-4691-3p, hsa-miR-6856-3p, hsa-let-7a-2-3p, hsa-let-7g-3p, hsa-miR-7159-3p, hsa-miR-4482-3p, hsa-miR-3160-5p, hsa-miR-188-3p, hsa-miR-3156-3p, hsa-miR-1260b, hsa-miR-1260a, hsa-miR-3160-5p, hsa-miR-6893-3p, hsa-miR-370-3p, hsa-miR-1976, hsa-miR-660-3p, hsa-miR-4667-3p, hsa-miR-6887-3p, hsa-miR-6802-3p, hsa-miR-6879-3p, hsa-miR-5589-5p, hsa-miR-4505, hsa-miR-5787, hsa-miR-6884-5p, hsa-miR-485-5p, hsa-miR-3975, hsa-miR-2467-5p, hsa-miR-3188, hsa-miR-4649-3p, hsa-miR-7160-5p, hsa-miR-646, hsa-miR-503-5p, hsa-miR-497-5p, hsa-miR-424-5p, hsa-miR-6838-5p, hsa-miR-15a-5p, hsa-miR-15b-5p, hsa-miR-4524b-5p, hsa-miR-4524a-5p, hsa-miR-195-5p, hsa-miR-16-5p, hsa-miR-4704-5p, hsa-miR-216b-3p, hsa-miR-342-3p, hsa-miR-4687-5p, hsa-miR-7977, hsa-miR-6734-3p, hsa-miR-5088-3p, hsa-miR-4685-3p, hsa-miR-4287, hsa-miR-6887-3p, hsa-miR-4313, hsa-miR-3133, hsa-miR-615-5p, hsa-miR-1915-3p, hsa-miR-6764-5p, hsa-miR-4726-3p, hsa-miR-6840-3p, hsa-miR-6887-3p, hsa-miR-6795-3p, hsa-miR-6826-3p, hsa-miR-6887-3p, hsa-miR-4640-3p, hsa-miR-6871-3p, hsa-miR-3065-3p, hsa-miR-545-3p, hsa-miR-8086, hsa-miR-664b-5p, hsa-miR-1273f, hsa-miR-4756-3p, hsa-miR-3913-3p, hsa-miR-489-3p, hsa-miR-4504, hsa-miR-542-3p, hsa-miR-146b-3p, hsa-miR-6779-3p, hsa-miR-1226-3p, hsa-miR-4691-3p, hsa-miR-7977, hsa-miR-4433a-5p, hsa-miR-4433b-5p, hsa-miR-2355-5p, hsa-miR-5588-3p, hsa-miR-2114-5p, hsa-miR-554, hsa-miR-4640-3p, hsa-miR-6798-3p, hsa-miR-323a-3p, hsa-miR-130a-5p, hsa-miR-23a-3p, hsa-miR-23c, hsa-miR-23b-3p, hsa-miR-4999-5p, hsa-miR-6882-3p, hsa-miR-6083, hsa-miR-4328, hsa-miR-4733-3p, hsa-miR-1226-3p, hsa-miR-6511b-3p, hsa-miR-6511a-3p, hsa-miR-3150a-5p, hsa-miR-3150b-5p |

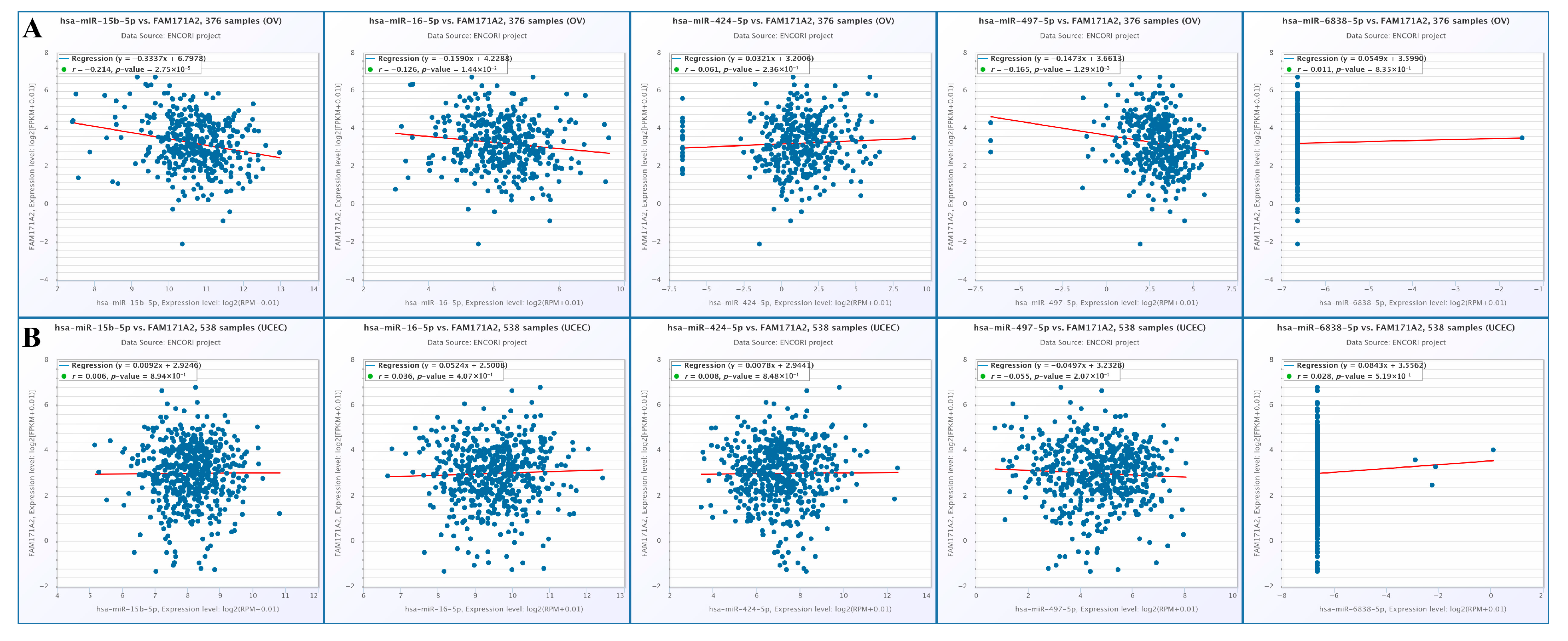
2.6. Correlation Analysis of FAM171A2 Expression with Selected miRNAs in Ovarian and Endometrial Cancers Using ENCORI
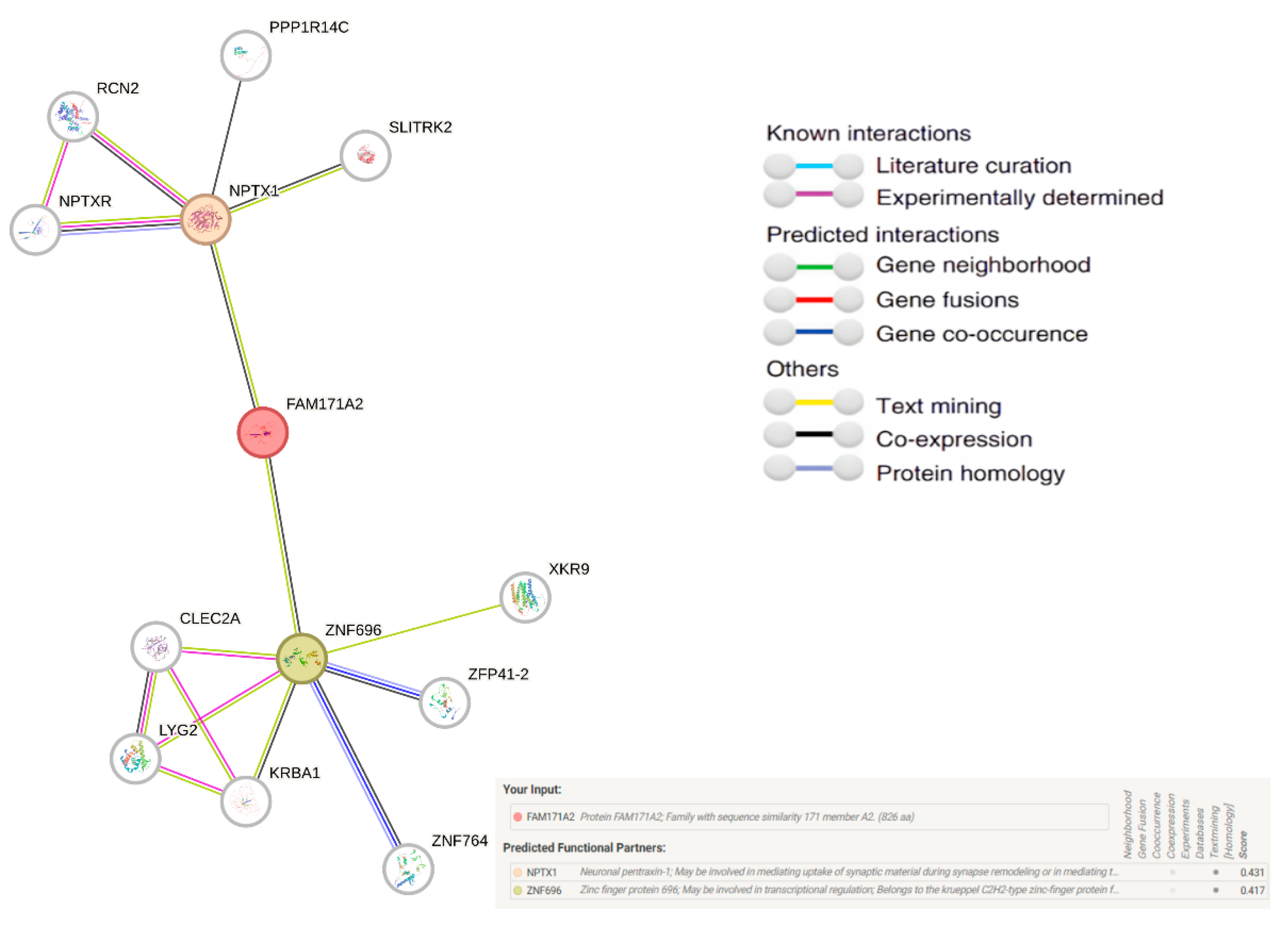
2.7. Comparative Network Analysis of FAM171A2: STRING
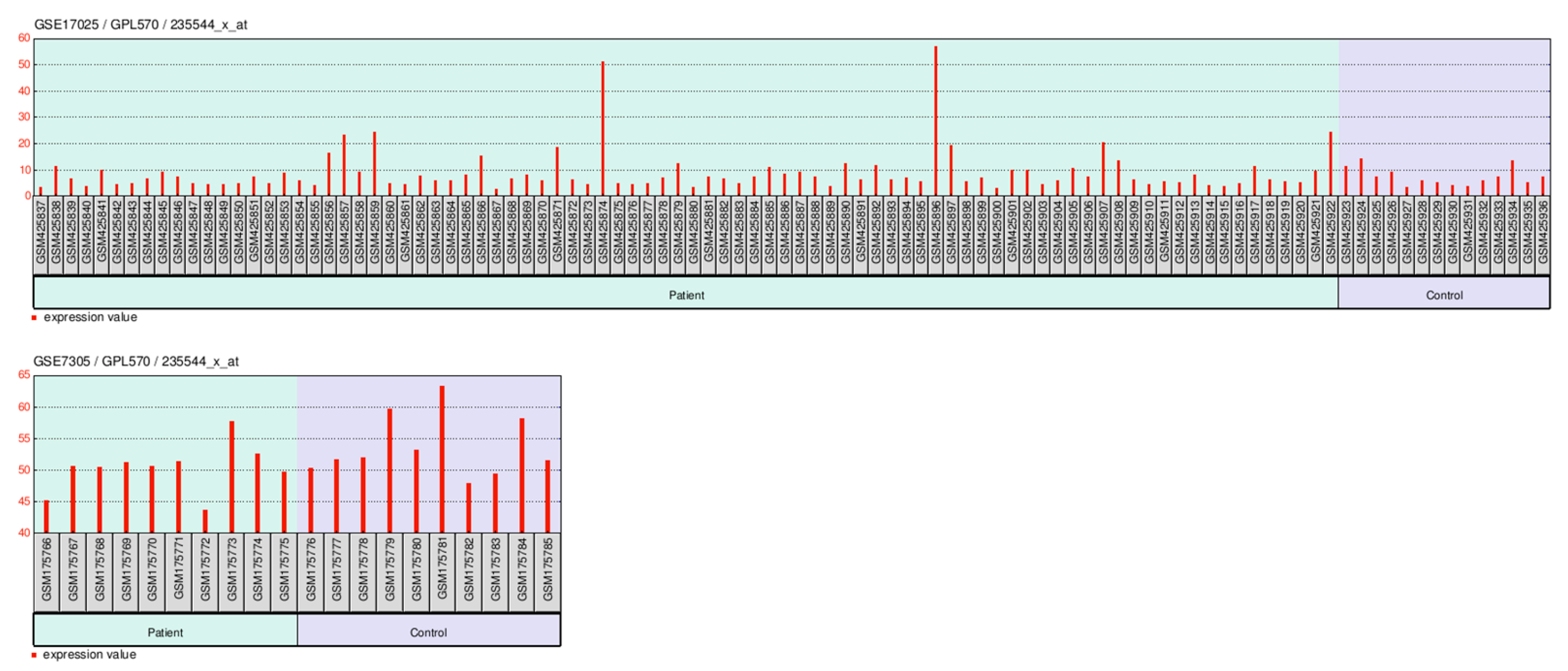
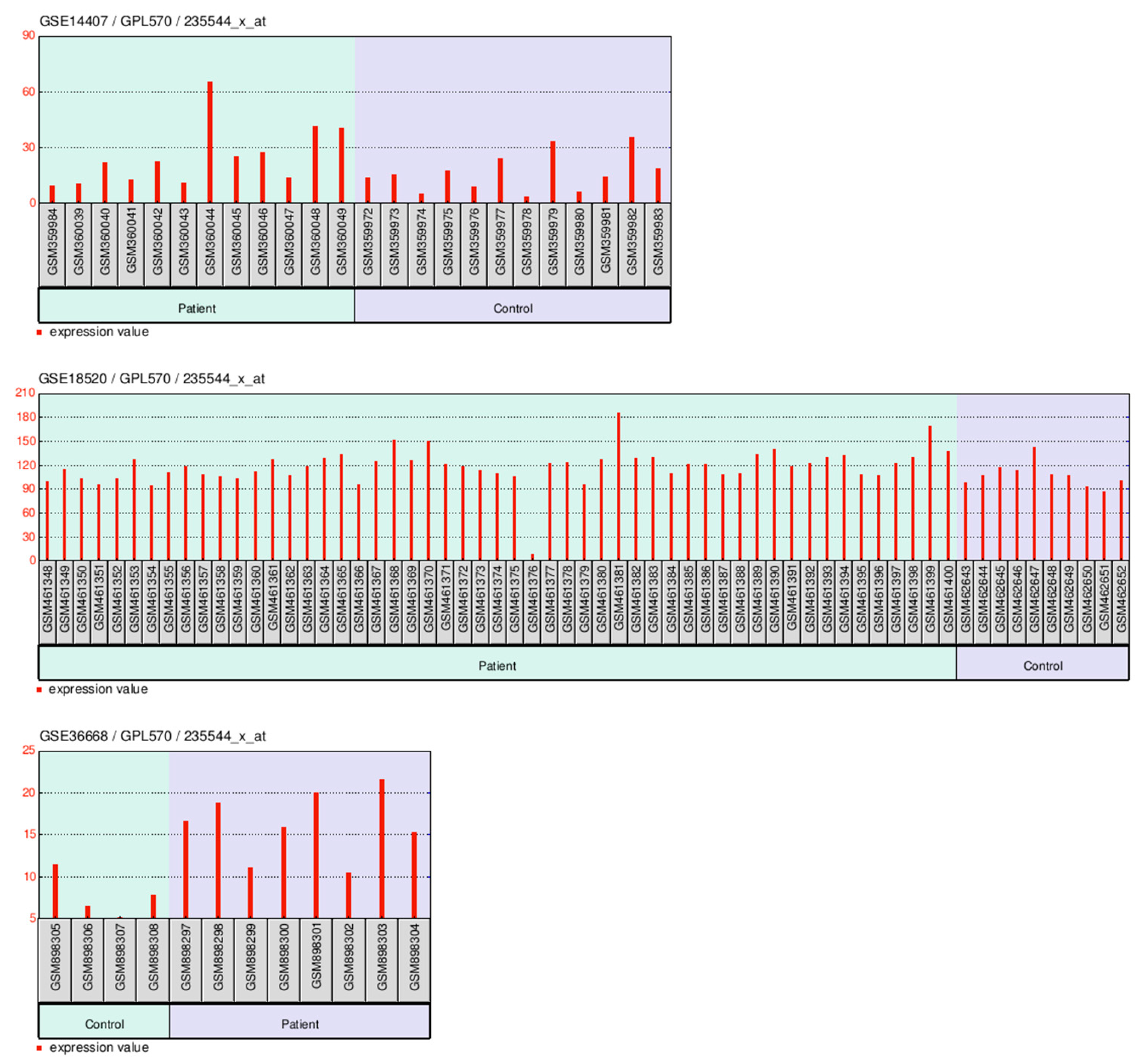
2.8. GEO-Based Expression Analysis FAM171A2 at Across Gynecologic Cancer Cohorts
2.8.1. GEO-Based Expression Analysis FAM171A2 for UCEC
2.8.2. GEO-Based Expression Analysis FAM171A2 for OV
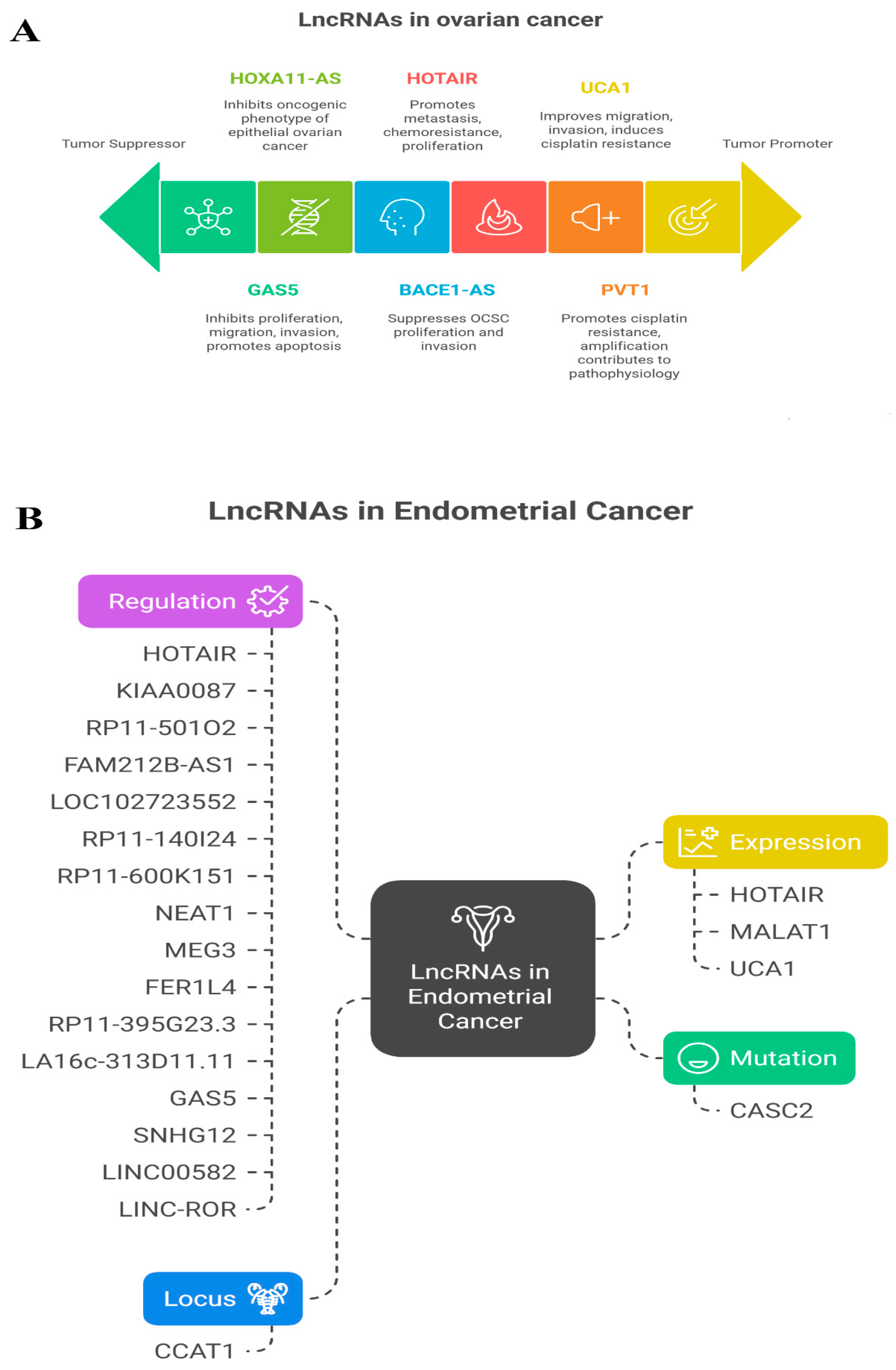
2.9. LncRNAs Associated with OV and UCEC
2.10. Correlation Analysis of FAM171A2 Expression with Selected lncRNAs at Across Gynecologic Cancer Cohorts
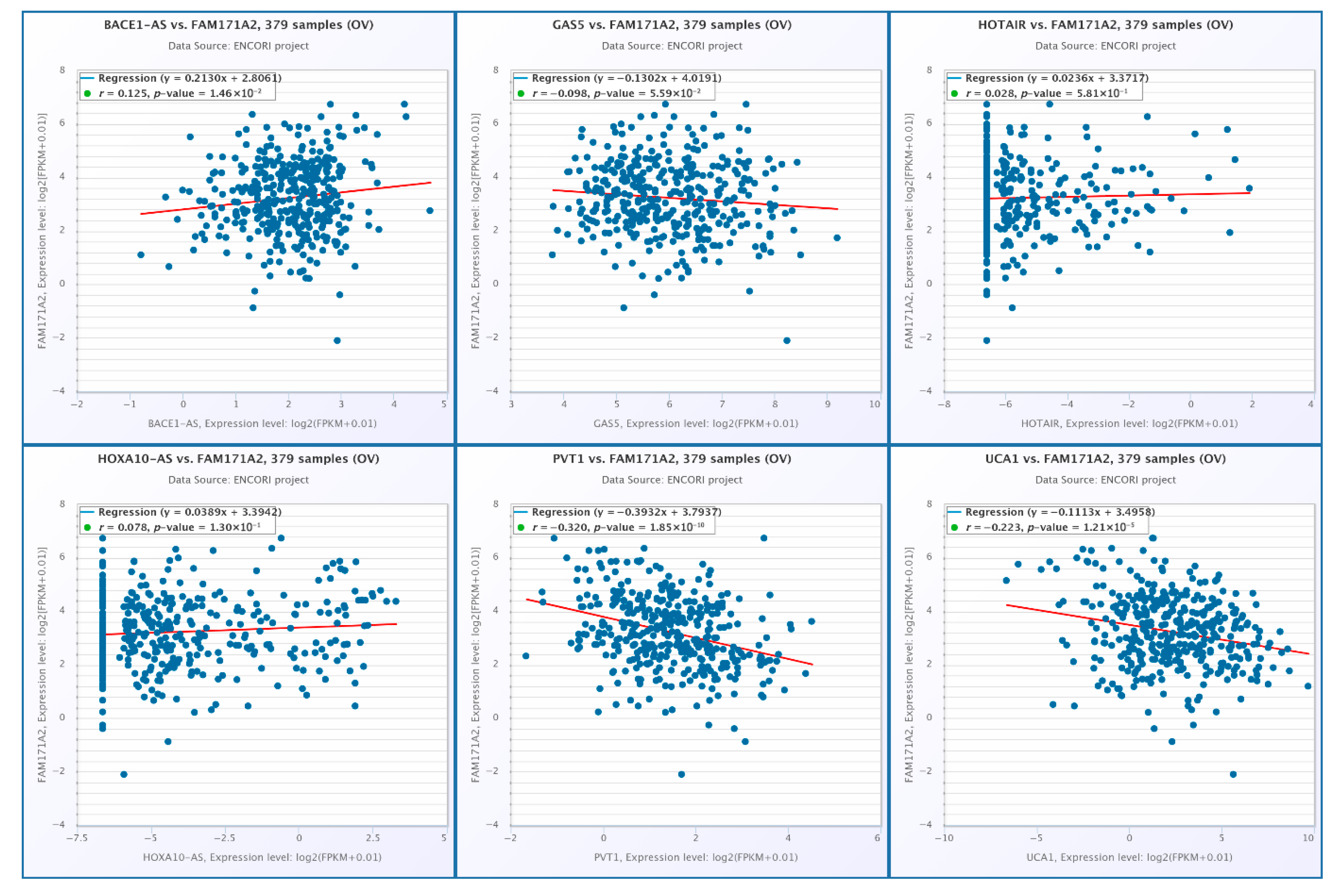
2.10.1. Correlation Analysis of FAM171A2 Expression with Selected miRNAs in OV
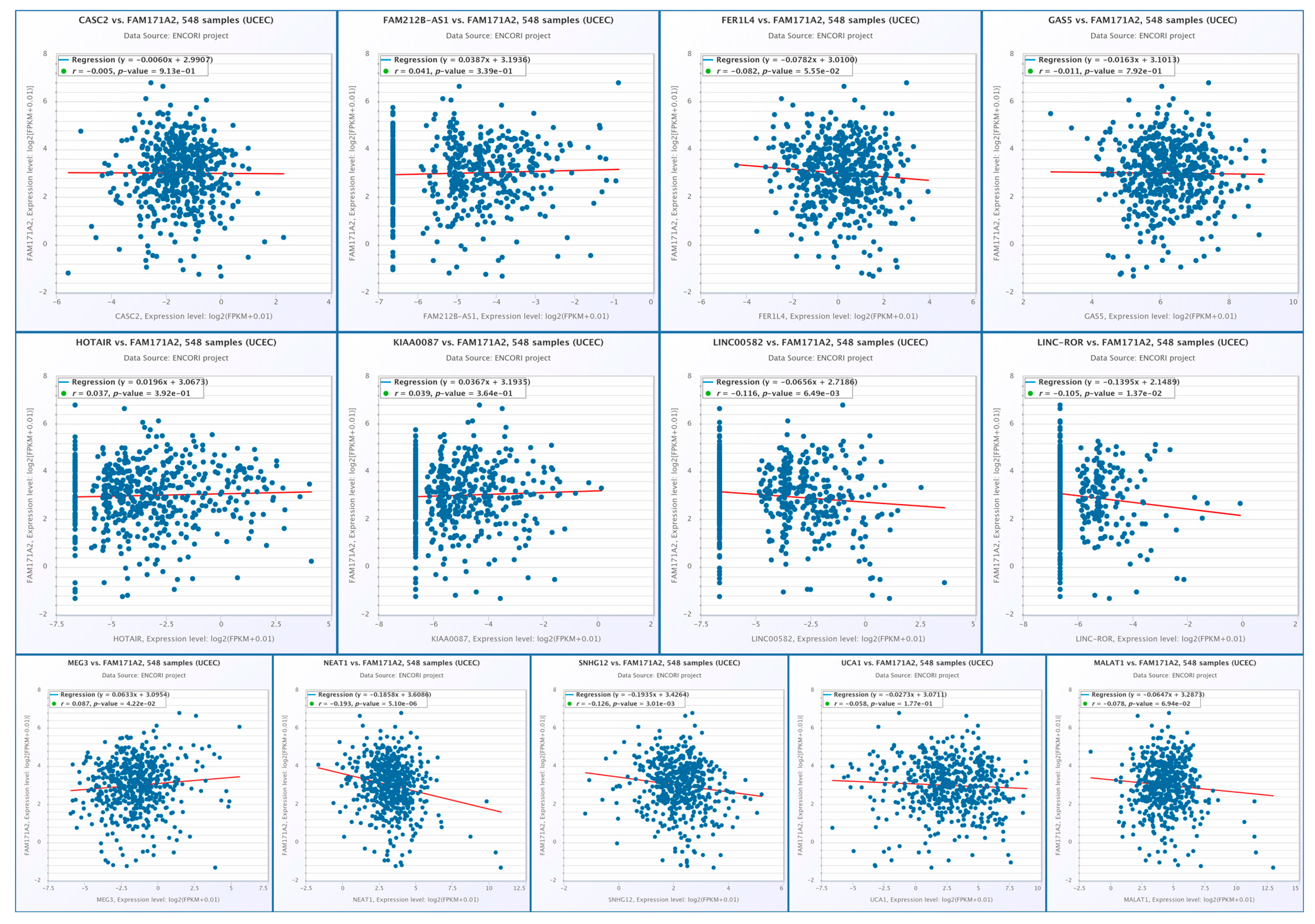
2.10.2. Correlation Analysis of FAM171A2 Expression with Selected lncRNAs in UCEC
3. Discussion
Limitations and Future Directions
4. Materials and Methods
4.1. Gene Expression Using the GEPIA2 Web Server
4.2. Normal and Tumor Comparisons via TNMplot
4.3. UALCAN-Based Expression Analysis in OV and UCEC
4.4. Kaplan–Meier Plotter Workflow for Gene-Expression–Survival Analyses
4.5. Prediction of FAM171A2 miRNA Interactions Using TargetScan 8.0
4.6. STRING Database-Based Analysis of FAM171A2 Interacting Proteins
4.7. Gene Expression Data Acquisition from GEO
4.8. Analysis of FAM171A2–miRNA and lncRNA Interactions Using the ENCORI Database
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AUC | Area Under the Curve |
| BMI | Body Mass Index |
| ceRNA | Competing Endogenous RNA |
| CI | Confidence Interval |
| CLIP-Seq | Cross-Linking Immunoprecipitation Sequencing |
| ECM | Extracellular Matrix |
| ENCORI | Encyclopedia of RNA Interactomes |
| EMT | Epithelial–Mesenchymal Transition |
| EV | Extracellular Vesicle |
| FC | Fold Change |
| FDR | False Discovery Rate |
| GEPIA2 | Gene Expression Profiling Interactive Analysis 2 |
| GEO | Gene Expression Omnibus |
| GTEx | Genotype-Tissue Expression |
| HGSC | High-Grade Serous Carcinoma |
| HPA | Human Protein Atlas |
| HR | Hazard Ratio |
| IHC | Immunohistochemistry |
| KMplot | Kaplan–Meier Plotter |
| lncRNA | Long Non-Coding RNA |
| miRNA | MicroRNA |
| mRNA | Messenger RNA |
| NF-κB | Nuclear Factor kappa-light-chain-enhancer of activated B cells |
| OV | Ovarian Cancer |
| PPI | Protein–Protein Interaction |
| PTX | Paclitaxel |
| RISC | RNA-Induced Silencing Complex |
| RNA-seq | RNA Sequencing |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins |
| TCGA | The Cancer Genome Atlas |
| TIMER2 | Tumor Immune Estimation Resource 2 |
| TNMplot | Tumor–Normal–Metastatic Plotter |
| TP53 | Tumor Protein 53 |
| TPM | Transcripts Per Million |
| UCEC | Uterine Corpus Endometrial Carcinoma |
| UALCAN | University of Alabama at Birmingham Cancer Data Analysis Portal |
| VEGFA | Vascular Endothelial Growth Factor A |
| ZNF696 | Zinc Finger Protein 696 |
References
- Jiang, Y.; Xu, Y.; He, J.; Sui, L.; Li, T.; Xia, N.; Yao, Q. Uncovering potential targets for antibody-drug conjugates in the treatment of gynecologic malignancies. Front. Pharmacol. 2025, 16, 1525733. [Google Scholar] [CrossRef] [PubMed]
- Bodriagova, O.; Previs, R.A.; Gaba, L.; Shankar, A.; Vidal, L.; Saini, K.S. Recent Advances in Gynecological Malignancies: Focus on ASCO 2023. Oncol. Ther. 2023, 11, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Aravantinou-Fatorou, A.; Georgakopoulou, V.E.; Dimopoulos, M.A.; Liontos, M. Precision medicine in gynecological cancer (Review). Biomed. Rep. 2025, 22, 43. [Google Scholar] [CrossRef] [PubMed]
- Rasila, T.; Saavalainen, O.; Attalla, H.; Lankila, P.; Haglund, C.; Hölttä, E.; Andersson, L.C. Astroprincin (FAM171A1, C10orf38): A Regulator of Human Cell Shape and Invasive Growth. Am. J. Pathol. 2019, 189, 177–189. [Google Scholar] [CrossRef]
- Wahab, A.; Almangush, A.; Andersson, L.C.; Nieminen, P.; Salo, T. Impact of Astroprincin (FAM171A1) Expression in Oral Tongue Cancer. Front. Oral Health 2020, 1, 599421. [Google Scholar] [CrossRef]
- Bao, C.; Lu, Y.; Chen, J.; Chen, D.; Lou, W.; Ding, B.; Xu, L.; Fan, W. Exploring specific prognostic biomarkers in triple-negative breast cancer. Cell Death Dis. 2019, 10, 807. [Google Scholar] [CrossRef]
- Sanawar, R.; Mohan Dan, V.; Santhoshkumar, T.R.; Kumar, R.; Pillai, M.R. Estrogen receptor-α regulation of microRNA-590 targets FAM171A1—A modifier of breast cancer invasiveness. Oncogenesis 2019, 8, 5. [Google Scholar] [CrossRef]
- Deng, H.; Xu, X.; Zhang, Y.; Li, Y. The complex role and molecular mechanism of family with sequence similarity genes in cancer: A comprehensive review. Discov. Oncol. 2025, 16, 1443. [Google Scholar] [CrossRef]
- UniProt. UniProt n.d. Available online: https://www.uniprot.org/uniprotkb/Q8N8K9/entry (accessed on 13 October 2025).
- FAM171A2 Protein Expression Summary—The Human Protein Atlas n.d. Available online: https://www.proteinatlas.org/ENSG00000161682-FAM171A2 (accessed on 13 October 2025).
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Kaprio, T.; Lindström, A.M.; Rasila, T.; Saavalainen, O.; Beilmann-Lehtonen, I.; Mustonen, H.; Haglund, C.; Andersson, L.C. Elevated tumor expression of Astroprincin (FAM171A1) is an independent marker of poor prognosis in colon cancer. BMC Gastroenterol. 2021, 21, 341. [Google Scholar] [CrossRef]
- Xu, W.; Han, S.-D.; Zhang, C.; Li, J.-Q.; Wang, Y.-J.; Tan, C.-C.; Li, H.-Q.; Dong, Q.; Mei, C.; Tan, L.; et al. The FAM171A2 gene is a key regulator of progranulin expression and modifies the risk of multiple neurodegenerative diseases. Sci. Adv. 2020, 6, eabb3063. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Kośla, K.; Orzechowska, M.; Jędroszka, D.; Baryła, I.; Bednarek, A.K.; Płuciennik, E. A Novel Set of WNT Pathway Effectors as a Predictive Marker of Uterine Corpus Endometrial Carcinoma–Study Based on Weighted Co-expression Matrices. Front. Oncol. 2019, 9, 360. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-X.; Xu, X.-X.; Tan, B.-Z.; Zhang, Z.; Zhou, X.-D. MicroRNA-29b Inhibits Angiogenesis by Targeting VEGFA through the MAPK/ERK and PI3K/Akt Signaling Pathways in Endometrial Carcinoma. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 41, 933–946. [Google Scholar] [CrossRef] [PubMed]
- Diéguez-Martínez, N.; Espinosa-Gil, S.; Yoldi, G.; Megías-Roda, E.; Bolinaga-Ayala, I.; Viñas-Casas, M.; Gorgisen, G.; Domingo-Ortí, I.; Pérez-Montoyo, H.; Bayascas, J.R.; et al. The ERK5/NF-κB signaling pathway targets endometrial cancer proliferation and survival. Cell. Mol. Life Sci. 2022, 79, 524. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shan, J.; Zhang, L.; Wang, R.; Wu, M.-Y.; Li, H.-M.; Xu, H.-M. The role of FAM171A2-GRN-NF-κB pathway in TBBPA induced oxidative stress and inflammatory response in mouse-derived hippocampal neuronal HT22 cells. Ecotoxicol. Environ. Saf. 2025, 289, 117445. [Google Scholar] [CrossRef]
- Cho, A.; Howell, V.M.; Colvin, E.K. The Extracellular Matrix in Epithelial Ovarian Cancer—A Piece of a Puzzle. Front. Oncol. 2015, 5, 245. [Google Scholar] [CrossRef]
- Loret, N.; Denys, H.; Tummers, P.; Berx, G. The Role of Epithelial-to-Mesenchymal Plasticity in Ovarian Cancer Progression and Therapy Resistance. Cancers 2019, 11, 838. [Google Scholar] [CrossRef]
- Fang, D.; Chen, H.; Zhu, J.Y.; Wang, W.; Teng, Y.; Ding, H.; Jing, Q.; Su, S.; Huang, S. Epithelial–mesenchymal transition of ovarian cancer cells is sustained by Rac1 through simultaneous activation of MEK1/2 and Src signaling pathways. Oncogene 2017, 36, 1546–1558. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.; Ma, H.; Zhang, J.; Zhou, X. Downregulation of miR-195 correlates with lymph node metastasis and poor prognosis in colorectal cancer. Med. Oncol. 2012, 29, 919–927. [Google Scholar] [CrossRef]
- Zhao, X.; Dai, L.; Yue, Q.; Wang, H.; Wang, X.U.; Li, Y.; Chen, R. MiR-195 inhibits migration, invasion and epithelial-mesenchymal transition (EMT) of endometrial carcinoma cells by targeting SOX4. J. Biosci. 2019, 44, 146. [Google Scholar] [CrossRef]
- Zare, E.; Yaghoubi, S.M.; Khoshnazar, M.; Jafari Dargahlou, S.; Machhar, J.S.; Zheng, Z.; Duijf, P.H.G.; Mansoori, B. MicroRNAs in Cancer Immunology: Master Regulators of the Tumor Microenvironment and Immune Evasion, with Therapeutic Potential. Cancers 2025, 17, 2172. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Jiang, Y.; Wan, Y.; Zhang, L.; Qiu, J.; Zhou, S.; Cheng, W. UCA1 functions as a competing endogenous RNA to suppress epithelial ovarian cancer metastasis. Tumor Biol. 2016, 37, 10633–10641. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.L.; Zhao, Z.S.; Zhang, M.Y.; Han, L.J.; Dong, Y.J.; Xu, B. Long Noncoding RNA PVT1 Facilitates Cervical Cancer Progression via Negative Regulating of miR-424. Oncol. Res. 2017, 25, 1391–1398. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Liu, Y.J.; Wang, C. A review of the regulatory mechanisms of extracellular vesicles-mediated intercellular communication. Cell Commun. Signal. 2023, 21, 77. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615, Correction in Nature 2012, 490, 292. [Google Scholar] [CrossRef]
- Polajžer, S.; Černe, K. Precision Medicine in High-Grade Serous Ovarian Cancer: Targeted Therapies and the Challenge of Chemoresistance. Int. J. Mol. Sci. 2025, 26, 2545. [Google Scholar] [CrossRef]
- Győrffy, B.; Surowiak, P.; Budczies, J.; Lánczky, A. Online Survival Analysis Software to Assess the Prognostic Value of Biomarkers Using Transcriptomic Data in Non-Small-Cell Lung Cancer. PLoS ONE 2013, 8, e82241. [Google Scholar] [CrossRef] [PubMed]
- Van De Vijver, M.J.; He, Y.D.; Van’t Veer, L.J.; Dai, H.; Hart, A.A.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. A Gene-Expression Signature as a Predictor of Survival in Breast Cancer. N. Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Daza, J.; Itzel, T.; Betge, J.; Zhan, T.; Marmé, F.; Teufel, A. Prognostic Cancer Gene Expression Signatures: Current Status and Challenges. Cells 2021, 10, 648. [Google Scholar] [CrossRef]
- Xu, X.; Yin, F.; Guo, M.; Gan, G.; Lin, G.; Wen, C.; Wang, J.; Song, P.; Wang, J.; Qi, Z.Q.; et al. Quantitative proteomic analysis of exosomes from umbilical cord mesenchymal stem cells and rat bone marrow stem cells. Proteomics 2023, 23, 2200204. [Google Scholar] [CrossRef]
- Liang, B.; Peng, P.; Chen, S.; Li, L.; Zhang, M.; Cao, D.; Yang, J.; Li, H.; Gui, T.; Li, X.; et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J. Proteom. 2013, 80, 171–182. [Google Scholar] [CrossRef]
- Ghasemzadeh, T.; Rajabi, A.; MalekAbbaslou, E.; Najari, P.; Akbarzadeh, S.; Tayefeh-Gholami, S.; Teimourian, S.; Hosseinpourfeizi, M.; Safaralizadeh, R. Evaluation of the expression of the long non-coding RNAs, LOWEG and MINCR, and their clinical significance in human gastric cancer. Egypt. J. Med. Hum. Genet. 2023, 24, 87. [Google Scholar] [CrossRef]
- Wu, J.; Liu, G.; An, K.; Shi, L. NPTX1 inhibits pancreatic cancer cell proliferation and migration and enhances chemotherapy sensitivity by targeting RBM10. Oncol. Lett. 2022, 23, 154. [Google Scholar] [CrossRef]
- Peng, X.; Pan, K.; Zhao, W.; Zhang, J.; Yuan, S.; Wen, X.; Zhou, W.; Yu, Z. NPTX1 inhibits colon cancer cell proliferation through down-regulating cyclin A2 and CDK2 expression. Cell Biol. Int. 2018, 42, 589–597. [Google Scholar] [CrossRef]
- Maino, B.; Ciotti, M.T.; Calissano, P.; Cavallaro, S. Transcriptional Analysis of Apoptotic Cerebellar Granule Neurons Following Rescue by Gastric Inhibitory Polypeptide. Int. J. Mol. Sci. 2014, 15, 5596–5622. [Google Scholar] [CrossRef]
- Apóstolo, N.; Smukowski, S.N.; Vanderlinden, J.; Condomitti, G.; Rybakin, V.; Ten Bos, J.; Trobiani, L.; Portegies, S.; Vennekens, K.M.; Gounko, N.V.; et al. Synapse type-specific proteomic dissection identifies IgSF8 as a hippocampal CA3 microcircuit organizer. Nat. Commun. 2020, 11, 5171. [Google Scholar] [CrossRef]
- Vescio, M.; Paracchini, L.; Beltrame, L.; D’Incalci, M.; Marchini, S.; Pattini, L. Modulation of gene expression associated with copy number variation identifies key regulatory programs in high-grade serous ovarian carcinoma. Adv. Cancer Biol. Metastasis 2023, 7, 100088. [Google Scholar] [CrossRef]
- Toulza, E.; Mattiuzzo, N.R.; Galliano, M.F.; Jonca, N.; Dossat, C.; Jacob, D.; De Daruvar, A.; Wincker, P.; Serre, G.; Guerrin, M. Large-scale identification of human genes implicated in epidermal barrier function. Genome Biol. 2007, 8, R107. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.M.; Xu, Q.H.; Liu, Y.Q.; Feng, Y.-W.; Han, S.-D.; Zhang, Y.-R.; Chen, S.-D.; Guo, Y.; Wu, B.-S.; Ma, L.-Z.; et al. Neuronal FAM171A2 mediates α-synuclein fibril uptake and drives Parkinson’s disease. Science 2025, 387, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Magnon, C.; Hall, S.J.; Lin, J.; Xue, X.; Gerber, L.; Freedland, S.J.; Frenette, P.S. Autonomic nerve development contributes to prostate cancer progression. Science 2013, 341, 1236361. [Google Scholar] [CrossRef]
- Zahalka, A.H.; Frenette, P.S. Nerves in cancer. Nat. Rev. Cancer 2020, 20, 143–157. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef]
- Schwarzenbach, H. Clinical significance of miR-15 and miR-16 in ovarian cancer. Transl. Cancer Res. 2016, 5, S50–S53. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949, Correction in Proc. Natl. Acad. Sci. USA 2005, 103, 2464. [Google Scholar] [CrossRef]
- Zhang, S.-Y.; Lu, Z.-M.; Lin, Y.-F.; Chen, L.-S.; Luo, X.-N.; Song, X.-H.; Chen, S.-H.; Wu, Y.-L. miR-144-3p, a tumor suppressive microRNA targeting ETS-1 in laryngeal squamous cell carcinoma. Oncotarget 2016, 7, 11637–11650. [Google Scholar] [CrossRef]
- Yan, J.-J.; Zhang, Y.-N.; Liao, J.-Z.; Ke, K.; Chang, Y.; Li, P.-Y.; Wang, M.; Lin, J.-S.; He, X.-X. MiR-497 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting VEGFA and AEG-1. Oncotarget 2015, 6, 29527–29542. [Google Scholar] [CrossRef]
- Luo, G.; He, K.; Xia, Z.; Liu, S.; Liu, H.; Xiang, G. Regulation of microRNA-497 expression in human cancer. Oncol. Lett. 2020, 21, 23. [Google Scholar] [CrossRef]
- Chen, Y.; Du, H.; Bao, L.; Liu, W. LncRNA PVT1 promotes ovarian cancer progression by silencing miR-214. Cancer Biol. Med. 2018, 15, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-Y.; Wang, X.-L.; Dang, Y.; Zhu, X.-Z.; Zhang, Y.-H.; Cai, B.-X.; Zheng, L. Long non-coding RNA UCA1 promotes the progression of paclitaxel resistance in ovarian cancer by regulating the miR-654-5p/SIK2 axis. Eur. Rev. 2020, 24, 591–603. Available online: https://www.europeanreview.org/article/20035 (accessed on 16 October 2025).
- Tang, Z.-L.; Zhang, K.; Lv, S.-C.; Xu, G.-W.; Zhang, J.-F.; Jia, H.-Y. LncRNA MEG3 suppresses PI3K/AKT/mTOR signalling pathway to enhance autophagy and inhibit inflammation in TNF-α-treated keratinocytes and psoriatic mice. Cytokine 2021, 148, 155657. [Google Scholar] [CrossRef] [PubMed]
- Pu, Z.; Wu, L.; Guo, Y.; Li, G.; Xiang, M.; Liu, L.; Zhan, H.; Zhou, X.; Tan, H. LncRNA MEG3 contributes to adenosine-induced cytotoxicity in hepatoma HepG2 cells by downregulated ILF3 and autophagy inhibition via regulation PI3K-AKT-mTOR and beclin-1 signaling pathway. J. Cell. Biochem. 2019, 120, 18172–18185. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, X.; Li, X.; Ma, Y.; Goel, A. Long non-coding RNAs in colorectal cancer: Novel oncogenic mechanisms and promising clinical applications. Cancer Lett. 2021, 504, 67–80. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, Y.; Liu, Z.; Zhang, Z.; Xiong, L.; Wen, Y. Recent advances of NEAT1-miRNA interactions in cancer. Acta Biochim. Biophys. Sin. 2022, 54, 153–162. [Google Scholar] [CrossRef]
- Adriaens, C.; Standaert, L.; Barra, J.; Latil, M.; Verfaillie, A.; Kalev, P.; Boeckx, B.; Wijnhoven, P.W.G.; Radaelli, E.; Vermi, W.; et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016, 22, 861–868. [Google Scholar] [CrossRef]
- Wu, Y.-C.; Ku, M.-H.; Huang, W.-L.; Dias, L.; Chen, C.-W. NEAT1 in Ovarian Cancer: A Key Regulator of Tumor Progression, Follicular Fluid Dynamics, and Therapeutic Resistance. Anticancer Res. 2025, 45, 825–842. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Bartha, Á.; Győrffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Li, Q.; Birkbak, N.J.; Gyorffy, B.; Szallasi, Z.; Eklund, A.C. Jetset: Selecting the optimal microarray probe set to represent a gene. BMC Bioinform. 2011, 12, 474. [Google Scholar] [CrossRef]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D661. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Day, R.S.; McDade, K.K.; Chandran, U.R.; Lisovich, A.; Conrads, T.P.; Hood, B.L.; Kolli, V.K.; Kirchner, D.; Litzi, T.; Maxwell, G.L. Identifier mapping performance for integrating transcriptomics and proteomics experimental results. BMC Bioinform. 2011, 12, 213. [Google Scholar] [CrossRef] [PubMed]
- Day, R.S.; McDade, K.K. A decision theory paradigm for evaluating identifier mapping and filtering methods using data integration. BMC Bioinform. 2013, 14, 223. [Google Scholar] [CrossRef] [PubMed]
- Hever, A.; Roth, R.B.; Hevezi, P.; Marin, M.E.; Acosta, J.A.; Acosta, H.; Rojas, J.; Herrera, R.; Grigoriadis, D.; White, E.; et al. Human endometriosis is associated with plasma cells and overexpression of B lymphocyte stimulator. Proc. Natl. Acad. Sci. USA 2007, 104, 12451–12456. [Google Scholar] [CrossRef] [PubMed]
- Bowen, N.J.; Walker, L.D.; Matyunina, L.V.; Logani, S.; Totten, K.A.; Benigno, B.B.; McDonald, J.F. Gene expression profiling supports the hypothesis that human ovarian surface epithelia are multipotent and capable of serving as ovarian cancer initiating cells. BMC Med. Genom. 2009, 2, 71. [Google Scholar] [CrossRef]
- Mok, S.C.; Bonome, T.; Vathipadiekal, V.; Bell, A.; Johnson, M.E.; Wong, K.-K.; Park, D.-C.; Hao, K.; Yip, D.K.P.; Donninger, H.; et al. A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: Microfibril-associated glycoprotein 2. Cancer Cell 2009, 16, 521–532. [Google Scholar] [CrossRef]
- Elgaaen, B.V.; Olstad, O.K.; Sandvik, L.; Odegaard, E.; Sauer, T.; Staff, A.C.; Gautvik, K.M. ZNF385B and VEGFA are strongly differentially expressed in serous ovarian carcinomas and correlate with survival. PLoS ONE 2012, 7, e46317. [Google Scholar] [CrossRef]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2013, 42, D92–D97. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soylemez, S.; Ayan, D. The Emerging Role of FAM171A2 in Gynecological Malignancies: Bioinformatic Insights from UCEC and Ovarian Cancer. Int. J. Mol. Sci. 2025, 26, 11126. https://doi.org/10.3390/ijms262211126
Soylemez S, Ayan D. The Emerging Role of FAM171A2 in Gynecological Malignancies: Bioinformatic Insights from UCEC and Ovarian Cancer. International Journal of Molecular Sciences. 2025; 26(22):11126. https://doi.org/10.3390/ijms262211126
Chicago/Turabian StyleSoylemez, Sibel, and Durmus Ayan. 2025. "The Emerging Role of FAM171A2 in Gynecological Malignancies: Bioinformatic Insights from UCEC and Ovarian Cancer" International Journal of Molecular Sciences 26, no. 22: 11126. https://doi.org/10.3390/ijms262211126
APA StyleSoylemez, S., & Ayan, D. (2025). The Emerging Role of FAM171A2 in Gynecological Malignancies: Bioinformatic Insights from UCEC and Ovarian Cancer. International Journal of Molecular Sciences, 26(22), 11126. https://doi.org/10.3390/ijms262211126





